Abstract
Background
Gestational diabetes mellitus (GDM) affects the metabolism of both the mother and fetus during and after pregnancy. Genetic factors are important in the pathogenesis of GDM, and associations vary by ethnicity. However, related studies about the relationship between the susceptibility genes and glucose traits remain limited in China. This study aimed to identify genes associated with GDM susceptibility in Chinese Han women and validate those findings using clinical data during pregnancy and postpartum period.
Methods
A genome-wide association study (GWAS) of 398 Chinese Han women (199 each with and without GDM) was conducted and associations between single nucleotide polymorphisms (SNPs) and glucose metabolism were identified by searching public databases. Relationships between filtered differential SNPs and glucose metabolism were verified using clinical data during pregnancy. The GDM group were followed up postpartum to evaluate the progression of glucose metabolism.
Results
We identified five novel SNPs with genome-wide significant associations with GDM: rs62069863 in TRPV3 gene and rs2232016 in PRMT6 gene were positive correlated with 1 h plasma glucose (1hPG) and 2 h plasma glucose (2hPG), rs1112718 in HHEX/EXOC6 gene and rs10460009 in LPIN2 gene were positive associated with fasting plasma glucose, 1hPG and 2hPG, rs927316 in GLIS3 gene was negative correlated with 2hPG. Of the 166 GDM women followed up postpartum, rs62069863 in TRPV3 gene was positively associated with fasting insulin, homoeostasis model assessment of insulin resistance.
Conclusions
The variants of rs62069863 in TRPV3 gene, rs2232016 in PRMT6 gene, rs1112718 in HHEX/EXOC6 gene, rs927316 in GLIS3 gene, and rs10460009 in LPIN2 gene were newly-identified susceptibility loci for GDM in the Chinese Han population. TRPV3 was associated with worse insulin resistance postpartum.
Trial registration
This study was registered in the Chinese Clinical Trial Registry. Trial registration number: ChiCTR2100043762. Date of first registration: 28/02/2021.
Similar content being viewed by others
Background
Gestational diabetes mellitus (GDM) is defined as abnormal glucose tolerance that is first detected during pregnancy [1]. It threatens both the mother and offspring during pregnancy and postpartum [2,3,4,5,6]. Previous studies have suggested that women with a history of GDM were at high risk of developing diabetes, dyslipidemia, and cardiovascular events postpartum [4,5,6]. Further, children whose mothers had GDM, have an increased risk of insulin resistance, obesity, and diabetes [1, 2]. In recent years, the prevalence of GDM in China has undergone a clear increase [7]. Early prediction and treatment could mitigate the maternal and fetal complications associated with GDM [8]. An understanding of the pathogenesis and genetic factors contributing to GDM could facilitate early prediction and treatment of the condition.
To date, epidemiological studies have identified a number of risk factors for GDM, including maternal age, pre-pregnancy body mass index (p-BMI), ethnicity, obesity, family history of type 2 diabetes mellitus (T2DM), and lipid disorders [3, 9]. Additionally, recent studies have also indicated that inflammatory factors, gut microbiome composition, and fatty acid levels were involved in the development of GDM [10,11,12].
Previous genetic investigations have identified several single nucleotide polymorphisms (SNPs) related to the risk of developing GDM. However, susceptibility varies substantially among women of different ethnicities. With increasing sample sizes and advances in sequencing technology, genome-wide association studies (GWAS) have become the main drivers for a range of discoveries in recent years. Several GWAS analyses have identified SNPs significantly associated with GDM at genome-wide significance, including two genetic variants (rs10830962 in MTNR1B gene and rs7754840 in CDKAL1 gene) identified in a Korean population [13]. In 2013, a GWAS focused on a European population found that HKDC1 and BACE2 were associated with GDM [14]. Given the variation in the results of those studies, susceptibility genes are highly likely to vary with ethnicity. However, related studies remain limited in China and the relationship between these genes and glucose traits requires further research. Thus, we conducted this study to search for genes associated with susceptibility to GDM in Chinese Han women and validated our findings using clinical data during pregnancy and postpartum period.
Methods
Study population
The study population was conducted in the first affiliated Hospital of Sun Yat-sen University between June and December 2021. The inclusion criteria were as follows: (1) age 18 to 45years; (2) Chinese Han pregnant women; (3) pregnant women received the 75-g oral glucose tolerance test (OGTT). In addition, the exclusion criteria were as follows: (1) diagnosis with diabetes before pregnancy; (2) multiple pregnancy; (3) chronic kidney disease or hepatic dysfunction; (4) a history of organ transplantation, hormonal therapy, or medication with anti-inflammatory agents; (5) malignancy. Diagnosis of GDM was in accordance with the International Association of Diabetes and Pregnancy Study Group criteria [15], based on any of the following after 75-g OGTT: fasting plasma glucose (FPG) level ≥ 5.1 mmol/L and < 7.0 mmol/L, 1 h plasma glucose (1hPG) level ≥ 10.0 mmol/L, or 2 h plasma glucose (2hPG) level ≥ 8.5 mmol/L and < 11.1 mmol/L. The GDM and control groups (pregnant women with normal glucose tolerance) were matched according to age and p-BMI. Control subjects were recruited during the same period as those in the GDM group.
This study was approved by the Medical Ethical Review Committee of the First Affiliated Hospital of Sun Yat-sen University (No. [2017]124 and No. [2021]309) and conducted according to the Declaration of Helsinki. Written informed consent to take part in the study was obtained from all participants. This study was registered in the Chinese Clinical Trial Registry (No. ChiCTR2100043762, first trial registration 28/02/2021).
Biochemical measurements
Between 24 and 28 weeks of pregnancy, participants were asked to undergo the 75-g OGTT after at least 8 h of fasting. FPG, 1hPG, and 2hPG were performed. Blood glucose and lipid measurements were made using fresh plasma samples. An additional sample was obtained for extraction of total genomic deoxyribonucleic acid (DNA). GDM women received ≥ 30 min of intensive lifestyle education provided by qualified nurses at 1 month after delivery. This education included physical exercise, smoking cessation, limiting alcohol intake, and keeping a healthy diet. In addition, 75-g OGTT were performed at 1–2 years after delivery for GDM women, as the average follow-up time was 17.31 ± 3.44 months. The 33 people who missed the postpartum follow-up include 26 refused postpartum participation (only phone interview) and 7 refused to have blood drawn for further evaluation. FPG, fasting insulin (FINS), 30 min plasma glucose (30minPG), 30 min insulin (30minINS), 2hPG, and 2 h insulin (2hINS) were tested. Glucose levels were determined using a glucose oxidase assay and insulin levels were analyzed by chemiluminescent assays. Blood glucose, insulin and lipids levels were measured using standard enzymatic procedures on an automatic chemistry analyzer (Abbott Aeroset, Chicago, IL, USA). All samples were measured in the laboratory of the Department of Biochemistry of the First Affiliated Hospital of Sun Yat-sen University.
The homoeostasis model assessment of β-cell function (HOMA-β) and insulin resistance (HOMA-IR) were calculated using the following formulae: HOMA-β = 20×FINS (µU/mL)/(FPG [mmol/L] − 3.5[mmol/L]), and HOMA-IR=(FPG [mmol/L] ×FINS [µU/mL]/22.5 [16, 17].
Genotyping and GWAS analysis
Genomic DNA was extracted using an established genomic DNA kit (Accurate Biotechnology Co., Ltd, Changsha, Hunan, China). The extracted genomic DNA was examined using 1% agarose gel electrophoresis. Each subject was genotyped using an Asian Screening Array bead chip from Illumina (San Diego, CA, USA), following the manufacturer’s protocol. A total of 659,184 SNPs were genotyped for each individual.
A series of quality control criteria were applied. The data exclusion criteria were as follows: (1) per-person missing rate > 5%, (2) per-SNP missing rate > 5%, (3) Hardy–Weinberg disequilibrium P value < 1E-6, and (4) minor allele frequency < 5%. Population structure was determined using principal components analysis, and the expected and observed distributions of the P-values were compared using Quantile-Quantile plots (Supplementary Figs. 1–2).
Database analysis
As described in previous studies [18,19,20], a secondary GWAS signal was identified (P < 5 × 10− 4) and the top 50 SNPs with the lowest P values in this study were selected for analysis. PubMed, GWAS Catalog, and Genotype-Tissue Expression (GTEx) databases were searched to identify SNPs relevant to glucose metabolism up to January 2022. SNPs were converted into genes by searching the NCBI SNP database. For PubMed and GWAS Catalog searches, the keywords were: “gestational diabetes mellitus” or “GDM” or “type 2 diabetes mellitus” or “T2DM” or “insulin” or “glucose”. The Version 7 release of the expression quantitative trait loci (eQTL) summarized data from the GTEx project was used for analysis of the association between SNPs and glucose metabolism-related tissues, including pancreas, liver, small intestine (terminal ileum), adipose (subcutaneous), and muscle (skeletal) [21].
Statistical analysis
Normally distributed data were expressed as the mean ± standard deviation. Qualitative data were presented as frequencies, and rate or composition ratios. Comparisons between two groups were conducted using the two-tailed unpaired Student’s t-test. P values < 0.05 were considered statistically significant. Statistical analyses were performed using SPSS 24.0. Associations between SNP genotypes and GDM or glucose traits were analyzed under the additive model using PLINK v1.9 software. Logistic and linear regression modelling were performed assuming an additive genetic model using the PLINK. Linear regression models about SNP genotypes and postpartum glucose traits were adjusted by postpartum obesity. A Manhattan plot of –log10 P values derived from logistic regression analysis plotted against chromosomal position was generated using the R statistical package. Log10 transformation was applied to FPG data, which were non-normally distributed.
Results
Baseline characteristics
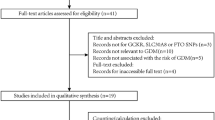
The study population comprised 398 participants, including 199 women with GDM and 199 women without GDM (Fig. 1). The native place of the participants was composed of 16 different provinces (Supplementary Table 1). Women in the GDM group aged 32.13 ± 3.65 years, and those in the control group aged 31.57 ± 3.78 years (P = 0.135). There were no differences in baseline characteristics between the two groups, as shown in Table 1.
GWAS for GDM
A total of 301,518 SNPs were retained from all individuals after quality control. A Manhattan plot of the GWAS was presented in Fig. 2. The top 50 SNPs identified by GWAS in increasing order of P value were shown in Supplementary Table 2. We converted the top 50 SNPs into genes and 32 genes were screened for relationships with glucose metabolism after searching the PubMed, GWAS catalog, and GTEx eQTL databases. Variants in TRPV3 gene (rs62069863, P = 1.95E-07) and PRMT6 gene (rs2232016, P = 1.20E-06) showed the strongest associations with GDM (Table 2 and Supplementary Table 2). Further, PRMT6, HHEX, EXOC6, GLIS3, and LPIN2 genes were strongly associated with glucose metabolism in at least two databases (Supplementary Fig. 3) and were therefore included in the next stage of the analysis. Although TRPV3 was only detected as glucose-sensitive in PubMed, it was also included in subsequent analyses, as it exhibited the most significant association with GDM in our population. The associations between these SNPs and GDM were presented in Table 2.
Manhattan plot of genome wide association analysis. Single nucleotide polymorphism locations were plotted on the x-axis according to their chromosomal position. Negative log10P values derived from logistic regression analysis under an additive model were plotted on the y-axis. The dashed line indicates the threshold of secondary GWAS signal
Relationships between identified SNPs and glucose metabolism
To further investigate the roles of these variants in GDM, we first performed an association analysis between the identified SNPs and FPG, 1hPG, and 2hPG during pregnancy. Five SNPs associated with glycemic traits in nongravid populations also demonstrated genome-wide significant associations in pregnant women (Table 3). Specifically, we found positive associations with FPG and SNPs in HHEX/EXOC6 and LPIN2 gene; 1hPG and SNPs in TRPV3 gene, PRMT6 gene, HHEX/EXOC6 gene, and LPIN2 gene; 2hPG and SNPs in TRPV3 gene, PRMT6 gene, HHEX/EXOC6 gene, and LPIN2 gene. The variant of rs927316 in GLIS3 gene was significantly negatively correlated with 2hPG. The positive associations between rs62069863 in TRPV3 gene and both 1hPG and 2hPG reached P = 2.60E-04 and P = 8.12E-05, with beta values ranging from 0.357 to 0.632 mmol/L and 0.367–0.614 mmol/L per A allele, respectively. The variant of rs2232016 in PRMT6 gene was strongly positively associated with 1hPG (P = 9.12E-06; beta range, 0.593–0.931 mmol/L per T allele) and 2hPG (P = 3.28E-04; beta range, 0.417–0.734 mmol/L per T allele). The SNP most strongly positively associated with FPG in HHEX/EXOC6 gene was rs1112718 (P = 1.38E-03; beta value range 0.008–0.016 log10 (mmol/L) per A allele). Further, rs10460009 in LPIN2 gene was positive associated with FPG (beta value range, 0.009–0.019 log10 (mmol/L) per A allele), while rs10460009 in LPIN2 gene was positive associated with 1hPG and 2hPG (P = 9.48E-03; beta range, 0.394–0.884 mmol/L per A allele; and P = 3.13E-04; beta range, 0.595–1.047 mmol/L per A allele, respectively).
Further, the relationships between identified SNPs and glucose metabolism during the postpartum period were analyzed. No significant difference was found between the above indices and PRMT6 or HHEX/EXOC6 (Supplementary Table 3), but significant positive relationships were found between TRPV3 and FINS, 30minINS, HOMA-IR and HOMA-β (Table 4 and Supplementary Table 4). Besides, marginally negative correlation was found between GLIS3 gene and FPG, while marginally positive correlation was found between LPIN2 gene and FPG (Supplementary Table 3). The associations between rs62069863 in TRPV3 gene and FINS reached P = 5.31E-05, with beta values ranging from 2.397 to 3.973 µlU/mL per A allele. In addition, strongly positive associations were observed between rs62069863 in TRPV3 gene and both HOMA-IR and HOMA-β (β = 0.701, P = 4.54E-05; β = 45.120, P = 2.48E-04).
Discussion
Here we reported the first China-based GWAS of maternal metabolic traits during pregnancy and postpartum. The main findings of this study were that variants of rs62069863 in TRPV3 gene, rs2232016 in PRMT6 gene, rs1112718 in HHEX/EXOC6 gene, rs927316 in GLIS3 gene, and rs10460009 in LPIN2 gene were associated with GDM and glucose traits. Further, we found that TRPV3 was positively related with worse insulin resistance of GDM women during the postpartum period.
Six genes at five loci exhibited genome-wide significant associations with maternal metabolic traits, including two genes (HHEX and GLIS3) that have demonstrated association with GDM in multiple studies [13, 22]. HHEX was positively associated with GDM risk in a Korean population and was previously reported as positively associated with T2DM in a Chinese population [23]. HHEX participates in pancreas development [24]. Some other studies found that SNPs in HHEX were associated with altered β cells secretion [25, 26]. GLIS3 was associated with GDM in European women [27] and with T2DM in Chinese women. GLIS3 might control insulin gene transcription and played a role in insulin secretion in β cells [28], while deficiency of GLIS3 induced apoptosis of the β cells of pancreatic islets and caused diabetes [27, 29, 30]. Hence, our results were consistent with those of previous studies.
We also found evidence for genome-wide significant associations of genes and loci with maternal metabolic traits that have not been previously reported in GDM populations. First, rs62069863 in TRPV3 gene was the most significant variant with GDM in Chinese Han population in our study and was associated with 1hPG, and 2hPG during pregnancy. Further, our results showed a significant correlation between TRPV3 and insulin resistance in GDM women after delivery. Cheung et al. reported that activation of TRPV3 could suppress adipogenesis by inhibiting the insulin receptor substrate 1/phosphoinositide 3-kinase/Akt/forkhead box protein O (FoxO) 1 axis [31], which was an essential glucose metabolism pathway. The biological role of TRPV3 in glucose and insulin metabolism is unknown. It has been reported that TRPV channels can modulate calcium homeostasis and insulin secretion in insulin-producing cells [32,33,34], which indicated that TRPV3 may be able to regulate glucose and insulin levels in a similar manner. However, this hypothesis requires further investigations to confirm.
A second novel finding of this study was the association between the rs2232016 T allele of PRMT6 gene and GDM, 1hPG, and 2hPG during pregnancy. The SNP rs2232016 is located in the only exon of PRMT6 gene and plays an important role in regulation of PRMT6 expression. It is highly likely that PRMT6 participates in glucose metabolism. Arginine methylation by PRMT6 was critical in maintaining adenosine cyclophosphate response element binding protein-regulated transcriptional coactivator 2 interaction on gluconeogenic promoters, and knockdown of hepatic PRMT6 in mice would lead to FPG decreasing [35]. Furthermore, Choi et al. reported that PRMT1 regulated FoxO3 through modulation of PRMT6 [36]. The absence of PRMT1 in skeletal muscle increased PRMT6 specifically methylates FoxO3, leading to enhanced expression of the autophagic marker. In addition, according to GTEx database, significant differences in the expression levels of the three genotypes of rs2232016 in PRMT6 gene were found in liver. It was reported that PRMT6 could regulated lipid transport [37]. Thereby, rs2232016 in PRMT6 gene might affect hepatic lipid metabolism. However, these issues still require further explorations.
In addition, we reported the first association between rs1112718 in HHEX/EXOC6 gene and GDM, although this SNP was previously reported as a T2DM susceptibility locus [38]. EXOC6 was associated with guanosine triphosphate binding protein-10 in adipocytes and participated in insulin signaling by influencing glucose transporter 4 (GLUT4) translocation [39]. GLUT4 controlled glucose transport into fat and muscle tissue in response to insulin and also into muscle during exercise [40]. However, rs1112718 was located in an intergenic region between HHEX and EXOC6, and its function remained unclear. Another locus, rs10460009 in LPIN2 gene, which was associated with FPG, 1hPG, and 2hPG in the current study, has not been previously associated with GDM risk or metabolic traits in humans. Aulchenko et al. found that LPIN2 was associated with T2DM and fat distribution [41], but studies about the role of LPIN2 in glucose metabolism are still limited.
Although the distributions of five new SNPs in Chinese Han population of our study were consistent with other population worldwide [42,43,44], the relationship between these five SNPs and GDM was not found in previous studies. There were two GWAS studies in other ethnicities, in Korea [13] and in European-American [14]. The genes found in this study were different from those two studies. However, those two studies reported different genes to each other. This revealed the genetic basis of GDM in different ethnicities. Therefore, it is necessary for different countries and regions to seek their own susceptibility genes.
This study demonstrated that five novel SNPs with genome-wide significant associations with GDM could contribute to the early prediction of GDM. The findings may provide new knowledge to our understanding of the causes of GDM. There are several limitations in our study. First, due to the relatively small sample size and slight lack of representative for Chinese Han population, our results require more Han women with GDM to expand representativeness and further verification by database mining and analysis of clinical factors. Moreover, this study did not include analysis of glycemic changes in offspring. A large-scale multicenter study with long-term follow-up should be performed in the future to validate our findings and expand representativeness. In addition, animal experiments are needed to further confirm the results of our study. Thus, further experiments are required to verify these findings.
Conclusions
In summary, our study was the first to show that rs62069863 in TRPV3 gene, rs2232016 in PRMT6 gene, rs1112718 in HHEX/EXOC6 gene, rs10460009 in LPIN2 gene and rs927316 in GLIS3 gene were susceptibility loci for GDM and were all significantly associated with blood glucose levels during pregnancy. Further, our data indicated that TRPV3 might participate in insulin resistance in GDM women postpartum.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- DNA:
-
Deoxyribonucleic acid
- eQTL:
-
Expression quantitative trait loci
- FINS:
-
Fasting insulin
- FoxO:
-
Forkhead box protein O
- FPG:
-
Fasting plasma glucose
- GDM:
-
Gestational diabetes mellitus
- GLUT4:
-
Glucose transporter 4
- GTEx:
-
Genotype-Tissue Expression
- GWAS:
-
Genome-wide association study
- HOMA-β:
-
Homoeostasis model assessment of β-cell function
- HOMA-IR:
-
Homoeostasis model assessment of insulin resistance
- OGTT:
-
Oral glucose tolerance test
- 1hPG:
-
1 h plasma glucose
- p-BMI:
-
Pre-pregnancy body mass index
- SNP:
-
Single-nucleotide polymorphism
- 2hINS:
-
2 h glucose load plasma insulin
- 2hPG:
-
2 h plasma glucose
- T2DM:
-
Type 2 diabetes mellitus
- 30minINS:
-
30 min glucose load plasma insulin
- 30minPG:
-
30 min plasma glucose
References
HAPO Study Cooperative Research Group. Hyperglycemia and adverse pregnancy outcomes. N Engl J Med. 2009;358:1991–2002.
Lowe WL Jr, Scholtens DM, Kuang A, Linder B, Lawrence JM, Lebenthal Y, et al. Hyperglycemia and adverse pregnancy outcome follow-up study (HAPO FUS): maternal gestational Diabetes Mellitus and childhood glucose metabolism. Diabetes Care. 2019;42:372–80.
McIntyre HD, Catalano P, Zhang C, Desoye G, Mathiesen ER, Damm P. Gestational Diabetes Mellitus. Nat Rev Dis Prim. 2019;5:47.
Goueslard K, Cottenet J, Mariet AS, Giroud M, Cottin Y, Petit JM, et al. Early cardiovascular events in women with a history of gestational Diabetes Mellitus. Cardiovasc Diabetol. 2016;15:15.
Pei L, Xiao HM, Lai FH, Li Z, Li Z, Yue S, et al. Early postpartum dyslipidemia and its potential predictors during pregnancy in women with a history of gestational Diabetes Mellitus. Lipids Health Dis. 2020;19:220.
Li Z, Cheng Y, Wang D, Chen H, Chen H, Ming WK, et al. Incidence rate of type 2 Diabetes Mellitus after gestational Diabetes Mellitus: a systematic review and meta-analysis of 170,139 women. J Diab Res. 2020;2020:3076463.
Wang C, Jin L, Tong MK, Zhang J, Yu J, Meng W, et al. Prevalence of gestational Diabetes Mellitus and its determinants among pregnant women in Beijing. J Matern Fetal Neonatal Med. 2020;35:1–7.
Sriboonvorakul N, Hu J, Boriboonhirunsarn D, Ng LL, Tan BK. Proteomics studies in Gestational Diabetes Mellitus: a systematic review and Meta-analysis. J Clin Med. 2022;11(10):2737. https://doi.org/10.3390/jcm11102737.
Zhang XM, Zhao X, Huo LL, Yuan N, Sun J, Du J, et al. Risk prediction model of gestational Diabetes Mellitus based on nomogram in a Chinese population cohort study. Sci Rep. 2020;10:21223.
Sun X, Sun H, Li P. Association of circulating inflammatory cells and platelets with gestational Diabetes and pregnancy outcomes. Clin Chim Acta. 2021;523:87–96.
Vitacolonna E, Masulli M, Palmisano L, Stuppia L, Franzago M. Inositols, Probiotics, and gestational Diabetes: clinical and epigenetic aspects. Nutrients. 2022;14:1543.
Liu Y, Xia YY, Zhang T, Yang Y, Cannon RD, Mansell T, et al. Complex interactions between circulating fatty acid levels, desaturase activities, and the risk of gestational Diabetes Mellitus: a prospective cohort study. Front Nutr. 2022;11:9:919357.
Kwak SH, Kim SH, Cho YM, Go MJ, Cho YS, Choi SH, et al. A genome-wide association study of gestational Diabetes Mellitus in Korean women. Diabetes. 2012;61:531–41.
Hayes MG, Urbanek M, Hivert MF, Armstrong LL, Morrison J, Guo C, et al. Identification of HKDC1 and BACE2 as genes influencing glycemic traits during pregnancy through genome-wide association studies. Diabetes. 2013;62:3282–91.
International Association of Diabetes and Pregnancy Study Groups Consensus Panel, Metzger BE, Gabbe SG, Persson B, Buchanan TA, Catalano PA, et al. International association of Diabetes and pregnancy study groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care. 2010;33:676–82.
He S, Ji D, Liu Y, Deng X, Zou W, Liang D, et al. Polymorphisms of mtDNA in the D-loop region moderate the associations of BMI with HOMA-IR and HOMA-beta among women with polycystic ovary syndrome: a cross-sectional study. J Assist Reprod Genet. 2023;40:1983–93.
Matli B, Schulz A, Koeck T, Falter T, Lotz J, Rossmann H, et al. Distribution of HOMA-IR in a population-based cohort and proposal for reference intervals. Clin Chem Lab Med. 2021;59:1844–51.
Huang YT, Heist RS, Chirieac LR, Lin X, Skaug V, Zienolddiny S, et al. Genome-wide analysis of Survival in Early-Stage non–small-cell Lung Cancer. J Clin Oncol. 2009;27:2660–7.
Ahn MJ, Won HH, Lee J, Lee ST, Sun JM, Park YH, et al. The 18p11.22 locus is associated with never smoker non-small cell Lung cancer susceptibility in Korean populations. Hum Genet. 2012;131:365–72.
Israel E, Lasky-Su J, Markezich A, Damask A, Szefler SJ, Schuemann B, et al. Genome-wide association study of short-acting β2-agonists. A novel genome-wide significant locus on chromosome 2 near ASB3. Am J Respir Crit Care Med. 2015;191:530–7.
GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; statistical methods groups—analysis Working Group. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13.
Ding M, Chavarro J, Olsen S, Lin Y, Ley SH, Bao W, et al. Genetic variants of gestational Diabetes Mellitus: a study of 112 SNPs among 8722 women in two Independent populations. Diabetologia. 2018;61:1758–68.
Jamaspishvili E, Zhang SJ, Tong HX, Eka Puspita O, Zein FMA. East Asian genome-wide association study derived loci in relation to type 2 Diabetes in the Han Chinese population. Acta Biochim Pol. 2019;66:679–86.
Jackson JT, Nutt SL, McCormack MP. The Haematopoietically-expressed homeobox transcription factor: roles in development, physiology and Disease. Front Immunol. 2023;14:1197490.
Park S, Liu M, Kang S. Alcohol intake interacts with CDKAL1, HHEX, and OAS3 genetic variants, Associated with the risk of type 2 Diabetes by lowering insulin secretion in Korean adults. Alcohol Clin Exp Res. 2018;42:2326–36.
Dimas AS, Lagou V, Barker A, Knowles JW, Mägi R, Hivert MF, et al. Impact of type 2 Diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63:2158–71.
Watanabe N, Hiramatsu K, Miyamoto R, Yasuda K, Suzuki N, Oshima N, et al. A murine model of neonatal Diabetes Mellitus in Glis3-deficient mice. FEBS Lett. 2009;583:2108–13.
Wen X, Yang Y. Emerging roles of GLIS3 in neonatal Diabetes, type 1 and type 2 Diabetes. J Mol Endocrinol. 2017;58:R73–85.
Yang YS, Chang BHJ, Samson SL, Li MV, Chan L. The kruppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 2009;37:2529–38.
Nogueira TC, Paula FM, Villate O, Colli ML, Moura RF, Cunha DA, et al. GLIS3, a susceptibility gene for type 1 and type 2 Diabetes, modulates pancreatic beta cell apoptosis via regulation of a splice variant of the BH3-only protein Bim. PLoS Genet. 2013;9:e1003532.
Cheung SY, Huang Y, Kwan HY, Chung HY, Yao X. Activation of transient receptor potential vanilloid 3 channel suppresses adipogenesis. Endocrinology. 2015;156:2074–86.
Skrzypski M, Billert M, Mergler S, Khajavi N, Nowak KW, Strowski MZ. Role of TRPV channels in regulating various pancreatic beta-cell functions: lessons from in vitro studies. Biosci Trends. 2017;11:9–15.
Zhong B, Ma S, Wang DH. TRPV1 mediates glucose-induced insulin secretion through releasing neuropeptides. In Vivo. 2019;33:1431–7.
Sawatani T, Kaneko YK, Doutsu I, Ogawa A, Ishikawa T. TRPV2 channels mediate insulin secretion induced by cell swelling in mouse pancreatic beta-cells. Am J Physiol Cell Physiol. 2019;316:C434–43.
Han HS, Jung CY, Yoon YS, Choi S, Choi D, Kang G, et al. Arginine methylation of CRTC2 is critical in the transcriptional control of hepatic glucose metabolism. Sci Signal. 2014;7:ra19.
Choi S, Jeong HJ, Kim H, Choi D, Cho SC, Seong JK, et al. Skeletal muscle-specific Prmt1 deletion causes muscle atrophy via deregulation of the PRMT6-FOXO3 axis. Autophagy. 2019;15:1069–81.
Cheng Y, Gao Z, Zhang T, Wang Y, Xie X, Han G, et al. Decoding m6A RNA methylome identifies PRMT6-regulated lipid transport promoting AML stem cell maintenance. Cell Stem Cell. 2023;30:69–85.
Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, et al. Fine-mapping type 2 Diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–13.
Sano H, Peck GR, Blachon S, Lienhard GE. A potential link between insulin signaling and GLUT4 translocation: Association of Rab10-GTP with the exocyst subunit Exoc6/6b. Biochem Biophys Res Commun. 2015;465:601–5.
Klip A, McGraw TE, James DE. Thirty sweet years of GLUT4. J Biol Chem. 2019;294:11369–81.
Aulchenko YS, Pullen J, Kloosterman WP, Yazdanpanah M, Hofman A, Vaessen N, et al. LPIN2 is associated with type 2 Diabetes, glucose metabolism, and body composition. Diabetes. 2007;56:3020–6.
Sudmant PH, Rausch T, Gardner EJ, et al. 1000 Genomes Project Consortium. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81.
Jung KS, Hong KW, Jo HY, Choi J, Ban HJ, Cho SB, et al. KRGDB: the large-scale variant database of 1722 koreans based on whole genome sequencing. Database (Oxford). 2020;2020:baaa030.
Major-Smith D, Heron J, Fraser A, Lawlor DA, Golding J, Northstone K. The Avon Longitudinal Study of parents and children (ALSPAC): a 2022 update on the enrolled sample of mothers and the associated baseline data. Wellcome Open Res. 2023;6:7:283.
Acknowledgements
The authors thanked the research assistants, paramedics and physicians of all participating centers for their supports and all women who participated in the study.
Funding
This work was supported by the Clinical Medical 5010 Project Foundation of Sun Yat-sen University (Grant number: 2017001) and the Science and Technology Foundation of Guangzhou City (Grant number:201803010101).
Author information
Authors and Affiliations
Contributions
XC conceived and designed the study. SY did the data analysis, interpreted the data and wrote the draft manuscript. FL helped with writing and revised the paper. SY, LP, ZR and HX collected the samples. ZL, WC, LC and HL collected the clinical data. YL and HX performed critical revision on the manuscript. All authors approved the final version before submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Medical Ethical Review Committee of the First Affiliated Hospital of Sun Yat-sen University (No. [2017]124 and No. [2021]309) and conducted according to the Declaration of Helsinki. Written informed consent to take part in the study was obtained from all participants. This study was registered in the Chinese Clinical Trial Registry (No. ChiCTR2100043762).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yue, S., Pei, L., Lai, F. et al. Genome-wide analysis study of gestational diabetes mellitus and related pathogenic factors in a Chinese Han population. BMC Pregnancy Childbirth 23, 856 (2023). https://doi.org/10.1186/s12884-023-06167-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12884-023-06167-3