Abstract
Background
Aberrant expression of microRNAs has been associated with migration of tumor cells. In this study, we aimed to investigate the biological significance of miR-944 whose function is unknown in breast cancer.
Methods
MiR-944 expression in breast cancer cells and tumors was evaluated by Taqman qRT-PCR assays. Transcriptional profiling of MDA-MB-231 cells expressing miR-944 was performed using DNA microarrays. Cell viability, migration and invasion were assessed by MTT, scratch/wound-healing and transwell chamber assays, respectively. The luciferase reporter assay was used to evaluate targeting of SIAH1, PTP4A1 and PRKCA genes by miR-944. SIAH1 protein levels were measured by Western blot. Silencing of SIAH1 gene was performed by RNA interference using shRNAs.
Results
Our data showed that miR-944 expression was severely repressed in clinical specimens and breast cancer cell lines. Suppression of miR-944 levels was independent of hormonal status and metastatic potential of breast cancer cells. Gain-of-function analysis indicated that miR-944 altered the actin cytoskeleton dynamics and impaired cell migration and invasion. Genome-wide transcriptional profiling of MDA-MB-231 cells that ectopically express miR-944 showed that 15 genes involved in migration were significantly repressed. Notably, luciferase reporter assays confirmed the ability of miR-944 to bind the 3´UTR of SIAH1 and PTP4A1 genes, but not PRKCA gene. Congruently, an inverse correlation between miR-944 and SIAH1 protein expression was found in breast cancer cells. Moreover, SIAH1 was upregulated in 75 % of miR-944-deficient breast tumors. Finally, SIAH1 gene silencing by RNA interference significantly impaired cell migration of breast cancer cells.
Conclusions
Our results pointed out that miR-944 is a novel upstream negative regulator of SIAH1 and PTP4A1 genes and provided for the first time evidence for its functional role in migration and invasion of breast cancer cells. They also suggest that miR-944 restoration may represent a potential strategy for breast cancer therapy.
Similar content being viewed by others
Background
Cancer is a major public health problem worldwide. Based on GLOBOCAN estimates, about 14.1 million new cancer cases and 8.2 million deaths occurred in 2012 around the world [1]. Notably, breast cancer is a leading cause of death in women with 1.38 million new cases diagnosed in 2008 worldwide ([2]. However, despite significant advances in screening, diagnosis, and personalized therapies, this disease still remains largely incurable. This situation is aggravated by the lack of relevant clinical molecular determinants and classifiers associated to prognostic and biological variables of patients. Therefore the search for novel biomarkers representative of the molecular features of tumors is required to better understand the mechanisms that contribute to disease progression and identify novel therapeutic targets.
MicroRNAs are evolutionary conserved small non-coding RNAs that function as negative regulators of gene expression by either inhibiting translation or inducing degradation of a set of specific messenger RNAs [3]. MicroRNAs regulate multiple physiological processes, including development, differentiation, growth, and cell death. In cancer cells, microRNAs may function either as oncogenes or tumor-suppressors (oncomiRs) [4]. Therefore, the altered expression of microRNAs may greatly contribute to the heterogeneous behavior of diverse human neoplasia and in some cases, may correlate with clinic-pathological features of tumors. Consequently, they represent novel potential prognostic biomarkers and therapeutic targets in cancer [5]. One of the most deadly hallmarks of cancer cells is their ability to metastasize to other tissues and organs [6]. This property can be promoted by a specific set of microRNAs named metastamiRs that target multiple transcripts related to cell migration [4]. It has been shown that several microRNAs target genes that drive cytoskeleton remodeling and promote tumor cell invasion [7], however, postranscriptional regulatory mechanisms involving microRNAs still remain poorly understood in cancer. Recently we performed a microRNAs profiling of breast carcinomas and found that miR-944 was significantly repressed in clinical specimens [8]. In the present study, we aimed to further investigate the biological significance of miR-944 in breast cancer. Here we identified multiple genes that are modulated by miR-944 and revealed that the cell migration-related SIAH1 and PTP4A1 genes are two novel targets of miR-944. Altogether, our data contribute for the understanding of the molecular mechanisms controlling cell migration and invasion of breast cancer cells.
Methods
Cell lines
Human MDA-MB-231, MCF-7, MDA-MB-453, ZR-75 and T457-D breast cancer cell lines and MCF-10A non-tumorigenic breast cells were obtained from the American Type Culture Collection and routinely grown in Dulbecco’s modified of Eagle’s medium (DMEM) supplemented with10 % fetal bovine serum and penicillin-streptomycin (50 unit/ml; Invitrogen). Cell lines were maintained at 37 °C in 5 % CO2.
Tissue collection
Locally invasive breast tumors and normal tissues were provided by the Institute of Breast Diseases-FUCAM, Mexico, following the regulations approved by the FUCAM ethics committee. A written informed consent was obtained from each participant prior to release for research use. None of the enrolled patients received any antineoplastic therapy before surgery. After tumor resection, specimens were embedded in Tissue-Tek and snap frozen in liquid nitrogen at -80 °C. Pathologist confirmed the existence of at least 80 % tumor cells in clinical specimens.
Quantitative reverse transcription and polymerase chain reaction (qRT-PCR)
The expression of miR-944 was measured by microRNA assays as implemented by manufacturer (ThermoFisher) and the comparative Ct (2 − ΔΔCt) method using an automatic baseline and a threshold of 0.2 to determine the Ct raw data. Total RNA (100 ng) of cells and tissues was obtained using the Trizol reagent (Invitrogen) and reverse transcribed using the looped-RT specific primer for miR-944, dNTPs (100 mM), reverse transcriptase MultiScribe (50 U/μl), 10X buffer, RNase inhibitor (20 U/μl) and RNase-free water. Then, retrotranscription reaction (1:15) was mixed with 10 μl master mix TaqMan (Universal PCR Master Mix, No AmpErase UNG, 2X), 7.67 μl RNase free water, and 1.0 μl PCR probe. PCR reaction was performed using a GeneAmp System 9700 (Applied Biosystems) as follows: 95 °C for 10 min, and 40 cycles at 95 °C for 15 s and 60 °C for 1 min. RNU44 was used as a control for normalization of data.
Transfection assays
The miR-944 precursor (4464066; Life Technologies), and scramble sequence (AM17110; Life Technologies) used as negative control, were transfected into MDA-MB-231 and MCF-7 cells using siPORT amine transfection agent (Ambion, Inc., Austin, TX, USA). Briefly, pre-miR-944 was diluted in 25 μl Opti-MEM to obtain a concentration range from 50 nM to 200 nM and added to wells containing 1x107 cells grown in 450 μl DMEM for 48 h. Expression of miR-944 was evaluated by qRT-PCR as described.
Cell viability assays
MDA-MB-231 and MCF-7 cells (2x104), transfected or not with miR-944 precursor (50 nM) or scramble sequence as described above, were incubated with 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium bromide (MTT, 1 mg/ml) at 37 °C for 4 h. The formazan dye crystals were solubilized with 500 μl isopropanol, 4 mM HCl, NP-40 0.1 % for 5 min. Absorbance was measured using a spectrophotometer at 540 nm wavelength. Experiments were performed three times by triplicate and results were represented as mean ± Standard Deviation (SD).
Cell migration and invasion assays
MDA-MB-231 and MCF-7 cells were transfected with miR-944 precursor (50 nM) or scramble as described above. Twenty-four hours postransfection, a vertical wound was traced in the cell monolayer. At 4 and 24 h, cells were fixed with 4 % paraformaldehyde and the scratched area was determined to quantify cell migration. In transwell assays, chambers (Corning) with 6.5-mm diameter and 8-μm pore size polycarbonate membrane were used. MDA-MB-231 and MCF-7 cells (1 × 105) were transferred to 0.5 ml serum-free medium and placed in the upper chamber, whereas the lower chamber was loaded with 0.8 ml medium containing 10 % fetal bovine serum. The total number of cells that migrated into the lower chamber was counted after 24 h incubation at 37 °C. Cell invasiveness was evaluated using transwell chambers coated with a layer of extracellular matrix (BD Biosciences). MDA-MB-231 cells were treated with pre-miR-944 (50 nM) or scramble and 24 h postransfection, the invasive cells were quantified. Non-transfected cells were used as control. Each experiment was performed three times by triplicate and results were represented as mean ± S.D.
Western blot analyses
Proteins obtained from breast tumors or MDA-MB-231 and MCF-7 cells transfected with miR-944 precursor (50 nM) or scramble as described above, were separated on 10 % polyacrylamide gels and transferred to PVDF membrane (Millipore). Membrane was incubated overnight at 4 °C with α-actinin-1 (sc-17829, Santa Cruz Biotechnology) or SIAH1 (ab2237 Abcam) primary antibodies, and then incubated with horseradish peroxidase–conjugated anti-mouse IgG or anti-goat IgG secondary antibodies (1:8,500, Zymed), respectively. Signal was detected and developed using the ChemiLucent (Chemicon) system.
Indirect immunofluorescence
MDA-MB-231 and MCF-7 cells transfected with miR-944 precursor were seeded on coverslips (1x103 cells/cm2). After 48 h, cells were rinsed with cytoskeleton buffer (10 mM MES pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 0.32 M sucrose) at 37 °C and fixed with 3 % cytoskeleton buffer for 15 min at 37 °C to maintain the integrity of the cytoskeleton. Then, cells were permeabilized with 0.1 % Triton-X 100-CB (Sigma-Aldrich) for 5 min, blocked with 0.5 % fish skin gelatin in PBS, and incubated with phalloidin-rhodamine (0.1 μg/μl) or alpha-actinin 1 antibodies for 1 hr at room temperature (Sigma-Aldrich). Finally, slides were assembled with vectashield® mounting media (Vector) containing DAPI and cells were observed under an Olympus FluoView FV1000 Confocal Microscope with an attached MRC1024 LSCM system (Bio-Rad). Cells were imaged from top to bottom in the Z-plane; images from the basal plane of the cells were captured and stored as digital images.
Microarrays analysis
Global gene expression analysis was done for MDA-MB-231 cells transfected with miR-944 precursor (50 nM) or scramble (30 nM) using the NimbleGen array (Roche). RNA samples were used to synthesize double-stranded labeled cDNA using SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen) and NimbleGen One-Color DNA Labeling Kit. Samples were hybridized in NimbleGen array 12x135K (12 x 135,000 features). After hybridization and washing, the processed slides were scanned using a NimbleGen MS200 Microarray Scanner. Raw data were extracted as pair files by NimbleScan software (version 2.5), background was corrected and data were normalized. The probe level files and gene summary files were produced and imported into ANAIS software (Analysis of NimbleGen Arrays Interface) for further analysis. The Student test with Varmixt package was used and raw P values were adjusted by the Benjamini and Yekutieli method to control the false discovery rate (FDR). Only genes with a Benjamini/Yekutieli value <0.05, and expression fold change >1.5 were considered as being differentially expressed.
Luciferase assays
The 3´UTR region of SIAH1, PTP4A1 and PRKCA genes was cloned downstream of luciferase gene into p-miR-report vector (Ambion). Then, recombinant plasmids (2 μg) were transfected into MDA-MB-231 cells. At 24 h pre-miR-944 (50 nM) or pre-miR-negative control (scramble) were co-transfected using lipofectamine RNAi max (Invitrogen). After 24 h, firefly and Renilla luciferase activities were measured by the Dual-Glo Luciferase Assay (Promega, Charbonnieres, France) using a Fluoreskan Ascent FL (Thermo Scientific). Data were normalized with respect to Renilla activity and p-values for differences were determined by the two-tailed Student’s t test.
Targeted inhibition of SIAH1
Two oligonucleotides pairs (21-23 nt length) corresponding to two short hairpin RNAs (shRNA) targeting the SIAH1 gene were designed (Additional file 1). To minimize the possibility of shRNAs off targeting effects, a nucleotide BLAST search was carried out. Each oligonucleotide pair was cloned into the pSilencer 5.1 U6 retro plasmid (ThermoFisher) and sequences were confirmed by automatic sequencing. The resulting plasmids were transfected into MDA-MB-231 cells and SIAH1 expression was evaluated by Western blot assays at 48 h post-transfection.
Statistical analysis
Experiments were performed three times by triplicate and results were represented as mean ± S.D. One-way analysis of variance (ANOVA) followed by Tukey’s test were used to compare the differences between means. A p < 0.05 was considered as statistically significant.
Results
MiR-944 is suppressed in breast cancer cell lines and clinical tumors
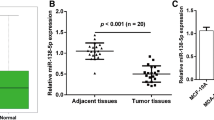
In order to confirm the clinical relevance of miR-944 in breast cancer, we quantified its expression by qRT-PCR in a set of clinical specimens obtained from a cohort of 40 patients (discovery cohort) from the FUCAM institution. Clinical features of breast tumors including hormonal receptor status, tumor size, histology, clinical stage, and tumor grade are summarized in Table 1. Results indicated that miR-944 expression was significantly (p < 0.05) diminished in tumors in comparison with adjacent normal tissues (Fig. 1a). Our results were validated by the analysis of 776 matched normal/tumor samples at The Cancer Genome Atlas (TCGA) (validation cohort), since the average expression of miR-944 was 8.16 in normal tissues versus 3.04 in tumors (Fig. 1b). To strengthen these data, we further analyzed the TGCA data for miR-204 and miR-10b, two miRNAs that have been previously reported as down-regulated and up-regulated, respectively, in breast cancer. As expected, miR-204 was suppressed, whereas miR-10b was overexpressed in the validation cohort (Additional file 2). On the other hand, miR-944 expression was significantly lower (8 to 9-fold) in MCF-7, MDA-MBD-231, MDA-MB-45, ZR-45, and T47-D breast cancer cell lines in comparison with non-tumorigenic MCF-10A breast cell line (Fig. 1c). Taken all together, our results confirmed that miR-944 was significantly suppressed in breast tumors.
MiR-944 is suppressed in clinical tumors and breast cancer cell lines. (a) MiR-944 expression measured by qRT-PCR in breast normal adjacent and tumor tissues (discovery cohort). (b) MiR-944 expression in 776 matched normal/tumor samples from The Cancer Genome Atlas (TCGA) (validation cohort). (c) MiR-944 expression measured by qRT-PCR in breast cancer cell lines and MCF-10A non-tumorigenic cell line. Data were normalized with the endogenous small-nucleolar RNU44. Bars represent the mean of three independent experiments performed three times ± S.D. Asterisks indicate p < 0.05
MiR-944 inhibits cell migration and invasion
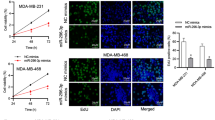
To define the functions of miR-944 we restored its expression using RNA mimics in triple negative MDA-MB-231 (highly metastatic) and oestrogen responsive MCF-7 (poorly invasive) breast cancer cells (Additional file 3). First, the effect of diverse concentrations of miR-944 precursor on cell viability was evaluated by MTT assays. Results showed minimal changes (less than 5 %) in cell viability of MDA-MB-231 transfected with 50 nM miR-944 precursor in comparison with scramble transfected and non-transfected controls. Using 100 nM and 200 nM miR-944 precursor, we observed a 10 % reduction on cell viability relative to controls (Fig. 2a). Similar results were obtained in MCF-7 cells (Fig. 2e). Then, we performed scratch/wound-healing assays in both breast cancer cell lines to evaluate the contribution of miR-944 in tumor cell migration. Data indicated that cell monolayers restoration was delayed in both MDA-MB-231 and MCF-7 cells transfected with miR-944 precursor (50 nM) when compared with non-treated and scramble-transfected cells at 24 h (Fig. 2b and f). In addition, transwell chamber assays showed that the number of migratory cells was significantly (p < 0.05) reduced in MDA-MB-231 (4-fold) and MCF-7 (8-fold) cells that ectopically express miR-944 (Fig. 2c and g) in comparison with control cells. Moreover, miR-944 significantly (p < 0.05) inhibited the ability of metastatic MDA-MB-231 cells to invade matrigel in vitro (Fig. 2d).
MiR-944 suppresses cell migration and invasion. (a and e) MTT cell viability assays of MDA-MB-231 (a) and MCF-7 (e) cells transfected with miR-944 precursor (50 nM to 200 nM). (b and f) Scratch/wound-healing assays of MDA-MB-231 (b) and MCF-7 (f) cells monolayers treated with miR-944 precursor (50 nM). (c and g) Transwell assays of MDA-MB-231 (c) and MCF-7 (g) cells transfected with miR-944 precursor (50 nM). (d) Matrigel invasion assays of MDA-MB-231 cells transfected with miR-944 precursor (50 nM). Non-transfected cells were used as controls. Bars represent the mean of three independent experiments performed three times ± S.D. Asterisks indicate p < 0.05
MiR-944 alters cytoskeleton organization
As cell migration may involve the coordinated expression and association of proteins driving the epithelial-mesenchymal transition (EMT), cytoskeleton organization and reinforcement of focal adhesions, we decided to determine if miR-944 contributes to these cellular processes. We first analyzed the expression of proteins modulating the EMT, including SIP1, ZEB1 and BMP2, by Western blot assays. Results showed no significant changes in the expression of these proteins in miR-944 expressing cells (data not shown). Then, we examined the organization of cytoskeleton in MDA-MB-231 and MCF-7 cells by analyzing the distribution of F-actin labeled with rhodamine-phalloidin using confocal microscopy. As α-actinin-1 is an actin-crosslinking protein that reinforces focal adhesions its subcellular distribution was also examined. As depicted in Fig. 3a (upper panels), MDA-MB-231 control cells were featured by an axial F-actin cytoskeleton organization, and the presence of structures, such as membrane ruffles (MR) and filopodia (F) associated to a migrating phenotype were evident. Interestingly, the ectopic expression of miR-944 induced a dramatic effect on overall cell morphology since spread area was increased (Fig. 3a bottom panel). Moreover, F-actin was redistributed in a radial mode towards the periphery of the cell, as well as in the central zone; and the membrane ruffles and filopodia structures were lost. Based on these morphological differences, we next analyzed the strengthening of adhesion-related structures. MDA-MB-231 cells transfected with miR-944 precursor exhibited a robust signal of α-actinin-1 and an increase in the number of contact points with F-actin in multiple points of cell body, indicative of the reinforcement of focal adhesions. Remarkably, these cells displayed enrichment in α-actinin-1-rich blebs at the rear end of the cell suggesting a strong adhesive process (Fig. 3b bottom panel). Likewise, restoration of miR-944 expression in MCF-7 cells induced changes in actin cytoskeleton organization and loss of the axial pattern in a similar manner as in MDA-MB-231 cells (Fig. 3c bottom panel). In addition, α-actinin-1 was redistributed and accumulated in focal points at the end or front of cells indicative of focal adhesions formation (Fig. 3d, bottom panel), although in a less extend in comparison with MDA-MB-231 cells transfected with miR-944 precursor.
MiR-944 alters cytoskeleton and focal adhesions. MDA-MB-231 and MCF-7 cells were treated for indirect immunofluorescence with rhodamine phalloidin to visualize F-actin (red) or with α-actinin1 antibody labeled with FITC (green). Nuclei were counterstained with DAPI (blue). (a) Phase contrast and immunofluorescence images show actin organization in non-transfected (control) and miR-944 expressing MDA-MB-231 (top panels) and (c) MCF-7 cells (bottom panels). Arrowheads indicate representative actin-rich membrane ruffling (MR); asterisk indicates filopodia (f). (b) Representative x-z confocal images of α-actinin-1 (green) and F-actin (red) organization in MDA-MB-231 (top panels) and (d) MCF-7 cells (bottom panels) non-transfected (control) or transfected with miR-944 precursor
MiR-944 modulates genes involved in cell adhesion and migration
In order to identify potential target genes of miR-944 that may explain the phenotypic changes described above, we carried out a transcriptional profiling of MDA-MB-231 cells that ectopically express miR-944 using DNA microarrays. Results evidenced that 1197 genes were significantly downregulated and 144 were upregulated (fold change >1.5; Additional file 4). Some of these modulated genes are well known cancer-related genes including MAPK1, IGF1R, SIAH1, PRKCA, RAC1, NOTCH2, MMP14, PAK1 and PTP4A1, among others. Classification of the set of repressed genes based on GO categories showed that 15 genes are involved in cell migration and invasion processes (Table 2).
MiR-944 targets SIAH1 and PTP4A1 genes
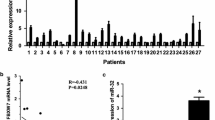
Data from DNA microarrays led us to the identification of potential new target genes for miR-944. Surprisingly, no genes involved in EMT and focal adhesions were found as directly modulated, thus we focused on genes involved in cell migration and cytoskeleton dynamics. Interestingly, the cell migration-related SIAH1, PTP4A1 (also known as PRL-1), and PRKCA genes were repressed after transfection of miR-944 precursor. These genes are key regulators of cell migration and cancer progression in diverse types of cancer [9–11]. Therefore, we investigated if SIAH1, PTP4A1 and PRKCA genes are direct targets of miR-944 using luciferase reporter assays. We identified the complementary site for miR-944 in the 3´UTR sequence of each gene and cloned it downstream of the luciferase coding region in the pmiR-report vector (Fig. 4a). Results showed that forced expression of miR-944 and co-transfection of pmiR-LUC-PRKCA-3´UTR did not result in significant differences in luciferase activity (Fig. 4b). In contrast, the co-transfection of miR-944 and pmiR-LUC-SIAH1-3´UTR or pmiR-LUC-PTP4A1-3´UTR plasmids significantly reduced the luciferase activity (p < 0.001 and p < 0.05, respectively) in comparison with controls (Fig. 4c and d). Because of its relevant role in migration of cancer cells we next focused in the analysis of the SIAH1 protein. Western blot assays revealed that SIAH1 protein levels were reduced in MDA-MB-231 cells transfected with miR-944 in comparison to non-transfected control cells (Fig. 4e). Congruently, the expression of SIAH1 was significantly increased in 53 % of miR-944 deficient breast tumors in comparison with normal adjacent tissues (Fig. 4f and 4g).
SIAH1 and PTP41A genes are miR-944 targets. (a) Schematic representation indicating the 3´UTR sequence of PRKCA, PTP4A1 and SIAH1 genes cloned in pmiR-report vector. Boxes indicate the miR-944 binding sites in target genes. (b, c, d) Luciferase reporter assays. MDA-MB-231 cells were co-transfected with miR-944 (or scramble as control) and pmiR-LUC-PRKCA-3´UTR (b), pmiR-LUC-PTP4A1-3´UTR (c) or pmiR-LUC-SIAH1-3´UTR (d) plasmids and relative luciferase activity was measured as described in methods. Results are expressed in light units. Bars represent the mean of three independent experiments performed three times ± S.D. (e) Immunodetection of SIAH1 by Western blot assays in MDA-MB-231 cells. Lane 1, MDA-MB-231 control cells; lane 2, MDA-MB-231 cells transfected with miR-944. (f) Immunodetection of SIAH1 in breast tumors and normal mammary tissues. β-tubulin was used as internal control. (g) Densitometry analysis of immunodetected bands in F. Pixels corresponding to β-tubulin were used to normalize SIAH1 expression. NS, non- significant. *p < 0.05; **p < 0.01; ***p < 0.001
Knockdown of SIAH1 impairs cell migration
To determine if targeted inhibition of SIAH1 affects cell migration we proceeded to knock-down its expression using RNA interference. Two specific short hairpin RNAs (dubbed as shSIAH1.1 and shSIAH1.2) targeting the human SIAH1 gene were designed and cloned into the pSilencer vector (Additional file 1). Both constructs were individually introduced into MDA-MB-231 cells and SIAH1 expression was analyzed by Western blot 48 h after transfection. Results showed that shSIAH1.2 sequence down-regulated the SIAH1 expression (Fig. 5a), whereas no significant effect was observed with shSIAH1.1 interfering sequence (data not shown). The expression of GADPH used as a control, did not show significant changes between treatments. Densitometric analysis of immunodetected bands showed that silencing induced by shSIAH1.2 construct was effective since this sequence suppressed SIAH1 expression by 42 % (Fig. 5b). The effect of SIAH1 silencing in cell migration was evaluated in MDA-MB-231 cells by scratch/wound-healing assays. Results showed that restoration of cell monolayers was significantly (p > 0.05) delayed in SIAH1-deficient cells when compared with scramble-transfected cells and non-treated control cells at 24 h (Fig. 5c).
SIAH1 silencing inhibits cell migration of breast cancer cells. (a) Western blot assays for SIAH1 knock-down in MDA-MB-231 cells using shSIAH1.2 interfering sequence. Scramble sequence was transfected as negative control. GAPDH was used as internal loading control. (b) Densitometric analysis of immunodetected bands in panel A. (c) Quantification of scratch/wound healing assays in non-transfected control, scramble transfected and SIAH1-deficient cells. Data represents the mean of three independent assays ± SD. (*p < 0.05)
Discussion
One of the most devastating hallmarks in breast cancer is represented by metastasis that is related to alterations in cell adhesion and migration. Evidence is now emerging indicating that microRNAs might constitute a regulatory event in cell migration [12]. Here, we described the biological significance and the effects of miR-944 dysregulation on cell migration in human breast cancer cells. Interestingly, miR-944 gene is located in the intron of the tumor suppressor protein p63 gene, which is a transcription factor frequently suppressed in breast cancer [13]. A feedback between p63 and several microRNAs has been observed in cancer. Tucci et al. [14] reported that loss of p63 and its miR-205 target results in increased cell migration and metastasis in prostate cancer. In order to elucidate the relevance of miR-944 in breast cancer, we first characterized MDA-MB-231 and MCF-7 cells that ectopically express miR-944. According to wound healing, transwell, and matrigel experiments, the restoration of miR-944 expression resulted in a significant reduction in cell migration and invasion. Intriguingly, impaired cell migration was featured by an increased association of α-actinin-1 with F-actin cytoskeleton on focal adhesion points, and loss of membrane ruffling and filopodia. These data suggested that miR-944 plays a significant role in the control of breast cancer cell morphology as cells lost the elongated shape associated with motile and mesenchymal cells, and adopted a spread, and unpolarized shape. During the preparation of this manuscript, an interesting study about miR-944 in cervical cancer was published. Xie et al. [15] showed that miR-944 is overexpressed in human cervical cancer cells and promotes cell proliferation, migration and invasion, while it has no effect on apoptosis. These, and our data, reflect the heterogeneous nature of tumors and indicate that miR-944 functions are tumor-specific.
In order to identify genes modulated by miR-944 that could be relevant in the underlying mechanism of cell migration, we defined the transcriptional profile of MDA-MB-231 cells that ectopically express miR-944. Bioinformatics analyses of modulated genes identified novel potential targets involved in cellular pathways related to cytoskeletal remodeling and cell migration. One interesting gene was SIAH, an E3 ubiquitin-protein ligase that belongs to a family of RING-domain proteins, including the ubiquitin ligases targeting proteins for proteasomal degradation. In diverse types of cancer, SIAH1 has a dual function in RAS, estrogen, DNA-damage, and hypoxia pathways therefore it is considered as an attractive anticancer drug target [16]. However, the proteosome inhibitor bortezomib used in clinical practice inhibits all the proteosome-mediated proteolysis without specificity causing systemic toxicity and resistance; thus the search for more specific E3 ubiquitin ligases is needed [17] In mouse models, the inhibition of SIAH proteins impairs tumor growth and metastasis of breast tumors [18]. Moreover, a number of studies have linked SIAH1 expression with disease progression in human cancer [19]. However, these studies reported opposite results indicating that SIAH1 may function both as an oncogene or a tumor suppressor depending on tumor type. Behling et al. [20] reported that SIAH levels were significantly increased in ductal carcinoma in situ compared with normal tissues. Moreover, tumors from patients with disease recurrence had higher SIAH expression than those from patients without recurrence. In patients with hepatocellular carcinoma (HCC), nuclear accumulation of SIAH1 was correlated with carcinogenesis, tumor proliferation and migration [21]. Furthermore, reduction of SIAH1 expression levels using RNA interference in HCC decreased tumor cell viability [22]. In our study, we observed that SIAH1 expression was decreased in almost half of breast tumors analyzed, which agreed with previous studies. Importantly, we demonstrated that miR-944 was able to down-regulate SIAH1 in vitro. Moreover, miR-944 and SIAH1 expression showed an inverse correlation in breast tumors. In addition, targeted silencing of SIAH1 using shRNAs confirmed the role of this protein in breast cancer cells migration. These findings suggested that the effects of miR-944 in cell migration may occur, at least in part, through targeting of SIAH1.
Another validated target of miR-944 in this study was the protein tyrosine phosphatase 4A1 (PTP4A1, also known as PRL-1). Interestingly, it was reported that PRL-1 promotes cell migration and invasion by regulating filamentous actin dynamics of A549 lung cancer cells [23]. PRL-1 also decreased the expression of focal adhesion proteins. Moreover, reduction in PRL-1 was associated to decrease cell membrane protrusions with a reduction in actin fiber extensions, which could reflect reduced adhesion turnover [24]. Tumor migration and metastasis are dynamic cellular processes that continuously exploit phospho-relay signaling systems. Overexpression of PRL-1 has been identified in pancreatic cancer cell lines [25]. Zheng et al. [26] demonstrated that PRL-1 promotes cell motility, invasion, and metastasis in ovarian cells. In addition, PRL-1 induced metastatic tumor formation in mice. In light of these findings, PRL-1 has been considered as a therapeutic target in cancer [27]. Here, we showed that miR-944 was able to bind the 3´UTR of PTP4A1 downregulating its expression at mRNA level. Moreover, miR-944 expressing cells exhibited morphological changes associated to alterations in actin cytoskeleton and focal adhesions that were similar to those describe in PLR-1-deficient cells. In summary, our findings showed for the first time that miR-944 expression was dramatically suppressed in breast cancer cell lines and tumors independently of hormonal status or metastatic potential. Thus, we cannot in the present study establish a correlation between the low expression of miR-944, the metastatic potential and hormonal receptors expression. The effects of miR-944 in cell migration inhibition may occur, at least in part, through targeting of SIAH1 and PTP4A1. In addition, our data pointed out that knockdown of gene expression by miR-944 could represent a molecular tool to specifically inhibit relevant druggable targets such as SIAH1 and PTP4A1 in breast cancer.
Conclusions
Our data provided evidences about the role of miR-944 as a novel upstream negative regulator of PTP4A1 and SIAH1 and contributed for the understanding of the molecular mechanisms controlling cell migration and invasion in breast cancer. This study also suggested that miR-944 restoration may represent a potential novel strategy for breast cancer therapy.
Abbreviations
DMEM, Dulbecco’s modified of Eagle’s medium; EMT, Epithelial-mesenchymal transition; MR, Membrane ruffles; PRKCA, Protein kinase C alpha; PTPA41, Protein tyrosine phosphatase 4A1; SIAH1 E3, ubiquitin-protein ligase
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics 2012. CA Cancer J Clin. 2015;65(2):87–108.
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917.
Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Lopez-Camarillo C, Marchat LA, Arechaga-Ocampo E, Perez-Plasencia C, Del Moral-Hernandez, Castaneda-Ortiz EJ, et al. MetastamiRs: Non-Coding MicroRNAs Driving Cancer Invasion and Metastasis. Int J Mol Sci. 2012;13:1347–79.
Hernando E. MicroRNAs and cancer: Role in tumourigenesis, patient classification and therapy. Clin Transl Oncol. 2007;9:155–60.
Schmidt-Kittler O, Ragg T, Daskalakis A, Granzow M, Ahr A, Blankenstein TJ, et al. From latent disseminated cells to overt metastasis: Genetic analysis of systemic breast cancer progression. Proc Natl Acad Sci USA. 2003;100:7737–42.
Valastyan S, Weinberg RA. Roles for microRNAs in the regulation of cell adhesion molecules. J Cell Sci. 2011;124:999–1006.
Flores-Pérez A1, Marchat LA2, Rodríguez-Cuevas S3, Bautista-Piña V3, Streber ML, et al. Dual targeting of ANGPT1 and TGFBR2 genes by miR-204 controls angiogenesis in breast cancer. In, Scientific Reports. 2016.
Shin EY, Shim ES, Lee CS, Kim HK, Kim EG. Phosphorylation of RhoGDI1 by p21-activated kinase 2 mediates basic fibroblast growth factor-stimulated neurite out growth in PC12 cells. Biochem Biophys Res Commun. 2009;6:384–89.
Zhang L, Ma P, Sun LM, Han YC, Li BL. MiR-107 down-regulates SIAH1 expression in human breast cancer cells and silencing of miR-107 inhibits tumor growth in a nude mouse model of triple-negative breast cancer. Mol Carcinog. 2015;55(5):768–77.
Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS One. 2013;10:8(9):e73985.
Pellegrino L, Stebbing J, Braga VM, Frampton AE, Jacob J, Buluwela L, et al. miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013;1:5400–12.
Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18(9):1487–99.
Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vosden KH, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;18:15312–17.
Xie H, Lee L, Scicluna P, Kavak E, Larsson C, Sandberg R, et al. Novel functions and targets of miR-944 in human cervical cancer cells. Int J Cancer. 2015;1:E230–41.
House CM, Möller A, Bowtell DD. Siah proteins: novel drug targets in the Ras and hypoxia pathways. Cancer Res. 2009;69:8835–38.
Lakshmanan M, Bughani U, Duraisamy S, Diwan M, Dastidar S, Ray A. Molecular targeting of E3 ligases–a therapeutic approach for cancer. Expert Opin Ther Targets. 2008;12:855–70.
Moller A, House CM, Wong CS, Scanlon DB, Liu MC, Ronai Z, et al. Inhibition of Siah ubiquitin ligase function. Oncogene. 2009;28:289–96.
Wong CS, Möller A. Siah: a promising anticancer target. Cancer Res. 2013;15:2400–06.
Behling KC, Tang A, Freydin B, Chervoneva I, Kadakia S, Schwartz GF, et al. Increased SIAH expression predicts ductal carcinoma in situ (DCIS) progression to invasive carcinoma. Breast Cancer Res Treat. 2011;29:717–24.
Brauckhoff A, Malz M, Tschaharganeh D, Malek N, Weber A, Riener MO, et al. Nuclear expression of the ubiquitin ligase seven in absentia homolog (SIAH)-1 induces proliferation and migration of liver cancer cells. J Hepatol. 2011;55:1049–57.
Brauckhoff A, Ehemann V, Schirmacher P, Breuhahn K. Reduced expression of the E3-ubiquitin ligase seven in absentia homologue (SIAH)-1 in human hepatocellular carcinoma. Verh Dtsch Ges Pathol. 2007;91:269–77.
Nakashima M, Lazo JS. Phosphatase of regenerating liver-1 promotes cell migration and invasion and regulates filamentous actin dynamics. J Pharmacol Exp Ther. 2010;334(2):627–33.
Achiwa H, Lazo JS. PRL-1 tyrosine phosphatase regulates c-Src levels, adherence, and invasion in human lung cancer cells. Cancer Res. 2007;15:643–50.
Stephens B, Han H, Hostetter G, Demeure MJ, Von Hoff DD. Small interfering RNA-mediated knockdown of PRL phosphatases results in altered Akt phosphorylation and reduced clonogenicity of pancreatic cancer cells. Mol Cancer Ther. 2008;7:202–10.
Zeng Q, Dong JM, Guo K, Li J, Tan HX, Koh V, et al. PRL-3 and PRL-1 promote cell migration, invasion, and metastasis. Cancer Res. 2003;1:2716–22.
Stephens BJ, Han H, Gokhale V, Von Hoff DD. PRL phosphatases as potential molecular targets in cancer. Mol Cancer Ther. 2005;4(11):1653–61.
Acknowledgements
We acknowledge to Universidad Autónoma de la Ciudad de México for support. The authors thank to the Microscopy Core at Instituto Fisiologia UNAM for their microscopy services.
Funding
This work was supported by CONACyT grants (Nos. 222335 and 233370). AFP was supported by ICyTDF-UACM fellowships (SRI/PB/64/2011).
Availability of data and materials
All supporting data for the conclusions is available as additional files.
Authors’ contributions
Conception and design (CLC, LAM, SRC, ERG, HAV, MAF). Development (AFP, CLC, LAM, SRC, ERG, HAV, JZR, MAF). Acquisition of data (AFP, MAFS, AFP, DRZ, AMAD, OHC, LFM). Drafted the manuscript (CLC and LAM). All authors critically revised and approved the manuscript for scientific content.
Competing interest
The authors have no conflict of interest to declare.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Tumour specimens were obtained following patient’s informed consent and the use of patient’s tissue was approved by the local ethics committee of the Institute of Breast Diseases, FUCAM Mexico.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1:
Oligonucleotides sequences used to silencing SIAH1 gene expression. (PDF 6 kb)
Additional file 2:
Comparative expression of miR-944 vs miR-204 and miR-944 vs miR-10b in TGCA validation cohort. (PDF 818 kb)
Additional file 3:
Taqman microRNA assay for miR-944 expression in MDA-MB-231 breast cancer cell line. (PDF 20 kb)
Additional file 4:
Raw data obtained from gene expression studies using DNA microarrays. (XLS 2.36 mb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Flores-Pérez, A., Marchat, L.A., Rodríguez-Cuevas, S. et al. Suppression of cell migration is promoted by miR-944 through targeting of SIAH1 and PTP4A1 in breast cancer cells. BMC Cancer 16, 379 (2016). https://doi.org/10.1186/s12885-016-2470-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-016-2470-3