Abstract
Background
Researches have manifested that the disorder of iron metabolism is participated in Gastric cancer (GC), but whether iron metabolism-relevant genes (IMRGs) is related to the survival outcome of GC remain unknown.
Methods
Eleven tumor as well as nine adjacent normal tissues from GC patients were underwent mRNA sequencing, and the The Cancer Genome Atlas Stomach Cancer (TCGA-STAD) datasets were acquired from the TCGA database. Cox analyses and least absolute shrinkage and selection operator (LASSO) regression were applied to build a IMRGs signature. The relationship between signature genes and the infiltration profiling of 24 immune cells were investigated using single-sample GSEA (ssGSEA). Meanwhile, the potential biological significance, genes that act synergistically with signature genes, and the upstream regulatory targets were predicted. Finally, the abundance of the signature genes were measured via the quantitative real-time PCR (qRT-PCR).
Results
A IMRGs signature was constructed according to the expression and corresponding coefficient of DOHH, P4HA3 and MMP1 (The Schoenfeld individual test showed risk score was not significant with P values = 0.83). The prognostic outcome of patients in the high-risk group was terrible (p < 0.05). Receiver operating characteristic (ROC) curves confirmed that the IMRGs signature presented good efficiency for predicting GC prognosis (AUC > 0.6). The nomogram was performed well for clinical utilize (C-index = 0.60), and the MMP1 expression significantly increased in the cohorts at age > 60 and Stage II-IV (p < 0.05). The positive correlation of P4HA3 and MMP1 expression as well as the negative correlation of DOHH expression with risk score (p < 0.0001) and worse prognosis (p < 0.05) were detected as well. Furthermore, 11 differential immune cells were associated with these signature genes (most p < 0.01). Finally, qRT-PCR revealed that the abundance of DOHH, P4HA3 and MMP1 were high in tumor cases, indicating the complex mechanism between the high expression of DOHH as a protective factor and the high expression of P4HA3 and MMP1 as the risk factors in the development of GC.
Conclusion
An iron metabolism-related signature was constructed and has significant values for foretelling the OS of GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer (GC) is a malignancy springing from the mucosal epithelial cells of stomach wall, which is one of the common digestive tract tumors. By 2020, the incidence of GC ranks fifth, and the mortality rate of GC ranks fourth [1]. At present, the treatment of gastric cancer is still mainly surgical treatment, combined with systemic chemotherapy, immunotherapy, targeted therapy, radiotherapy, and other auxiliary treatment mode. However, these treatments did not raise the five-year survival rate of patients with advanced GC [2]. In addition, GC patients are characterized by low early diagnosis rate, low radical resection rate and low 5-year survival rate [4,5,6]. Relevant research results show that advanced GC patients' median survival time is less than 1 year, and the five-year survival rate is about 18% [5,6,7,8]. The prognosis of metastatic GC patients is very poor, as the median of survival time is just 4–9 months [9]. Therefore, we need to find biomarkers with good prognosis prediction for GC, and to provide a new basis for clinical gene detection, targeted therapy and individualized treatment of GC.
Iron is an essential nutrient for promoting cell metabolism, proliferation and growth. The hydrogen peroxide was catalyzed by ferrous iron to generate reactive oxygen species through Fenton reavtion, which not only causes lipid and protein damage, but also oxidative damage DNA, inducing mutations and facilitating the emergence and preservation of tumors. Iron metabolism disorder is engaged in tumor occurrence, angiogenesis, invasion as well as metastasis, which is a general appearance in many tumors. Related studies have found that there are abnormal iron metabolism in lung cancer, prostate cancer, liver cancer, breast cancer, and, kidney cancer [10,11,12,13]. Some researches have also manifested that iron metabolism disorder is engaged in the process of gastric cancer, but whether it is linked to the prognosis of GC and the specific molecular mechanism are still unknown.
In this study, we first screened the DE-IMRGs from the GC expression profile of The Cancer Genome Atlas (TCGA) database, then constructed the prognostic signature of iron metabolism related GC, and probed the linkage between the immune cells and signature genes. Finally, the expression of signature genes was confirmed through external datasets and quantitative real-time PCR (qRT-PCR), which is a great significance for GC to explore potential therapeutic targets and molecular mechanisms.
Materials and methods
Data sourse
This research was allowed by the ethical committee of The First Affiliated Hospital of Kunming Medical University. All patients in this study signed written informed consent documents.
Specimen acquisition & sequencing: 11 tumor tissues and 9 normal tissues from GC patients were included in mRNA-seq analysis, where normal tissues were selected at least 1.0 cm from the tumor margin. These specimens were all gathered from The First Affiliated Hospital of Kunming Medical University and were freshly frozen and reserved at -80 °C immediately after surgery. Afterward, the total RNA from tissue cases was extracted using TRIzol reagent (Invitrogen, CA, USA). The RNA integrity was assessed by Bioanalyzer 2100 (Agilent, CA, USA) and then a final cDNA library with the average insert size of 300 + 50 bp by PCR was estimated. Finally, the mRNA sequencing was run relying on the illumina Novaseq™ 6000 (LC Bio Technology CO.,Ltd. Hangzhou, CHN).
Collection of TCGA data and iron metabolism-relevant genes (IMRGs): Additionally, the mRNA data, clinicopathological data and DNA methylation information from the TCGA Stomach Cancer (STAD) datasets were regained from the TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga, accessed on 13 October 2017), including 373 tumor cases and 32 normal cases. The 428 IMRGs cohorts (v7.4) was downloaded from the Molecular Signatures Database (MSigDB v2023.1.Hs) database (https://www.gsea-msigdb.org/gsea/msigdb) with the keywords of Iron metabolism and we retrieved 428 IMRGs.
Identification of differentially expressed genes (DEGs)
The mRNA data from the sequencing data were selected for identification of the DEGs through edgeR package [14, 15]. Similarly, the mRNA data from the 373 tumor and 32 normal cases of TCGA database were used to identify the DEGs via edgeR package [14, 15]. The DEGs were displayed in the volcano map by ggplot2 package. The overlapping genes of the IMRGs, the DEGs of TCGA and the DEGs of sequencing data were selected by Venn tool. These overlapping genes were DE-IMRGs.
The correlation between DEGs and CpG site methylation
The CpG site methylation levels of DE-IMRGs were extracted from the TCGA, and the relationship were estimated between the DE-IMRGs and CpG site methylation levels via psych [16] in R (|cor|> 0.25).
Construction and evaluation of a IMRGs signature
The univariate and multivariate cox regression analyses were firstly run to select survival-related genes using survminer R package (version 0.4.8). The prognostic genes were further confirmed using LASSO regression algorithm via glmnet R package (version 4.1–3). After that, the 345 cancer cases with complete survival data in the TCGA database were classified into two cohorts, namely training cohort (242) and validation cohort (103), according to 7:3. The formula for calculation of risk score was ‘h0(t)*exp(β1X1 + β2X2 + … + βnXn)’ via survival in R. The Global Schoenfeld residual test was employed for checking Proportional Hazard (PH) assumptions and estimating the partial residuals estimated by the Cox proportional hazards model via a residual plot. Further, the cases were separated into low- and high-risk group based on the median of risk-score. Besides, the Kaplan–Meier survival curve as well as the ROC curve were produced to evaluate the signature, and were drawn via survival and the survivalROC in R software.
Independent prognostic analysis of IMRGs signature
All of the clinicopathological factors of 345 cancers with complete clinical information were subjected to the model for COX independent prognostic analysis to assess the independent clinical prognostic factors. Afterward, the nomogram was built for GC patients to predict the survival of via the R package rms.
Correlation analysis between the IMRGs signature and clinical characteristics
The clinical information (Age, gender, pathological M, N, T stage, tumor stage and treatment type), survival information of GC data in the TCGA database were collected, as well as the classification of low- and high-risk groups and signature genes. Heatmap of signature genes expression between different clinical subgroups was displayed using pheatmap (version 1.0.12) and magrittr (version 2.0.1) and the statistic results were exhibited with the box plots using Wilcoxon test. And meanwhile, the survival differences of 345 patients with GC in high- and low- expression patterns of signature genes was analyzed using Kaplan–Meier survival analysis.
Immune infiltration analysis
The ssGSEA algorithm and Wilcoxon test were applied to estimate the infiltration profile of immune cell and the differences in immune infiltration between two risk subgroups. Meanwhile, the relation between the 24 immune cells and signature genes were investigated based on the ssGSEA algorithm.
Gene Set Enrichment Analysis (GSEA)
GSEA were conducted for the potential biological significance and classical functions involving signature genes in GC. Using ‘c5.go.v7.4.entrez.gmt’ (GO) and ‘c2.cp.kegg.v7.4.entrez.gmt’ (KEGG) downloaded from the GSEA website (http://www.gsea-msigdb.org/gsea/msigdb, accessed on 6 September 2023) as the background gene set, the high- and low- expression groups of each signature gene were divided based on the median value of the expression values for GSEA using clusterProfiler [17] (version 4.0.2) and org.Hs.eg.db (version 3.13.0), and threshold was set to | NES |> 1, NOMP < 0.05, q < 0.25.
Collaborative gene analysis of signature
In order to study the genes that cooperate with the signature genes, R package psych [16] was run to count the pearson correlation between the three signature genes with all genes, and then performed the correlation analysis according to |Pearson coefficient value|> 0.6 with FDR < 0.05 threshold.
The function annotation of different collaborative genes were analyzed via the ClusterProfiler [17,18,19,20]. Additionally, enrichment analysis was performed separately for each collaborative gene based on a significance threshold p < 0.05 and visualized by ggplot2 in R.
Transcription factors (TFs)-miRNA-mRNA network of signature genes
In order to reveal which miRNAs targeting signature genes and which TF interacting with signature genes may be involved in the prognosis in GC, TRRUST database (http://www.grnpedia.org/trrust, accessed on 6 September 2023) and miRWalk database (http://mirwalk.umm.uni-heidelberg.de/, accessed on 6 September 2023) were utilized for prediction analysis. The TF-miRNA-mRNA network was constructed and visualized using Cytoscape software.
qRT-PCR
The total RNA of the tumor cases (10) and the normal cases (10) were extracted using TRIzol Reagent (Invitrogen, CA, USA). The sweScript RT I First strand cDNA SynthesisAll-in-OneTM First-Strand cDNA Synthesis Kit (Servicebio, WuHan, CHN) was used for reverse transcription. The primer sequences were showed in the Table S1. The reference gene GAPDH was used in qRT-PCR experiments. Finally, the relative abundance of signature genes was detected by the 2xUniversal Blue SYBR Green qPCR Master Mix (Servicebio, WuHan, CHN) and standardized with the 2−ΔΔct method.
Results
Identification of DEGs
In the sequencing data, 11,481 DEGs were screened from the tumor and normal cases (Fig. 1A). In TCGA database, 5062 DEGs were screened from the tumor and normal cases (Fig. 1B). Finally, 58 DE-IMRGs were obtained from 5062 DEGs of TCGA, 11,481 DEGs of sequencing data and 428 IMRGs (Fig. 1C).
The linkage between DEGs and methylation of CpG site
Ten DEGs whose expression were associated with CpG sites were obtained to explore the potential regulation of DNA methylation on gene expression (Table 1), including SLC11A1, FANCG, CCND1, ALKBH2, CDK5RAP1, CDO1, HMBS, NTHL1, FXN and KIF23.
Construction and evaluation of a IMRGs signature
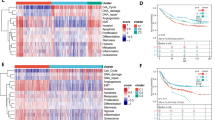
The 58 DE-IMRGs of the TCGA database were imported to univariate COX regression and five prognosis related genes (P4HA3, DOHH, POLD1, MMP1 and FANCE) were obtained (Fig. 2A). These five genes were further imputed in multivariate Cox regression analysis, indicating DOHH (coef = -0.38, HR = 0.68, p = 0.05), P4HA3 (coef = 0.30, HR = 1.36, p = 0.09) and MMP1 (coef = 0.08, HR = 1.08, p = 0.11) were selected as signature genes (Fig. 2B). Further, the LASSO algorithm confirmed the importance of three prognostic genes, and DOHH, P4HA3, and MMP1 were involved in the Cox proportional hazards model for construction of the IMRGs signature (Fig. 2C). The Schoenfeld individual test showed risk score was not significant with P values = 0.83 (Fig. 2D), suggesting the PH assumption of the IMRGs signature remained inviolate.
Construction and evaluation of a IMRGs signature. A Univariate Cox regression analysis selected 5 prognosis related genes (P4HA3, DOHH, POLD1, MMP1 and FANCE); B Multivariate Cox regression analysis selected 3 prognosis related genes (DOHH, P4HA3 and MMP1) as signature genes. C LASSO coefficients profiles (left) to determine the number of factors and cross-validation diagram (right) for tuning parameter selection in the least absolute shrinkage and selection operator (LASSO) model. From left to right along the x-axis, with the increases of lambda, the compression parameter increases and the absolute value of the coefficient decreases. The number on top are the number of variables with non-zero regression coefficients in the LASSO model. Variables with non-zero coefficients are important features for our screening. D The Schoenfeld individual test showed P values = 0.83
The patients in training cohort were classified into low- and high-risk groups (Fig. 3A). In addition, the signature genes of P4HA3 and MMP1 were positively linked to risk-score, but the DOHH was negatively associated with risk-score (Fig. 3B). The Kaplan–Meier curve showed that patients with higher risk had a poorer prognosis with p < 0.01 (Fig. 3C). In details, the AUCs for OS (1-, 2-, 3-, 4- and 5-years) were 0.617, 0.604, 0.629, 0.653 and 0.711 (Fig. 3D).
Construction of the signature genes prognostic signature based on signature genes in the training cohort. A The distribution of the risk score, OS. B Heatmap of the signature genes correlated with risk-score. C Kaplan Meier analysis of two risk groups. D AUC value of prediction performance of the model for survival rate (1, 2, 3, 4 and 5 year)
Likewise, the GC patients of validation cohort were segmented into low- and high-risk groups (Fig. 4A). The correlation of risk-score and signature genes (Fig. 4B), and the result of Kaplan–Meier curve were similar with training cohort (p = 0.01) (Fig. 4C). Moreover, the AUCs for 1-year was 0.675; 2-years was 0.649; 3-years was 0.609; 4-years was 0.602 and 5-years was 0.602 (Fig. 4D).
Construction of the signature genes prognostic signature in the validation cohort. A The distribution of the risk score, OS. B Heatmap of the signature genes correlated with risk-score. C Kaplan Meier analysis of two risk groups. D AUC value of prediction performance of the model for survival rate (1, 2, 3, 4 and 5 year)
Independent prognostic analysis of IMRGs signature
The 345 patients of TCGA with complete information including treatment type, risk-score, ajcc pathologic stage, gender, ajcc pathologic m, age at index and ajcc pathologic t were included for univariate Cox regression analysis. As shown in Fig. 5A, treatment type and risk-score were related with risk model. Moreover, treatment type and risk-score were selected as independent prognostic factors by multivariate Cox regression analysis (Fig. 5B). Next, treatment type and risk-score were included to establish a nomogram and the C-index of nomogram model was 0.60 (Fig. 5C). Furthermore, the calibration effect of 1- and 3-years in calibration curve were performed well (Fig. 5D). Decision curve analysis (DCA) shows that the nomogram model achieves better net benefit than 1-year OS rate (Fig. 5E).
Building and evaluation of a nomogram model linked to signature. A Univariate Cox regression analysis; B Multivariate Cox regression analysis to select independent prognostic factors. C The nomogram model comprised of independent prognostic factors; D Calibration curves of OS (1-, 3-, and 5-year) predicted by the nomogram; E The DCA curves to show the expected net benefits based on the nomogram prediction at different threshold probabilities. None: assume an event will occur in no patients (horizontal black line); All: assume an event will occur in all patients (green line)
Correlation analysis between the IMRGs signature and clinical characteristics
The heatmap and box plots showed that the expression of the P4HA3 and MMP1 were high in GC patients of high-risk group. However, the abundance of the DOHH was high in GC of low-risk group (p < 0.0001) (Fig. 6A-B). And meanwhile, patients with high expression of P4HA3 and MMP1 had worse prognosis than that with low expression, otherwise, individuals with high expression of DOHH had a high survival probability (p < 0.05) (Fig. 6C), consist with the results generated above. Further, it can be seen that the expression of MMP1 was related to the age and tumor stage of GC patients, that is, the MMP1 expression significantly increased in the cohorts at age > 60 and Stage II-IV (p < 0.05) (Fig. 6B).
The linkage between the IMRGs signature and clinical characteristics. A Heatmap for the expression of three signature genes in different clinical sub-groups. B The boxplot for the expression of three signature genes in different clinical sub-groups (age, risk score, tumor pathologic stage); C Kaplan Meier analysis of high- and low- expression groups of each signature gene
Immune infiltration analysis
For studying the imparity of immune infiltration in patients with differing risks, the spearman correlations for 24 immune cells were calculated. The 11 immune cells were different between the two risk GC subgroups (Fig. 7A). The relationship between the signature genes and 24 immune cells and signature genes shown that P4HA3 was significantly associated with 17 immune cells; MMP1 was significantly associated with 12 immune cells; DOHH was significantly related to 18 immune cells (Fig. 7B).
GSEA of signature genes
For the biological functions and signaling pathways involving signature genes, GSEA were employed under the GO and KEGG analysis (Fig. 8A-B). It was indicated that P4HA3 was mainly enriched in the cytokine-cytokine receptor, interaction pathways in cancer and the functions of extracellular matrix structural constituent. DOHH is mainly functionally related to the KEGG pathways of cell cycle, calcium signaling pathway, as well as the GO functions of DNA replication and muscle contraction. MMP1 might be relevant to the enrichment of Toll-like/Nod-like receptor signaling pathways and various catabolic processes. Besides, it was observed that ascular smooth muscle contraction term was common to DOHH and MMP1.
Collaborative gene analysis of signature genes
To investigate genes that act synergistically with signature genes, the pearson correlations were calculated between pairwise genes with the criteria of |Cor|> 0.6, FDR < 0.05 (Fig. 9A). The P4HA3 were associated with 49 genes, such as ADAMTS2, ANTXR1 and APCDD1L (Fig. 9A). The DOHH were associated with 33 genes, such as ADAMTS2, ANTXR1 and APCDD1L (Fig. 9A). The MMP1 were associated with 2 genes including IL24 and MMP3 (Fig. 9A). The GO and KEGG results of 84 collaborative genes were shown in the Fig. 9B-C. The top 8 GO terms were collagen-containing extracellular matrix and extracellular matrix organization etc. The KEGG pathways of enrichment were ‘Protein digestion and absorption’ as well as ‘AGE-RAGE signaling pathway in diabetic complications’.
Construction of the TF-miRNA-Gene network targeting signature genes
In order to further elaborate the upstream regulatory miRNAs and TFs effecting signature genes, the TF-miRNA-mRNA network was predicted and pictured in Fig. 10, where 24 TF targeting MMP1 (such as NF-κB, STAT3, ETS1, CITED2, BACH1) as well as 53 miRNAs interacting with three signature genes were predicted. It can be seen that the hsa-miR-6815-3p and hsa-miR-331-3p were common to regulate the expression of P4HA3 and DOHH.
qRT-PCR
The expression of DOHH, P4HA3 and MMP1 were detected via qRT-PCR. The abundance of these genes were higher in tumor cases than normal cases (Fig. 11).
Discussion
In order to investigate the prognostic IMRGs signature in GC, DOHH, P4HA3 and MMP1 was selected using univariate and multivariate Cox regression analyses. The Kaplan Meier curves manifested the worse outcome of patients in the high-risk group. ROC curves confirmed that the IMRGs signature presented good efficiency for predicting GC prognosis. Further, the positive correlation of P4HA3 and MMP1 expression as well as the negative correlation of DOHH expression with risk score and worse prognosis were detected as well, and the MMP1 expression significantly increased in the cohorts at age > 60 and Stage II-IV. Finally, qRT-PCR revealed that the abundance of DOHH, P4HA3 and MMP1 were high in tumor cases, indicating the complex mechanism between the high expression of DOHH as a protective factor and the high expression of P4HA3 and MMP1 as the risk factors in the development of GC. We make the case that this study has valuable significance for foretelling the OS of GC.
Iron is required for cell proliferation and growth, and it promote the formation of toxic-free radical species. Besides, iron metabolism-relevant pathways, containing uptake-export, storage, and regulation processes, may be aberrantly mediated during the course of cancer progression [21]. For instance, transferrin receptor 1 (TFR1), which is a carrier in the regulation of cell growth as well as iron uptake, is aberrantly expressed in tumors and is intimately linked to tumour proliferation as well as metastasis [22,23,24]. This has led to TFR1 being an ideal target for cancer prevention and cure [25]. Besides, a recent study on GC shows that the combination of amino acid, lipid, and iron metabolism might play a role in malignancies by participating in ferroptosis-related metabolic regulation mechanisms, and the prediction model targeting ferroptosis in GC has also been widely studied [26]. Prior investigations have shown that iron metabolism probably contribute to a number of cancers, namely lung cancer [27], leukemia [28], prostate cancer [29], and kidney cancer [30]. Nevertheless, present researches has centered on the involvement of iron metabolism in cancer progression and management, with hardly ever discussing the value of iron metabolism-related genes in cancer prognosis foretelling. Thus, the prognostic signature of iron metabolism in GC required to be adequately probed.
As a HEAT-repeat protein, DOHH has eight tandem helical hairpins on a symmetric dyad. DOHH encompasses two possible iron coordination sites (one on each dyad) which comprised of two rigorous conserved His-Glu motifs, and the activity of DOHH activity could be recovered nothing but by the appending of Fe2+ to the apoenzyme [31]. DOHH catalyzes the last process of maturation of eIF5A, an momentous protein in proliferation of eukaryotic cell [32,33,34,35]. The metal chelating compounds could efficiently suppress deoxyhypusine hydroxylation in eIF5A, and arrest the progression of cell cycle in mammalian cells, including human cancer cells and HUVEC cells, at the boundary of G1/S [35, 36]. P4HA3 is expressed at very low levels in normal adults and fetal tissues [37]. A recent research relying on the bioinformatic analysis and TCGA database found that up-regulation of P4HA3 was highly linked to genes responding to ECM generation in breast cancer, and higher expression of P4HA3 is relevant to worse prognosis [38]. Previous studies have shown P4HA3 is significantly up-regulated in GC, and up-regulation of P4HA3 is epigenetically activated by Slug, which is correlated with GC metastasis and poor survival [39]. MMP-1 have been authenticated to have agonist activity against PAR1, which is expressed in most human tissues, containing the majority of cell types in the blood vessel wall, platelets, and inflammatory cells, and is thought to be the primary enzyme responsible for collagen degradation [40,41,42,43,44]. Although basal expression of MMP-1 is widespread, some disease states lead to further increasing expression of MMP-1, a result that is usually linked to adverse outcomes. Combined with these studies, the prognosis prediction of GC was composed of three iron metabolism related genes, including DOHH, P4HA3 and MMP1. The external verification set further proved the good performance of three gene signature in predicting GC prognosis Jianming Wei et al. found that LC22A17 associated with poor overall survival in GC [45]. Yuehong Cui et al. found that NOTCH1 and NOTCH3 associated with poor overall survival and low expression of NOTCH2 associated with poor overall survival in GC [46]. In the current research, we postulated that the iron metabolism-relevant signature for OS can adequately foretell the clinical consequences of GC patients.
The concept of immunity promoting or suppressing the tumours is widely accepted, and one of the most influential anticancerous therapies that have been exploited in these years is the therapy targeting immune checkpoints. And the development of tumours is not only regulated by the intrinsic variations in cancerous cells but also dependent on the lymphocyte infiltration and activation [47]. Our results uncovered that the immune state was notably disparate between the two risk GC patients, containing the the proportions of Macrophages, dendritic cells (DCs), Eosinophils, Immature dendritic cells (DCi), Neutrophils, Natural killer cells (NK), Mast cells, Plasmacytoid dendritic cells (DCp), Tem, Gamma delta T cells (γδT) and Thi cells. Huang XM et al. found that ICOS( +) Tregs associated with poor overall survival in GC and pDCs play a underlying role in recruiting ICOS( +) Tregs [48]. Sammarco G et al. found that mast cells exerted a cancer promotion role in GC through the release of angiogenic and lymphangiogenic factors [49]. Eosinophils have either anti-tumor impacts or stimulate the development of tumour by secreting various cytokines and factors containing eosinophil-derived neurotoxin, peroxidase, eosinophil cationic protein, and major basic protein [50]. Macrophages was comprised of two main types, namely M1 and M2. Macrophage M2 exert an momentous role in tumor progression, facilitating pro-angiogenic and immunosuppressive signal in the tumor while M1 macrophage infiltration may be linked to a favorable survival rate [51]. People suffering from GC displayed a notably higher neutrophil infiltration in GC tumuor tissues. These tumour-infiltrating neutrophils revealed a phenotype of activated CD54+ and expressed high level programmed death-ligand 1 (PD-L1), which was an immunosuppressive molecule and relevant to disease progression and lowered GC patient survival [52]. As vital lymphocytes in innate immunity, NK cells exert important impact in restraining GC initiation, progression, and metastases, and can improve NK cells’ killing activity toward GC. Gene therapy have been uncovered to directly or indirectly activate NK cells [53]. The memory T cell (Tm) comprises of two populations, namely the effector memory T cells (Tem) and central memory T cells (Tcm), and the ratio of CD4( +)/CD8( +) Tem were notably increased in GC than healthy controls [54]. In addition, the prognostic genes (DOHH, P4HA3 and MMP1) have remarkable linkages with immune cell, suggesting the complexity between iron metabolism and immunity. Moreover, GO and KEGG analyses performed using these 84 collaborative genes suggested that these 84 collaborative genes were primarily engaged in ‘AGE-RAGE signaling pathway in diabetic complications’ and ‘protein digestion and absorption’. In fact, previous studies revealed that ‘AGE-RAGE signaling pathway in diabetic complication constitute the main mechanisms of vascular oxidative stress and enable the activation of NADPH oxidase (Nox) and NF-κB, thus incepting a vicious cycle of oxidative stress and inflammation [55,56,57,58,59,60]. Previous studies have demonstrated that the receptor activator of NF-κB ligand signaling pathway can promote the metastasis of tumor cells [61, 62]. The ‘protein digestion and absorption’ pathway had been found to be linked to pancreatic neuroendocrine tumours as well as breast cancer [63, 64]. Expression of DOHH, P4HA3 and MMP1 was assessed with qRT-PCR in tumor cases and the normal cases. The results demonstrated that the abundance of DOHH, P4HA3 and MMP1 were higher in tumor cases than normal cases. The experiment verified the aforementioned conclusion.
All in all, this study manifested that the IMRGs (DOHH, P4HA3 and MMP1) presented good efficiency for predicting GC prognosis, implying that this gene signature relevant to iron metabolism was a promising biomarker in foretelling the prognosis of GC, which would provide new idea on the digging of underlying predictive biomarkers for GC patients. Nevertheless, this study had certain limitations. For instance, public data may have certain boundedness when we analyze the prognostic performance of gene signatures. Additional data, including primary data from patients with GC, are required to affirm the predictive effect of these gene signature.
Availability of data and materials
The datasets analysed during the current study are available in the TCGA repository, [https://portal.gdc.cancer.gov/projects/TCGA-STAD]. The mRNA-seq datasets generated during the current study are available from the corresponding author on reasonable request.
Abbreviations
- GC:
-
Gastric cancer
- IMRGs:
-
Iron metabolism-relevant genes
- DE-IMRGs:
-
Differentially expressed IMRGs
- ROC:
-
Receiver operating characteristic
- ssGSEA:
-
Single-sample GSEA
- AUCs:
-
Area under the ROC curves
- OS:
-
Overall survival
- MSigDB:
-
Molecular Signatures Database
- DEGs:
-
Differentially expressed genes
- TFR1:
-
Transferrin receptor 1
- TCGA:
-
The Cancer Genome Atlas
- DC:
-
Dendritic cells
- DCi:
-
Immature dendritic cells
- NK:
-
Natural killer cells
- DCp:
-
Plasmacytoid dendritic cells
- ΓδT:
-
Gamma delta T cells
- PD-L1:
-
Programmed death-ligand 1
- Tm:
-
Memory T cell
- Tem:
-
Effector memory T cells
- Tcm:
-
Central memory T cells
- Nox:
-
NADPH oxidase
- TFs:
-
Transcription factors
- qRT-PCR:
-
Quantitative real-time PCR
- GSEA:
-
Gene Set Enrichment Analysis
References
Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Oh SC, Sohn BH, Cheong JH, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun. 2018;9(1):1777. https://doi.org/10.1038/s41467-018-04179-85.
Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010. https://doi.org/10.1016/s0140-6736(14)62038-9.
Shafabakhsh R, Yousefi B, Asemi Z, et al. Chitosan: Acompound for drug delivery system in gastric cancer–A review [J]. Carbohydr Polym. 2020;242: 116403.
Dan J, Wang J, Wang Y, et al. LncRNA-MEG3 inhibits pro-liferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed Pharmacother. 2018;99:931–8.
Catalano V, Labianca R, Beretta GD, et al. Gastric cancer. Crit Rev Oncol Hematol. 2005;54(3):209–41.
Asplund J, Kauppila JH, Mattsson F, et al. Survival trends in gastric adenocarcinoma: A population-based study in Sweden. Ann Surg Oncol. 2018;25(9):2693–702.
Lin JX, Lin JP, Xie JW, et al. Prognostic value and association of sarcopenia and systemic inflammation for patients with gastric cancer following radical gastrectomy. Oncologist. 2019;24(11):e1091–101.
van Cutsem E, Sagaert X, Topal B, et al. Gastric can⁃cer. Lancet. 2016;388(10060):2654–64.
Pinnix ZK, Miller LD, Wang W, et al. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2(43):43ra56.
Louandre C, Ezzoukhry Z, Godin C, et al. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int J Cancer. 2013;133(7):1732–42.
Greene CJ, Attwood K, Sharma NJ, et al. Transferrin receptor 1 upregulation in primary tumor and downregulation in benign kidney is associated with progression and mortality in renal cell carcinoma patients. Oncotarget. 2017;8(63):107052–75.
Tesfay L, Clausen KA, Kim JW, et al. Hepcidin regulation in prostate and its disruption in prostate cancer. Cancer Res. 2015;75(11):2254–63.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. https://doi.org/10.1093/bioinformatics/btp616. Epub 2009 Nov 11.
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40(10):4288–97. https://doi.org/10.1093/nar/gks042. Epub 2012 Jan 28.
Thornley B, Adams CE, Awad G. Chlorpromazine versus placebo for schizophrenia. Cochrane Database Syst Rev. 2003;(2):CD000284. https://doi.org/10.1002/14651858.CD000284.
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021;2(3):100141. https://doi.org/10.1016/j.xinn.2021.100141. PMID: 34557778; PMCID: PMC8454663.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28(11):1947–51.
Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587-d592.
Forciniti S, Greco L, Grizzi F, Malesci A, Laghi L. Iron Metabolism in Cancer Progression. Int J Mol Sci. 2020;21(6):2257. https://doi.org/10.3390/ijms21062257.
Adachi M, Kai K, Yamaji K, Ide T, Noshiro H, Kawaguchi A, et al. Transferrin receptor 1 overexpression is associated with tumour de-differentiation and acts as a potential prognostic indicator of hepatocellular carcinoma. Histopathology. 2019;75:63–73. https://doi.org/10.1111/his.13847.
Cui C, Cheng X, Yan L, Ding H, Guan X, Zhang W, et al. Downregulation of TfR1 promotes progression of colorectal cancer via the JAK/STAT pathway. Cancer Manage Res. 2019;11:6323–41. https://doi.org/10.2147/cmar.S198911.
Jeong SM, Hwang S, Seong RH. Transferrin receptor regulates pancreatic cancer growth by modulating mitochondrial respiration and ROS generation. Biochem Biophys Res Commun. 2016;471:373–9. https://doi.org/10.1016/j.bbrc.2016.02.023.
Clarke SL, Thompson LR, Dandekar E, Srinivasan A, Montgomery MR. Distinct TP53 mutation subtypes differentially influence cellular iron metabolism. Nutrients. 2019;11(9):9. https://doi.org/10.3390/nu11092144.
Wang L, Wang H. The putative role of ferroptosis in gastric cancer: a review. Eur J Cancer Prev. 2023. https://doi.org/10.1097/CEJ.0000000000000817. Epub ahead of print. PMID: 37318883.
Sukiennicki GM, Marciniak W, Muszyńska M, et al. Iron levels, genes involved in iron metabolism and antioxidative processes and lung cancer incidence. PLoS ONE. 2019;14(1): e0208610. https://doi.org/10.1371/journal.pone.0208610.
Tsuma-Kaneko M, Sawanobori M, Kawakami S, et al. Iron removal enhances vitamin C-induced apoptosis and growth inhibition of K-562 leukemic cells. Sci Rep. 2018;8(1):17377. https://doi.org/10.1038/s41598-018-35730-8.
Iron VD. Metabolism in prostate cancer; from basic science to new therapeutic strategies. Front Oncol. 2018;8:547. https://doi.org/10.3389/fonc.2018.00547.
Linehan WM, Schmidt LS, Crooks DR, et al. The metabolic basis of kidney cancer. Cancer Discov. 2019;9(8):1006–21. https://doi.org/10.1158/2159-8290.Cd-18-1354.
Kim YS, Kang KR, Wolff EC, Bell JK, McPhie P, Park MH. Deoxyhypusine hydroxylase is a Fe(II)-dependent, HEAT-repeat enzyme. Identification of amino acid residues critical for Fe(II) binding and catalysis [corrected]. J Biol Chem. 2006;281(19):13217–25.
Park MH. J Biochem (Japan). 2006;139:161–9. PubMed: 16452303.
Chen KY, Liu AY. Biol Signals. 1997;6:105–9. PubMed: 9285092.
Caraglia M, Marra M, Giuberti G, D’Alessandro AM, Baldi A, Tassone P, Venuta S, Tagliaferri P, Abbruzzese A. J Biochem (Tokyo). 2003;133:757–65. PubMed: 12869532.
Hanauske-Abel HM, Park MH, Hanauske AR, Popowicz AM, Lalande M, Folk JE. Biochim Biophys Acta. 1994;1221:115–24. PubMed: 8148388.
Clement PM, Hanauske-Abel HM, Wolff EC, Kleinman HK, Park MH. Int J Cancer. 2002;100:491–8. PubMed: 12115536.
Kukkola L, Hieta R, Kivirikko KI, Myllyharju J. Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. J Biol Chem. 2003;278(48):47685–93.
Winslow S, Lindquist KE, Edsjo A, Larsson C. The expression pattern of matrix-producing tumor stroma is of prognostic importance in breast cancer. BMC Cancer. 2016;16(1):841.
Song H, Liu L, Song Z, Ren Y, Li C, Huo J. P4HA3 is epigenetically activated by slug in gastric cancer and its deregulation is associated with enhanced metastasis and poor survival. Technol Cancer Res Treat. 2018;1(17):1533033818796485.
Galt SW, Lindemann S, Allen L, et al. Outside-in signals delivered by matrix metalloproteinase-1 regulate platelet function. Circ Res. 2002;90(10):1093–9.
Galis ZS, Sukhova GK, Lark MW, Libby P. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1994;94(6):2493–503.
Brinckerhoff CE, Rutter JL, Benbow U. Interstitial collagenases as markers of tumor progression. Clin Cancer Res. 2000;6(12):4823–30.
De Garavilla L, Vergnolle N, Young SH, et al. Agonists of proteinase-activated receptor 1 induce plasma extravasation by a neurogenic mechanism. Br J Pharmacol. 2001;133:975–87.
Giambernardi TA, Grant GM, Taylor GP, et al. Overview of matrix metalloproteinase expression in cultured human cells. Matrix Biol. 1998;16(8):483–96.
Wei J, Gao X, Qin Y, Liu T, Kang Y. An iron metabolism-related SLC22A17 for the prognostic value of gastric cancer. Onco Targets Ther. 2020;14(13):12763–75. https://doi.org/10.2147/OTT.S287811. PMID:33363382;PMCID:PMC7751842.
Cui Y, Li Q, Li W, Wang Y, Lv F, Shi X, Tang Z, Shen Z, Hou Y, Zhang H, Mao B, Liu T. NOTCH3 is a Prognostic Factor and Is Correlated With Immune Tolerance in Gastric Cancer. Front Oncol. 2021;5(10):574937. https://doi.org/10.3389/fonc.2020.574937. PMID:33479597;PMCID:PMC7814877.
Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, et al. Role of tumor microenvironment in tumorigenesis. J Cancer. 2017;5:761–73. https://doi.org/10.7150/jca.17648.
Huang XM, Liu XS, Lin XK, Yu H, Sun JY, Liu XK, Chen C, Jin HL, Zhang GE, Shi XX, Zhang Q, Yu JR. Role of plasmacytoid dendritic cells and inducible costimulator-positive regulatory T cells in the immunosuppression microenvironment of gastric cancer. Cancer Sci. 2014;105(2):150–8. https://doi.org/10.1111/cas.12327. Epub 2014 Jan 4. PMID: 24261990; PMCID: PMC4317822.
Sammarco G, Varricchi G, Ferraro V, Ammendola M, De Fazio M, Altomare DF, Luposella M, Maltese L, Currò G, Marone G, Ranieri G, Memeo R. Mast cells, angiogenesis and lymphangiogenesis in human gastric cancer. Int J Mol Sci. 2019;20(9):2106. https://doi.org/10.3390/ijms20092106. PMID:31035644;PMCID:PMC6540185.
Sakkal S, Miller S, Apostolopoulos V, Nurgali K. Eosinophils in cancer: favourable or unfavourable? Curr Med Chem. 2016;23(7):650–66. https://doi.org/10.2174/0929867323666160119094313. PMID: 26785997.
Wang XL, Jiang JT, Wu CP. Prognostic significance of tumor-associated macrophage infiltration in gastric cancer: a meta-analysis. Genet Mol Res. 2016;15(4). https://doi.org/10.4238/gmr15049040. PMID: 27966749.
Wang TT, Zhao YL, Peng LS, Chen N, Chen W, Lv YP, Mao FY, Zhang JY, Cheng P, Teng YS, Fu XL, Yu PW, Guo G, Luo P, Zhuang Y, Zou QM. Tumour-activated neutrophils in gastric cancer foster immune suppression and disease progression through GM-CSF-PD-L1 pathway. Gut. 2017;66(11):1900–11. https://doi.org/10.1136/gutjnl-2016-313075. Epub 2017 Mar 8. PMID: 28274999; PMCID: PMC5739867.
Du Y, Wei Y. Therapeutic potential of natural Killer cells in gastric cancer. Front Immunol. 2019;21(9):3095. https://doi.org/10.3389/fimmu.2018.03095.PMID:30719024;PMCID:PMC6348255.
Zhang R, Li F, Li H, Yu J, Ren X. The clinical significance of memory T cells and its subsets in gastric cancer. Clin Transl Oncol. 2014;16(3):257–65. https://doi.org/10.1007/s12094-013-1066-5. Epub 2013 Jun 21 PMID: 23793812.
Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107(9):1058–70. https://doi.org/10.1161/CIRCRESAHA.110.223545.
Goldin A, Beckman JA, Schmidt AM, Creager MA. Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation. 2006;114(6):597–605. https://doi.org/10.1161/CIRCULATIONAHA.106.621854.
Yan SF, Ramasamy R, Schmidt AM. The RAGE axis: a fundamental mechanism signaling danger to the vulnerable vasculature. Circ Res. 2010;106(5):842–53. https://doi.org/10.1161/CIRCRESAHA.109.212217.
Bierhaus A, Chevion S, Chevion M, et al. Advanced glycation end product-induced activation of NF-κB is suppressed by α-lipoic acid in cultured endothelial cells. Diabetes. 1997;46(9):1481–90. https://doi.org/10.2337/diab.46.9.1481.
Wautier M-P, Chappey O, Corda S, Stern DM, Schmidt AM, Wautier J-L. Activation of NADPH oxidase by AGE links oxidant stress to altered gene expression via RAGE. Am J Physiol Endocrinol Metab. 2001;280(5):E685–94. https://doi.org/10.1152/ajpendo.2001.280.5.E685.
Arai M, Nihonmatsu-Kikuchi N, Itokawa M, Rabbani N, Thornalley PJ. Measurement of glyoxalase activities. Biochem Soc Trans. 2014;42(2):491–4. https://doi.org/10.1042/BST20140010.
Lazennec G, Richmond A. Chemokines and chemokine receptors: new insights into cancer-related inflammation. Trends Mol Med. 2010;16(3):133–44. https://doi.org/10.1016/j.molmed.2010.01.003.
Chen Q, Sun W, Liao Y, et al. Monocyte chemotactic protein-1 promotes the proliferation and invasion of osteosarcoma cells and upregulates the expression of AKT. Mol Med Rep. 2015;12(1):219–25. https://doi.org/10.3892/mmr.2015.3375.
Akkiprik M, Peker I, Özmen T, Amuran GG, Güllüoğlu BM, Kaya H, Özer A. Identification of differentially expressed IGFBP5-related genes in breast cancer tumor tissues using cDNA microarray experiments. Genes (Basel). 2015;6:1201–14. https://doi.org/10.3390/genes6041201.
Wang DD, Liu ZW, Han MM, Zhu ZM, Tu YL, Dou CQ, Jin X, Cai SW, Du N. Microarray based analysis of gene expression patterns in pancreatic neuroendocrine tumors. Eur Rev Med Pharmacol Sci. 2015;19:3367–74.
Acknowledgements
We acknowledge the support from Department of Oncology First Affiliated Hospital of Kunming Medical University. We are grateful to Professor Wen Zhengqi and his team because they provided the data of cases from the General Surgery Department. We must give our sincere thanks to the GC patients who took part in this research.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Hongbin Zhang conceived the idea for the study. All authors approved the manuscript and provid-ed relevant suggestions.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of The First Affiliated Hospital of Kunming Medical University. Written informed consent was obtained from all participantsfor the use of tissue samples and clinical data for research purpose.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Table S1. Primer information.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, X., Ren, J., Zhou, R. et al. Construction of iron metabolism-related prognostic features of gastric cancer based on RNA sequencing and TCGA database. BMC Cancer 23, 1106 (2023). https://doi.org/10.1186/s12885-023-11569-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-023-11569-9