Abstract
Backgrounds
Currently, family with sequence similarity 65 member A (FAM65A) is reported as a pivotal regulator in various cancers. However, the effect of FAM65A in lung squamous cell carcinoma (LSCC) is still unclear, the prime objective of this research is to explore the role of FAM65A in LSCC.
Methods
Gene expression data and correlated clinical information were downloaded from the public database and the expression of FAM65A was detected. The expression of FAM65A was also detected in our collected clinical samples and LSCC cell lines. Survival package of R language was used to determine the survival significance of FAM65A. Proteins expression level was determined via western blot assay. Cell function experiments and in vivo experiments were performed to explore the effect of FAM65A on LSCC cell biological behaviors.
Results
FAM65A expression was significantly increased in LSCC clinical samples and cell lines. High FAM65A expression predicted poor prognosis in LSCC patients. After silencing FAM65A, the ability of LSCC cell proliferation, invasion and migration was decreased, and LSCC cell cycle was blocked. Moreover, in vivo experiments revealed that silencing FAM65A could inhibit LSCC cell proliferation.
Conclusions
High FAM65A expression could enhance proliferative, invasive and migratory abilities of LSCC. FAM65A might be a novel biomarker of LSCC.
Similar content being viewed by others
Introduction
Lung cancer is a primary malignant tumor that occurs in the lungs [1, 2]. The primary risk factors for lung cancer include smoking, long-term exposure to asbestos, certain radioactive gases (such as radon), and air pollution [3]. Globally, the morbidity and mortality of lung cancer remain high among malignant tumors, characterized by rapid disease progression, poor treatment effect and poor prognosis [4,5,6]. According to the 2022 reports from the International Agency for Research on Cancer (IARC) and the American Cancer Society (ACS), nearly 2.5 million new cases were diagnosed, accounting for 12.4% of the total global cancer cases [7]. Lung cancer is constantly threatening the survival and development of human beings, and the burden of lung cancer is heavy. At present, the research on lung cancer is still under constant exploration around the world.
In recent decades, with the rapid development of molecular biology technology and molecular diagnostic technology, lung tumor markers with important clinical diagnostic value have been continuously discovered [8, 9]. The diagnosis and treatment of advanced lung cancer has already entered the era of individualized and precise treatment guided by molecular typing from histological typing to determine treatment. In terms of immunotherapy, anti-PD-1 (programmed death molecule 1) / PD-L1 (Programmed cell Death-Ligand 1)/monoclonal antibodies are used to treat non-small cell lung cancer (NSCLC) and has achieved good curative effect in related malignant tumors such as and bladder cancer [10,11,12,13]. In terms of targeted therapy, the current common driver genes in the field of lung cancer mainly include epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), c-ros proto-oncogene 1 (ROS1) tyrosine kinase and so on [14,15,16]. For advanced lung adenocarcinoma, related targeted therapy drugs have been widely used in clinic, such as erlotinib (Erlotinib) in the treatment of patients with EGFR-mutant non-squamous NSCLC [17]. However, due to the lack of available gene mutations in the targeted therapy of lung squamous cell carcinoma (LSCC), its research progress is still relatively slow compared to lung adenocarcinoma. In addition, with the wide application of targeted drug therapy, drug resistance has gradually been found, such as in the process of epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKIs) drug treatment [18]. Gene mutation occurrence during treatment always results in decreased treatment effect [19]. Therefore, exploring the relevant mechanism of the process of LSCC and to find new gene therapy targets for LSCC is particularly important. Through the in-depth study of genomics, potential biomarkers can be found to discover new gene targets.
Cell migration is critically important in both normal physiological processes and abnormal pathological processes [20]. The establishment and maintenance of anterior-posterior polarity is essential for cell movement in a particular direction. In the process of cell polarization, the dynamic reorganization of the actin cytoskeleton, the spatial localization of specific proteins, and the redirection of secretion transport play a crucial role [21]. Cell polarization and migration are dependent on the close cooperation between several proteins, such as the small GTP-binding protein RHO GTPase [22]. Currently, there are 20 RHO GTPase family members, including 8 subfamilies, that have been discovered in mammals [23]. RHO GTPases control multiple cellular events by switching between an active GTP-binding state and an inactive GDP-binding state [24,25,26]. What is clear now is that RHO protein is involved in nearly every stage of tumorigenesis, and several studies have shown that reduced RHO protein function leads to the morphological changes observed in tumor cells. Inhibiting RHO protein may promote a more aggressive tumor phenotype [27]. The K-ras protein, through its intrinsic GTPase activity, acts as a molecular switch controlling cell proliferation, differentiation, and survival [28,29,30]. Research indicates that the co-expression of Vav1 and K-RasG12D in lung tissue significantly increased the malignancy of lung cancer, suggesting that these two oncogenes synergistically promote the development of lung tumors [31]. RHO family Interacting Cell Polarization regulator 1 (RIPOR1), also known as family with sequence similarity 65 member A (FAM65A), is a newly discovered RHO effector protein, which can regulate the polarity of migrating cells by binding to GTP-bound RHO protein through the N-terminal HR1 domain [32]. Moreover, it has been reported FAM65A can be regarded as a biomarker in patients with cholangiocarcinoma [33].
In this study, we analyzed the correlation between FAM65A expression in LSCC and clinical prognostic outcomes, and validated FAM65A expression in clinical samples and multiple LSCC cell lines. In addition, we investigated the effect of FAM65A on the biological behavior of LSCC cells and further validated in vivo experiments. This study may promote further research on FAM65A as a therapeutic target for LSCC.
Materials and methods
Data source and preprocessing
Gene expression profiles of GSE75037 [34], GSE67061 [35], GSE18842 [36], GSE2088 [37], GSE161533 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) datasets sourced from Gene Expression Omnibus (GEO) were downloaded to evaluate FAM65A expression in LSCC. RNA sequencing data of 502 lung cancer and 49 normal samples with relevant clinical information were accessed from The Cancer Genome Atlas (TCGA) database. For RNA sequencing (RNA-seq) data, counts data was to converted to transcripts per million (TPM) reads based on the qCML method using edgeR package of R language [38], then TPM was converted to log2 (TPM + 1) for further analysis. GEO2R was used to compare two or more groups of the microarray data in order to identify genes that are differentially expressed across experimental conditions.
Construction of survival model
Hazard ratio (HR) value of the cox risk regression model and the chi-square test p-value for the survival of the high and low groups based on TCGA data set was calculated using the survival package of R language. Based on the FAM65A expression median, LSCC samples were divided into two groups with high and low FAM65A expression. Kaplan-Meier survival analysis was performed based on the relevant clinical data accessed from TCGA, and finally the overall survival curve was drawn.
GSEA analysis
RNA sequencing data for LSCC was obtained from the TCGA database. The expression levels of all genes in LUSC were calculated in relation to the expression of FAM65A. The genes were then ranked according to their correlation coefficients. Gene set enrichment analysis (GSEA) was conducted using GSEA v4.0.3, available at https://www.gsea-msigdb.org/gsea/index.jsp.
PPI network analysis
The target protein FAM65A was entered into the STRING 11.5 database (https://cn.string-db.org) to obtain the PPI network and TSV dataset. A protein-protein interaction analysis for the FAM65A gene was conducted with a threshold of 0.7. Additionally, the TSV dataset was imported into CytoScape 3.8.0 for data analysis and network visualization. The constructed PPI network was then uploaded to the CytoScape 3.8.0 software for connectivity analysis. Genes interacting with FAM65A were identified, and the Pearson correlation of their expression levels with FAM65A in LSCC was calculated.
Survival analysis
The cancer samples in LSCC were divided into high and low expression groups based on the median expression level of FAM65A. Survival analysis was then performed using the survival package, and the differences in survival between the high and low expression groups were assessed using the log-rank test.
Tumor immune microenvironment analysis
ESTIMATE algorithm [39] was used to evaluate the tumor immune microenvironment scores of the samples accessed from TCGA. The tumor immune microenvironment scores included the score for tumor purity, the level of stromal cells presents, and the level of immune cell infiltration in tumor tissue. And then correlational analysis was performed between gene expression and tumor immune microenvironment scores.
Patients and clinical specimens
Fresh specimens from 36 LSCC patients were collected during surgery and the compared surrounding tissues were collected as controls. Every patient was required to sign a written informed consent. All aspects of this study have been approved by the hospital research ethics committee. All patients did not receive any anticancer chemotherapy, radiotherapy, drug treatment, and biological immunotherapy before surgery. The patient’s clinical information is listed in Table 1. Paraffin sections of lung tissues were dewaxed, rehydrated, and stained in hematoxylin dye solution at 25 °C for 5 min. Tap water was utilized to wash the sections. The washed sections were then immersed in 1% hydrochloric acid alcohol solution for several seconds and rinsed by tap water until the tissues were back to blue. The sections were stained by Eosin for 3–5 min and washed in tap water again. After dehydration, clearing and sealing, the nuclei presented purple blue or blue, the cytoplasm showed pink, and red blood cells showed red. In addition, some clinical fresh samples in vitro were stored at -80 °C for real-time quantitative PCR (RT-PCR) and western blot.
Immunohistochemical staining
Paraffin-embedded tissue was sliced and dewaxed. After antigen retrieval, primary antibodies were incubated with slides at 4 °C overnight. Herein, anti- FAM65A (#PA5-52228, Invitrogen, Waltham, MA, USA) was used. After washing, secondary antibodies were incubated at 37 °C for 30 min and washed. Then diaminobenzidine (DAB) was applied for color development. Lastly, all sections were scanned by Pannoramic DESK, P-MIDI, P250, P1000 (3D HISTECH, Budapest, Hungary) and were read by Pannoramic Scanner (3D HISTECH, Budapest, Hungary).
Cell culture
Human LSCC cell lines (LK2, H1703, H2170 and SK-MES-1) and the normal lung epithelial cell line BEAS-2B were purchased from the cell bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). BEAS-2B was cultured in BEBM medium (#CC-3171, LONZA, Basel, Switzerland) containing 0.01 mg/mL fibronectin, 0.03 mg/mL bovine collagen type I and 0.01 mg/mL bovine serum albumin (FBS, #10099158, Gibco, Waltham, MA, USA). The LSCC cell lines were maintained in RPMI-1640 media (#11875093, Gibco, Waltham, MA, USA) containing 10% FBS, 100 U/ml penicillin, and 100 mg/ml streptomycin (#15070063, Invitrogen, Carlsbad, CA, USA) in a humidified incubator with 5% CO2 at 37℃.
Cell transfection
To down-regulate FAM65A expression, FAM65A short hairpin RNA (5′-AAA CAG CCT CTC TGA AAA TGC ATC TTA ACT GTG AAA CTT CCC ACA AAG CTG GAA TAA CGC TAC CAG TTC T-3′) and negative control (5′-CCG GCA ACA AGA TGA TCA GCA CCA ACT GCG TTG GTG CTC TTC ATC TTG TTG TTT TT3′). After removing LSCC cells (SK-MES-1 and H1703 cell lines) from the incubator, trypsin digestion and counting were performed. The cells were seeded into 6-well plates at a density of 5 × 103 cells per well. After 12 h of culture, the cell density would reach 50-70% of the total density of the culture dish and then siRNA transfection was performed. For siRNA configuration, the EP tube containing siRNA was centrifuged at 1000 rpm for 1 min, and an appropriate amount of DEPC water was added to dissolve it, and then a pipette gun was used to resuspend it to mix thoroughly. The tubes containing siRNA (7.5 µL) + opti-MEM (180 µL) and Lipofectamine 2000 (7.5 µL, #11668019, Thermo Fisher Scientific, Waltham, MA, USA) + opti-MEM (180 µL, #31985070, Gibco, Waltham, MA, USA) separately were mixed upside down and let stand for 5 min. Lipofectamine 2000 (7.5 µL) + opti-MEM (180 µL) mixture was added to siRNA + opti-MEM mixture. After being thoroughly mixed, the mixture was placed at room temperature for 15 min, and then added to the cells in the culture dish. After 4–6 h, the mixture was changed into complete medium for further culture.
Real-time quantitative PCR (RT-PCR)
Total RNA was isolated from cells using the RNeasy Mini Kit (#74904, QIAGEN, Hilden, Germany). Reverse transcription PCR was performed using Superscript IV Reverse Transcriptase (#18090010, ThermoFisher, Waltham, MA, USA). Gene fold changes were determined by the 2−ΔΔCt algorithm. β-Actin was used as a reference gene to normalize 2−ΔΔCt based assessments. The specific primer sequences were designed as follows: FAM65A, forward 5′-GGC GAG TTT CAT CTC CGA AT-3′, reverse: 5′-AGA CAC TGC CCA CAA CCA CA-3′, β-Actin, forward 5′-GTG GCC GAG GAC TTT GAT TG-3′, reverse: 5′-CCT GTA ACA ACG CAT CTC ATA TT-3′.
Cell viability and cell cycle assay
Cell viability was detected using the Cell Counting Kit 8 (CCK-8, #CK04, Dojindo, Shanghai, China). Transfected SK-MES-1 and H1703 cell lines were seeded into 96-well plates at a density of 5 × 103 cells per well and incubated at 37 °C for 0, 24, 48, 72, and 96 h. Subsequently, CCK-8 solution (10 µl) was added into each well and incubated for 4 h at 37 °C. Optical density (OD) value was determined at 450 nm with a microplate reader (ThermoFisher, Waltham, MA, USA).
For tumor sphere formation assay, LSCC cells were cultured in 6-well ultralow-attachment plate, in serum-free DMEM/F12 supplemented with B27, 20 ng/mL EGF and 20 ng/mL bFGF, and 4 µg/mL heparin. After 2 weeks of culture, the spheres were photographed and counted. For cell cycle detection, 72 h after siRNA transfection, the cells were trypsinized and washed with PBS, then fixed overnight at 4 °C in 70% ethanol, and then incubated with RNase and propidium iodide (PI, # P1304MP, ThermoFisher, Waltham, MA, USA) for 10 min. At last these cells were analyzed by flow cytometry (ThermoFisher, Waltham, MA, USA).
Bromodeoxyuridine (BrdU) labeling
Cells were seeded onto coverslips, which were put into 24-well plates. 24 h later, the cells were incubated using BrdU for 1 h and subsequently probed using anti-BrdU antibody for 2 h (#19–160, Upstate Biotechnology, Lake Placid, New York, USA). Images of cells were collected under a laser scanning microscope (Carl Zeiss, Oberkochen, Germany).
Transwell invasion assay
LSCC cells (SK-MES-1 and H1703 cell lines) were seeded in 24-well plates (5 × 104 cells/well) and incubated in a cell incubator for 20 h. After transfection, the cells kept culturing for another 24 h. The cells were trypsinized and counted, and 1 × 105 cells were seeded into each transwell chamber coated with Matrigel (1:8, 80 µL) and 100 µL serum-free DMEM medium. The complete medium was added to the lower chamber of the Transwell chamber, and after 24 h of culture, the cells in the upper chamber were wiped with a cotton swab. Cells were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet for 10 min. Under the microscope, five fields of view were randomly selected to be photographed and counted. The experiment was repeated three times and the average was calculated.
Wound healing assay
LSCC cells (SK-MES-1 and H1703 cell lines) were seeded in 6-well plates (3 × 105 cells/well) cultured in a cell incubator for 20 h. On the back of the 6-well plate, a uniform horizontal line was drawn and cells were scratched with a pipette (10 µL) along a ruler perpendicular to the horizontal line on the back, and scraped cells were removed by rinsing with PBA. After transfection treatment, culture medium was added. Samples were collected at 0 h and 48 h, and the scratch healing area was calculated as: mobility = scratch healing area/scratch initiation area ×100%. The experiment was repeated in triplicate and the average value was calculated.
Co-immunoprecipitation
Cells were lysed in a lysis buffer containing protease inhibitors and incubated on ice for 30 min. The lysates were clarified by centrifugation at 12,000 g for 15 min at 4 °C, and the supernatant was collected. Antibodies specific to the protein of interest were added to the lysate and incubated overnight at 4 °C with gentle agitation. Protein A/G agarose beads were then added to the mixture and incubated for 2–4 h at 4 °C to capture the immune complexes. The beads were washed extensively with lysis buffer to remove non-specific binding. Bound proteins were eluted from the beads by boiling in SDS-PAGE sample buffer for 5 min. The samples were analyzed by SDS-PAGE and subsequent immunoblotting to detect the presence of co-precipitated proteins.
Western blot
RIPA lysis buffer (#P0013B, Beyotime Biotechnology, Shanghai, China) was used for protein extraction from lung tissue. SDS-PAGE was used to separate protein samples (60 µg) and PVDF membranes (#03010040001, Merck, Rahway, New Jersey, USA) were used for transfer. Extracted protein was then incubated on the membrane overnight at 4 °C after blocking using the primary antibody (FAM65A antibody, 1:1,000;#19325, Cell Signaling Technology, Danfoss, Massachusetts, USA ). Next, HRP-conjugated secondary antibodies (1:5,000; #7074 Cell Signaling Technology, Danfoss, Massachusetts, USA) were used for the incubation in the next day. Visualization was mainly achieved by a fluorescence imager (Alpha, SAN Antonio, California, USA) and the expression levels of specific proteins were normalized to β-actin levels.
Xenograft assays in nude mice
Male athymic BALB/c nude mice (6 weeks old) were purchased from Shanghai Laboratory Animals Center (Shanghai, China) and randomly divided into 3 groups with 5 mice in each group. SK-MES-1 cells were transfected with control vector and FAM65A shRNA according to the instructions of Lipofectamine 3000 (#L3000001, Invitrogen, Waltham, MA, USA). Cells were treated with 5 µg/mL Puromycin (#A1113802, Thermo Fisher Scientific, Waltham, MA, USA) for 2 weeks and injected into mice (106 cells/0.1 ml PBS per mouse) subcutaneously. The needle was inserted into the armpit of the left foreleg at a 45° angle and a 5-mm depth, midway down. The longest diameter (a) and the shortest diameter (b) of the tumor were measured every 3 days with digital calipers, and the tumor volume (V) was calculated according to formula: V = a × b2/2. The mice were killed and photographed 15 days after injection. The xenograft tumors were excised and analyzed. The experiments were conducted in accordance with the approved guidelines of the Laboratory Animal Ethical Committee at China-Japan Friendship Hospital (Approval No.2022-KY-127).
Statistical analysis
All statistical analyses and visualizations were performed using Microsoft Excel, GraphPad Prism 8.0 or R software (version 4.1.0) except as otherwise noted. Clinical characteristics were expressed as mean ± standard deviation or n (%). The Benjamini-Hochberg method was used to control the false discovery rate (FDR). Adjusted p-values below 0.05 were considered significant. Pearson correlation analysis was performed to determine correlations between identified features and clinical parameters.
Results
FAM65A was overexpressed in lung cancer
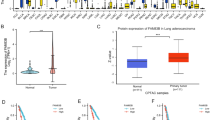
Through evaluating the data sourcing from GEO datasets (Fig. 1A) and TCGA database (Fig. 1B), FAM65A expression in LSCC samples was observed much higher than that in normal samples. In accordance with the results of bioinformatics analysis, the mRNA (Fig. 1C) and protein (Fig. 1D) expression of FAM65A were significantly higher in our collected caner samples compared with normal samples. These results suggested that FAM65A may play an oncogenic role in lung cancer. Through immunohistochemical analysis of clinical samples, lung squamous cell carcinoma tissue was visible with disordered cell arrangement, formation of keratinizing carcinoma nests, increased nuclear volume, increased nuclear-cytoplasmic ratio, deep nuclear staining, different nuclear size and shapes, and abnormal splitting phenomenon (Fig. 1E). FAM65A mRNA expression (Fig. 1F) and protein expression (Fig. 1G) were also observed highly expressed in 4 lung cancer cell lines (SK-MES-1, H1703, H2170 and LK2) compared with normal lung cell line BEAS-2B, and FAM65A had the highest expression in SK-MES-1 cell line.
FAM65A was highly expressed in LSCC as an oncogene. (A). FAM65A showed significantly high expression in cancer tissues GEO datasets GSE75037, GSE67061, GSE18842, GSE2088 and GSE161533; (B). FAM65A expression was increased in cancer tissues compared with ordinary tissues sourced from TCGA database; (C-D). RT-PCR (C) and WB (D) results showed that FAM65A expression in cancer tissues was significantly higher than that in ordinary tissues sourced from the clinical samples we collected. **p < 0.01, ***p < 0.001; (E). Immunohistochemical observation of histomorphology of LSCC tissues and corresponding adjacent tissues. Magnification ×400. (F). RT-PCR detection of FAM65A expression in normal cell line BEAS-2B, LSCC cell lines SK-MES-1, H1703, H2170 and LK2. **p < 0.01; (G). WB detection of FAM65A expression in normal cell line H520, lung cancer cell lines SK-MES-1, H1703, H2170 and LK2. **p < 0.01; ns, no significance
High FAM65A expression led to poor prognosis in LSCC patients
Based on the results of ANOVA analysis of gene expression in different pathological stages, FAM65A expression was found positively associated with pathologic stages and had the highest expression in Stage IV (Fig. 2A). The results of Kaplan-Meier survival analysis showed that FAM65A expression was positively correlated with poorer prognosis. Additionally, the higher the expression level of FAM65A associated with unfavorable prognosis in lung cancer (Fig. 2B). In addition, univariate COX regression analysis was conducted for FAM65A (Fig. 2C). Through systematically analyzing the clinical information of TCGA patient records, including Gender, Pathologic Stage and Race, were, it was found that Stage IV had the highest risk, followed by Stage III. We further conducted survival analysis, and the results showed that patients with high FAM65A expression had significantly lower disease-free survival (DFS) (Fig. 2D), disease-specific survival (DSS) (Fig. 2E), overall survival (OS) (Fig. 2F), and progression-free survival (PFS) (Fig. 2G) compared to patients with low expression. Subsequently, we evaluated the predictive ability for 1-year, 3-year, and 5-year survival (Fig. 2H), and performed both multivariate and univariate COX regression analyses on FAM65A (Fig. 2I-J). The findings indicated that FAM65A was closely associated with poor prognosis.
FAM65A expression was positively correlated with the overall survival rate of LSCC. (A). FAM65A expression in samples of different pathological stages; (B). The relationship between the FAM65A expression and the survival of patients; (C). Univariate cox regression analysis results of FAM65A. (D) Disease-free survival anlysis. (E) Disease-specific survival anlysis. (F) Overall survival anlysis. (G) Progression-free survival anlysis. (H) 1-year, 3-year, and 5-year survival anlysis. (I) Multivariate_cox_regression analysis results of FAM65A. (J) Univariate cox regression analysis results of FAM65A
Enrichment analysis of FAM65A in different biological pathways
Based on KEGG enrichment analysis, it was found that FAM65A was negatively correlated with pathways such as the NF-KAPPA B signaling pathway, proteasome, DNA replication, and mismatch repair, while it was positively correlated with pathways such as focal adhesion, dilated cardiomyopathy, and ECM-receptor interaction (Fig. 3A). Furthermore, according to NSE analysis, pathways such as focal adhesion, dilated cardiomyopathy, and ECM-receptor interaction were positively enriched, whereas pathways such as oxidative phosphorylation and ribosome were negatively enriched (Fig. 3B). Subsequently, through GO-BP analysis, it was discovered that FAM65A was negatively correlated with pathways such as the vascular endothelial growth factor receptor signaling pathway and postsynaptic signaling regulation, while it was positively correlated with pathways such as intracellular substance transport and oxidative phosphorylation (Fig. 3C). Moreover, pathways such as substrate adhesion-dependent cell spreading and neurite organization were positively enriched, while pathways such as oxidative phosphorylation and ATP synthesis coupled electron transport were negatively enriched (Fig. 3D). Further GO-CC analysis showed that FAM65A was negatively correlated with pathways involved in cell adhesion protein complexes and integrin complexes, while it was positively correlated with pathways such as the mitochondrial ribosomal small subunit and cytochrome complexes (Fig. 3E). Pathways such as actomyosin and stress fibers were positively enriched, whereas pathways such as the mitochondrial ribosomal large subunit and cytochrome complexes were negatively enriched (Fig. 3F). Finally, through GO-MF analysis, it was revealed that FAM65A was negatively correlated with pathways such as calcium release channel activity and ligand-gated calcium channel activity, while it was positively correlated with structural matrix proteins and NADH dehydrogenase complexes (Fig. 3G). Pathways such as ARF guanine nucleotide exchange factor activity and calcium release channel activity were positively enriched, while pathways such as the mitochondrial ribosomal large subunit and cytochrome c oxidase were negatively enriched (Fig. 3H).
FAM65A regulated cellular immune infiltration
The correlation between FAM65A expression and the level of immune cell infiltration, including B cells, CD4 + T cells, CD8 + T cells, myeloid dendritic cells, macrophages and neutrophil cells (Fig. 4). The results showed that the expression of FAM65A had a strong positive correlation with CD4 + T cells (R = 0.31, P = 4.3e-14), myeloid dendritic cells (R = 0.31, P = 2.1e-13) and neutrophils (R = 0.18, P = 1.9e-5), indicating that FAM65A maybe paly a certain role of cellular immune infiltration.
Analysis of FAM65A interaction protein
We identified seven genes interacting with FAM65A, namely FAU, PDCD10, RHOB, RHOC, STK24, STK25, and STK26, and constructed a PPI interaction network (Fig. 5A). We further conducted a correlation analysis between FAM65A and the interacting genes and found that the expression of FAM65A was negatively correlated with PDCD10, FAU, and STK26, while it was positively correlated with RHOB, STK25, RHOC, and STK24 (Fig. 5B). Subsequent scatter plot analyses and Pearson correlation analysis confirmed a significant negative correlation between FAM65A and FAU, PDCD10, and STK26, whereas a positive correlation was observed between FAM65A and RHOB, as well as STK25 (Fig. 5C-G).
Figure 4. FAM65A expression was related with cellular immune infiltration. (A) PPI protein interaction map. (B) Expression correlation between FAM65A and other genes. (C) Scatter plot of FAM65A and FAU gene expression values. (D) Scatter plot of FAM65A and PDCD10 gene expression values. (E) Scatter plot of FAM65A and RHOB gene expression values. (F) Scatter plot of FAM65A and STK25 gene expression values. (G) Scatter plot of FAM65A and STK26 gene expression values
Down-regulating FAM65A affected biological characteristics of LSCC cells
In order to further explore the effect of FAM65A expression on the biological behavior of LSCCC cells, we knocked down FAM65A in two LSCC cell lines (SK-MES-1 and H1703) and explored the effect of knocking down FAM65A on the biological behavior of LSCC cells. Firstly, the knockdown effect of FAM65A in SK-MES-1 and H1703 cells was confirmed. Western blot analysis revealed a significant decrease in FAM65A expression in the sh-FAM65A group (Fig. 6A-B). As shown in Fig. 6C-D, the BrdU labeling and CCK8 assays showed that the proliferation of LSCC cells was significantly suppressed after knocking down of FAM65A (p < 0.01). In addition, knock-down of FAM65A also affected the cell cycle of LSCC cells (Fig. 7). The percentage of LSCC cells at G2/M phase, where apoptosis might be induced, was increased (SK-MES-1, p < 0.05; H1703, p < 0.01). Next, the effect of FAM65A on the capacity of invasion and metastasis of lung cancer cells was detected. From the results of transwell assay and wound healing assay, it was found that the ability of invasion and metastasis of lung cancer cells was significantly weakened (Fig. 8A and B). The results of western blotting analysis showed that in SK-MES-1 and H1703 cell lines, the expression of N-cadherin, fibronectin and vimentin was significantly decreased and the expression of E-cadherin was significantly increased after silencing FAM65A compared with the control group (Fig. 8C). The decreased N-cadherin and vimentin protein expression indicated that FAM65A silencing impaired epithelial-mesenchymal transition (EMT) in LSCC cells. To further verify the interaction between FAM65A and RHOB, we conducted co-immunoprecipitation experiments and found a significant interaction between FAM65A and RHOB in the SK-MES-1 and H1703 cell lines (Fig. 8D).
FAM65A regulated the capacity of LSCC cells proliferation. (A). Detection of FAM65A protein expression levels using Western blot; (B) Quantitative analysis bar graph for the Western blot experiment; (C). BrdU labeling was used to detect the proliferation ability of LSCC cell line SK-MES-1 and H1703 after knockdown of FAM65A. Scale bar = 20 μm; (D). CCK8 was used to detect the proliferation ability of LSCC cell line SK-MES-1 and H1703 after knockdown of FAM65A. Each experiment was repeated three times. **p < 0.01; ***p < 0.001
FAM65A promoted the LSCC cells invasion and migration ability, and the expression of EMT related genes. (A). Knockdown of FAM65A inhibited the migration of SK-MES-1 and H1703 cells. Scale bar = 50 μm. **p < 0.01; (B). The wound healing assay showed that sh-KIF1A inhibited LSCC cells migration. Scale bar = 100 μm. **p < 0.01; ***p < 0.001; (C) RT-PCR results of the EMT related genes expression. (D) FAM65A and RHOB protein interactions. Each experiment was repeated three times. **p < 0.01
FAM65A promoted LSCC in vivo
To investigate the effect of FAM65A in vivo, xenograft assays were conducted using SK-MES-1 FAM65A KO cells. FAM65A depletion was demonstrated having profound effects on growth of SK-MES-1 xenograft tumor (Fig. 9A). Upon observing and measuring the tumor volume, it was found that the tumor size in the sh-FAM65A group of mice was significantly smaller compared to the sh-Vector group (Fig. 9B). This clarified the impact of FAM65A on tumor formation. Subsequently, we conducted Western blot experiments to examine EMT markers and discovered that knocking down FAM65A significantly inhibited the EMT process (Fig. 9C-D). Further, by collecting tumor tissue samples and performing immunohistochemical staining to detect N-cadherin expression, it was found that the expression level of N-cadherin significantly decreased after FAM65A knockdown (Fig. 9E). In conclusion, we found that the tumor incidence in cells with FAM65A knockdown was significantly lower than that in negative control cells, and the knockdown notably inhibited the EMT process.
FAM65A promoted LSCC in vivo. (A). Representative image of tumors at the end point. There were 5 nude mice in each group; (B). Growth curve of nude mice inoculated with sh-Vector and sh-FAM65A SK-MES-1cells; (C). Detection of epithelial-mesenchymal transition marker protein levels using Western blot; (D). Quantitative analysis of Western blot results presented as bar graphs; (E). Detection of N-cadherin expression using immunohistochemical staining. **p < 0.01
Discussion
As early as 1986, Dulbecco R had pointed out that the sequence analysis of the human genome was the turning point in cancer research [40]. With the popularization of high-throughput sequencing technology, genome and transcriptome research has played an important role in screening tumor molecular markers, determining tumor therapeutic targets, and exploring tumor transformation mechanisms [41,42,43]. In the present study, by analyzing the sequencing or microarray data in GEO and TCGA databases, it was concluded that FAM65A is upregulated in LSCC and associated with poor prognosis [44]. High expression of FAM65A was also validated in our collected clinical samples and LSCC cell lines. Further, we preliminarily explored the role of FAM65A in the occurrence and development of LSCC through in vitro and in vivo experiments. The results showed that FAM65A can promote the proliferation, invasion and metastasis of LSCC, and down-regulation of FAM65A can block the cell cycle of LSCC and may cause apoptosis.
Gene expression plays an important role in the pathogenesis of human tumors [45,46,47]. “Mutation” has been reported to be the predominant form of FAM65A DNA alteration in all tumors [44]. Notably, specific gene mutations are valuable in predicting prognosis in some cancer patients. For most proteins, phosphorylation is an extremely important regulatory mechanism [48, 49]. Phosphorylation at the S351 locus of FAM65A was experimentally confirmed as early as 2009 [50]. In a previous study, the total protein phosphorylated expression level of FAM65A at S351 was reported to be down-regulated in primary tumors [44]. The mutation of FAM65A in LSCC is of interest.
Although a lot of analysis has been done in the early stage of this paper, it is still necessary to further explore the specific mechanism of FAM65A promoting the process of LSCC. The role of FAM65A as a Rho effector protein is most noteworthy. FAM65A is an adaptor protein consisting of a C-terminal ARM domain and an N-terminal HR1 domain. The ARM domain binds to CCM3, which in turn binds to MST3 and MST4, whereas HR1 domain interacts with RHO proteins through a GTP-dependent manner. FAM65A and RHO activity has no effect on MST3 and MST4 kinase activity, but binding of active RHO to FAM65A results in a new localization of MST4 from the Golgi to some cytoplasmic puncta in a CCM3- and FAM65A-dependent manner. There were still limitations in this study, including the lack of in-depth exploration into the mechanisms by which FAM65A affects the progression of LSCC and the deficiencies in the animal experiments. In the future, we will further refine and supplement the experimental results to make the conclusion that ‘FAM65A plays a critical role in the progression of LSCC’ more specific and convincing. Considering the mutation of FAM65A in LSCC, the mechanism of FAM65A in LSCC deserves further exploration.
Conclusion
Collectively, our findings suggest that FAM65A plays a crucial role in the progression of LSCC. FAM65A is overexpressed in LSCC and predicts poor prognosis and promote LSCC invasion and metastasis. Our results provide a better understanding of the role of FAM65A in LSCC progression and a potential therapeutic target and prognostic predictor against this malignancy.
Data availability
The data used to support the findings of this study are included within the article. The raw data could be shared at https://www.jianguoyun.com/p/DcXrzaAQmv2MCxi8n-MEIAA.
Abbreviations
- LSCC:
-
Lung Squamous Cell Carcinoma
- EGFR:
-
Epidermal Growth Factor Receptor
- ALK:
-
Anaplastic Lymphoma Kinase
- ROS1:
-
c-Ros Proto-Oncogene 1
- EGFR-TKIs:
-
Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitor
- EMT:
-
Epithelial-Mesenchymal Transition
- RNA-seq:
-
RNA Sequencing
- TPM:
-
Transcripts Per Million
References
Schabath MB, Cote ML. Cancer Progress and priorities: Lung Cancer. Cancer Epidemiol Biomarkers Prev. 2019;28(10):1563–79.
Nasim F, Sabath BF, Eapen GA. Lung Cancer. Med Clin North Am. 2019;103(3):463–73.
Nooreldeen R, Bach H. Current and Future Development in Lung Cancer diagnosis. Int J Mol Sci 2021, 22(16).
Maomao C, He L, Dianqin S, Siyi H, Xinxin Y, Fan Y, Shaoli Z, Changfa X, Lin L, Ji P, et al. Current cancer burden in China: epidemiology, etiology, and prevention. Cancer Biol Med. 2022;19(8):1121–38.
Bade BC, Dela Cruz CS. Lung Cancer 2020: epidemiology, etiology, and Prevention. Clin Chest Med. 2020;41(1):1–24.
Seijo LM, Peled N, Ajona D, Boeri M, Field JK, Sozzi G, Pio R, Zulueta JJ, Spira A, Massion PP, et al. Biomarkers in Lung Cancer Screening: achievements, promises, and challenges. J Thorac Oncol. 2019;14(3):343–57.
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63.
Sears CR, Mazzone PJ. Biomarkers in Lung Cancer. Clin Chest Med. 2020;41(1):115–27.
Guo H, Zhang J, Qin C, Yan H, Liu T, Hu H, Tang S, Tang S, Zhou H. Biomarker-Targeted Therapies in Non-Small Cell Lung Cancer: Current Status and Perspectives. Cells 2022, 11(20).
Hoy H, Lynch T, Beck M. Surgical Treatment of Lung Cancer. Crit Care Nurs Clin North Am. 2019;31(3):303–13.
Yin J, Wu Y, Yang X, Gan L, Xue J. Checkpoint inhibitor Pneumonitis Induced by Anti-PD-1/PD-L1 therapy in Non-small-cell Lung Cancer: occurrence and mechanism. Front Immunol. 2022;13:830631.
Dantoing E, Piton N, Salaun M, Thiberville L, Guisier F. Anti-PD1/PD-L1 immunotherapy for Non-small Cell Lung Cancer with Actionable Oncogenic driver mutations. Int J Mol Sci 2021, 22(12).
Cimadamore A, Massari F, Santoni M, Lopez-Beltran A, Cheng L, Scarpelli M, Montironi R, Moch H. PD1 and PD-L1 inhibitors for the treatment of kidney Cancer: the role of PD-L1 assay. Curr Drug Targets. 2020;21(16):1664–71.
Fu K, Xie F, Wang F, Fu L. Therapeutic strategies for EGFR-mutated non-small cell lung cancer patients with osimertinib resistance. J Hematol Oncol. 2022;15(1):173.
Herrera-Juarez M, Serrano-Gomez C, Bote-de-Cabo H, Paz-Ares L. Targeted therapy for lung cancer: beyond EGFR and ALK. Cancer. 2023;129(12):1803–20.
Yu ZQ, Wang M, Zhou W, Mao MX, Chen YY, Li N, Peng XC, Cai J, Cai ZQ. ROS1-positive non-small cell lung cancer (NSCLC): biology, diagnostics, therapeutics and resistance. J Drug Target. 2022;30(8):845–57.
Giatromanolaki A, Sivridis E, Arelaki S, Koukourakis MI. Expression of enzymes related to glucose metabolism in non-small cell lung cancer and prognosis. Exp Lung Res. 2017;43(4–5):167–74.
Song T, Yu W, Wu SX. Subsequent treatment choices for patients with acquired resistance to EGFR-TKIs in non-small cell lung cancer: restore after a drug holiday or switch to another EGFR-TKI? Asian Pac J Cancer Prev. 2014;15(1):205–13.
Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol. 2021;33(1):40–6.
Trepat X, Chen Z, Jacobson K. Cell migration. Compr Physiol. 2012;2(4):2369–92.
Inagaki N, Katsuno H. Actin waves: origin of cell polarization and Migration? Trends Cell Biol. 2017;27(7):515–26.
Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol. 2015;36:103–12.
Narumiya S, Thumkeo D. Rho signaling research: history, current status and future directions. FEBS Lett. 2018;592(11):1763–76.
Clayton NS, Ridley AJ. Targeting rho GTPase signaling networks in Cancer. Front Cell Dev Biol. 2020;8:222.
Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40(6):1378–82.
Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9(11):846–59.
Sahai E, Marshall CJ. RHO-GTPases and cancer. Nat Rev Cancer. 2002;2(2):133–42.
Ni D, Li X, He X, Zhang H, Zhang J, Lu S. Drugging K-Ras(G12C) through covalent inhibitors: Mission possible? Pharmacol Ther. 2019;202:1–17.
Karmakar S, Kaushik G, Nimmakayala R, Rachagani S, Ponnusamy MP, Batra SK. MicroRNA regulation of K-Ras in pancreatic cancer and opportunities for therapeutic intervention. Semin Cancer Biol. 2019;54:63–71.
Shui B, La Rocca G, Ventura A, Haigis KM. Interplay between K-RAS and miRNAs. Trends Cancer. 2022;8(5):384–96.
Shalom B, Farago M, Salaymeh Y, Sebban S, Risling M, Pikarsky E, Katzav S. Vav1 accelerates ras-driven lung cancer and modulates its tumor microenvironment. Cell Signal. 2022;97:110395.
Faraz K, Mardakheh AS, Christopher J, Marshall. RHO binding to FAM65A regulates Golgi reorientation during cell migration. J Cell Sci. 2016;129(24):4466–79.
Kotawong K, Thitapakorn V, Roytrakul S, Phaonakrop N, Viyanant V, Na-Bangchang K. Plasma phosphoproteome and differential plasma phosphoproteins with opisthorchis viverrini-related cholangiocarcinoma. Asian Pac J Cancer Prev. 2015;16(3):1011–8.
Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba et al. II : An Expression Signature as an Aid to the Histologic Classification of Non-Small Cell Lung Cancer. Clin Cancer Res 2016, 22(19):4880–4889.
Tong R, Feng L, Zhang L, Zhang J, Mao Y, Zhang K, Gao Y, Wang G, Cheng S. Decreased Interferon Alpha/Beta Signature Associated with Human Lung Tumorigenesis. J Interferon Cytokine Res. 2015;35(12):963–8.
Sanchez-Palencia A, Gomez-Morales M, Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R, Farez-Vidal ME. Gene expression profiling reveals novel biomarkers in nonsmall cell lung cancer. Int J Cancer. 2011;129(2):355–64.
Fujiwara T, Hiramatsu M, Isagawa T, Ninomiya H, Inamura K, Ishikawa S, Ushijima M, Matsuura M, Jones MH, Shimane M, et al. ASCL1-coexpression profiling but not single gene expression profiling defines lung adenocarcinomas of neuroendocrine nature with poor prognosis. Lung Cancer. 2012;75(1):119–25.
Nayshool O, Kol N, Javaski E, Amariglio N, Rechavi G. SurviveAI: Long Term Survival Prediction of Cancer patients based on somatic RNA-Seq expression. Cancer Inf. 2022;21:11769351221127875.
Spiess M, Fernandez D, Nguyen T, Liu I. Generalized estimating equations to estimate the ordered stereotype logit model for panel data. Stat Med. 2020;39(14):1919–40.
Dulbecco R. A turning point in cancer research: sequencing the human genome. Science. 1986;231(4742):1055–6.
Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348–65.
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu J, Yu X, Shi S. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol. 2021;14(1):91.
Hong M, Tao S, Zhang L, Diao LT, Huang X, Huang S, Xie SJ, Xiao ZD, Zhang H. RNA sequencing: new technologies and applications in cancer research. J Hematol Oncol. 2020;13(1):166.
Liang W, Mo C, Wei J, Chen W, Gong W, Shi J, Hou X, Li C, Deng Y, Ou M. FAM65A as a novel prognostic biomarker in human tumors reveal by a pan-cancer analysis. Discov Oncol. 2021;12(1):60.
Group PTC, Calabrese C, Davidson NR, Demircioglu D, Fonseca NA, He Y, Kahles A, Lehmann KV, Liu F, Shiraishi Y, et al. Genomic basis for RNA alterations in cancer. Nature. 2020;578(7793):129–36.
Jin F, Yang Z, Shao J, Tao J, Reissfelder C, Loges S, Zhu L, Scholch S. ARID1A mutations in lung cancer: biology, prognostic role, and therapeutic implications. Trends Mol Med. 2023;29(8):646–58.
Ascione CM, Napolitano F, Esposito D, Servetto A, Belli S, Santaniello A, Scagliarini S, Crocetto F, Bianco R, Formisano L. Role of FGFR3 in bladder cancer: treatment landscape and future challenges. Cancer Treat Rev. 2023;115:102530.
Bilbrough T, Piemontese E, Seitz O. Dissecting the role of protein phosphorylation: a chemical biology toolbox. Chem Soc Rev. 2022;51(13):5691–730.
Zhang WJ, Zhou Y, Zhang Y, Su YH, Xu T. Protein phosphorylation: a molecular switch in plant signaling. Cell Rep. 2023;42(7):112729.
Chen RQ, Yang QK, Lu BW, Yi W, Cantin G, Chen YL, Fearns C, Yates JR 3rd, Lee JD. CDC25B mediates rapamycin-induced oncogenic responses in cancer cells. Cancer Res. 2009;69(6):2663–8.
Acknowledgements
Not applicable.
Funding
This work was supported by the Key Clinical Projects of Peking University Third Hospital [Grant No. BYSYRCYJ2023001] and the Fundamental Research Funds for the Central Universities [Grant No.3332020078].
Author information
Authors and Affiliations
Contributions
Fangjun Chen contributed to the conception of the study. Guangliang Qiang, Peng Ren and Rui Xu contributed to the bioinformatic analysis. Jin Zhang and Guangliang Qiang contributed to experiments and manuscript preparation. Chaoyang Liang and Guangliang Qiang contributed significantly to writing-reviewing and editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This project with animal experiments was approved by the Ethics Committee of China-Japan Friendship Hospital with approval No.2022-KY-127.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, F., Ren, P., Xu, R. et al. FAM65A promotes the progression and growth of lung squamous cell carcinoma in vivo and vitro. BMC Cancer 24, 944 (2024). https://doi.org/10.1186/s12885-024-12701-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12701-z