Abstract
Background
Autosomal dominant polycystic kidney disease (ADPKD), the predominant type of inherited kidney disorder, occurs due to PKD1 and PKD2 gene mutations. ADPKD diagnosis is made primarily by kidney imaging. However, molecular genetic analysis is required to confirm the diagnosis. It is critical to perform a molecular genetic analysis when the imaging diagnosis is uncertain, particularly in simplex cases (i.e. a single occurrence in a family), in people with remarkably mild symptoms, or in individuals with atypical presentations. The main aim of this study is to determine the frequency of PKD1 gene mutations in Iranian patients with ADPKD diagnosis.
Methods
Genomic DNA was extracted from blood samples from 22 ADPKD patients, who were referred to the Qaem Hospital in Mashhad, Iran. By using appropriate primers, 16 end exons of PKD1 gene that are regional hotspots, were replicated with PCR. Then, PCR products were subjected to DNA directional Sanger sequencing.
Results
The DNA sequencing in the patients has shown that exons 35, 36 and 37 were non- polymorphic, and that most mutations had occurred in exons 44 and 45. In two patients, an exon-intron boundary mutation had occurred in intron 44. Most of the variants were missense and synonymous types.
Conclusion
In the present study, we have shown the occurrence of nine novel missense or synonymous variants in PKD1 gene. These data could contribute to an improved diagnostic and genetic counseling in clinical settings.
Similar content being viewed by others
Background
One of the most prevalent inherited kidney disorders that affects both kidneys is autosomal dominant polycystic kidney disease (ADPKD), which leads to a progressive loss of kidney function and kidney failure [1]. About one to two infants in 1000 live at birth, and approximately 10% of people who undergo dialysis are affected by this disease [2, 3]. ADPKD occurs in two types including type I and type II, caused by PKD1 and PKD2 mutations, respectively [4, 5].
PKD2 mutation causes end-stage renal disease at an average age of 74 years, which occurs in 10–15% of cases; on the other hand, PKD1 mutation results in end-stage renal disease at an average age of 54 years which occurs in 80–90% of total cases of ADPKD. The latter is the more severe form of the disease [1, 3, 5]. Patients having end-stage kidney disease should receive renal replacement therapy (RRT) or dialysis to stay alive. However, dialysis has some limitations, including lack of vascular access, risks of vascular thrombosis, infections, diminished quality of life, and loss of the kidney biosynthetic functions [6]. Patients who were diagnosed with ADPKD before age of 30 and patients who have hypertension or hematuria before age of 35, have a worse renal outcome [7]. ADPKD diagnosis is typically carried out by kidney ultrasound imaging, computed tomography scan or magnetic resonance imaging; however, considering the similarity of ADPKD to other cystic kidney disorders, conventional imaging methods do not often lead to a definite diagnosis [1, 2]. Additionaly, molecular methods have an important role to confirm ADPKD diagnosis, especially in young kidney donors, patients with negative family history, individuals who present ADPKD with unusual symptoms in childhood and patients who have relatives suffering from this disorder [8, 9].
ADPKD is the most frequent genetic kidney disorder (frequency of about 0.1%), which results in 5–8% of end-stage renal diseases (ESRDs). ESRD is a progressive, disease with enlarged polycystic kidneys typically occuring in the late middle age [5]. Polycystin-1, is a large multidomain protein encoded by PKD1 gene. It has domains and regions that are homologous with a number of different proteins [10]. Polycystin- 1 has been proposed to act as a G protein–coupled receptor [11]. Instead, polycystin-2 (the protein coded by PKD2) is homologous to an ion-channel subunit [12, 13]. Most cases of ADPKD leading to ESRD are caused by PKD1 mutations [14]. Nevertheless, the genetic determination of the locus mutation has advanced slowly, due to the fact that PKD1 contains a 12,906-bp coding sequence divided into 46 exons and that the 5′ region of the gene, from upstream of exon 1 to exon 33, is inserted in a complex genomic area and repeated more than 4 times on the same chromosome [15]. The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Homologous sequences searches in a number of databases have found one partial cDNA and two genomic sequences with significant homology to both polycystin-1 and -2 [16].
The PKD1-like homologous gene (HG) has revealed a number of specific deletions and a low level of substitutions (about 2%) in comparison with PKD1 [17]. The HG locus analysis of PKD1 has been highly difficult. Thus, the quantity of identified PKD1 mutations is still incomplete, with 82 modifications described in the Online Human Gene Mutation Database (HGMD) [18]. A multiple number of methods have been used to screen the repeated region [19,20,21,22,23], however, the 3′ area has received insufficient attention, with 57.3% of all mutations found in the single-copy area covering 20% of the coding region. PKD2 (a less-complex gene) has revealed 41 mutations with potential effects of truncating and possibly inactivating the translated protein [24]. A discrete number of missense changes have also been described [19, 23,24,25,26]. Since numerous somatic mutations and a significant rate of formation of novel germline mutations are needed to explain cystogenesis [19], it has been proposed that infrequent mechanisms promote a high rate of PKD1 mutations. A long polypyrimidine region in IVS21, which could theoretically form triplex DNA structures [27, 28], has been considered as a possible cause of mutations in downstream exonsequences [22]. These multiple substitutions and other modifications were described to match HG sequences, possibly indicating a gene conversion with the remotely located HG loci [21, 29]. PKD1 gene (OMIM 601313) is located in the 16p13.3 chromosome region and consists of 46 exons. Exons 1–33 of PKD1 replicates around 6 times in HG, which has challenged PKD1 genetic analysis. Until January 2015, approximately 2322 PKD1 sequence variants and 278 PKD2 sequence variants were reported in ADPKD mutation databases, as well as 1177 and 211 human mutations in PKD1 and PKD2 sequences, respectively [16, 17]. Although mutation data for PKD genes of different populations are available, there are few reports for PKD mutations in the Iranian population. The main goal of this study was to establish the frequency of mutations in the PKD1 gene obtained by PCR (Polymerase Chain Reaction) and DNA Sanger sequencing [30] in the Iranian patients with ADPKD diagnosis.
Methods
Patient selection
Twenty-two ADPKD patients were obtained from the Ghaem Hospital; (Mashhad, Iran) between April 2012 to March 2013. They were included after diagnosis and disease characteristics as ADPKD. The study was approved by ethics committee of Mashhad University of Medical Sciences. Before the blood sample were collected, all patients provided their informed consents.
We excluded patients later clinically diagnosed by Von Hippel-Lindau disease and Tuberous Sclerosis. In addition, patients without symptoms of polycystic kidney disease or those who had other syndromes were also excluded in this study.
Amplification assay
Genomic DNA was extracted from 22 whole-blood samples using the standard salting-out method and it was quantified by NanoDrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA). Eight-specific primers within the region of the exon 31–46 were designed with the Primer 3 software (Table 1). Sequences were checked for self- or inter-molecular annealing with a nucleic-acid-folding software (OligoAnalyzer 3.1). We performed local-alignment analyses with the BLAST program to confirm the specificity of the designed primers (http://www-ncbi-nlm-nih-gov.acces.bibl.ulaval.ca/tools/primer-blast). Bidirectional sequence analysis was conducted for all PCR amplicons.
Amplification was performed in a thermal cycler, GeneAmp PCR System 9700 (Applied Biosystems, Massachusetts, USA), including 150 ng of genomic DNA, 10X PCR buffer, 2 mM MgCl2, 1 Unit Taq DNA polymerase (Genet Bio, South Korea), 0.2 mM dNTP mix, and 5 pmol of each primer in a final volume of 20 μl. Cycling parameters were as follows: an initial denaturation at 95 °C for 5 min, 35 cycles at 95 °C for 30 s, annealing for 30 s at 52 °C, 57 °C, 69 °C, 67 °C, 54 °C, 61 °C and 62 °C for primer#1 to #8 respectively, and a final extension step at 72 °C for 35 s, ended by a last extension at 72 °C for 5 min.
PCR products were analyzed by electrophoresis in a 1.5% agarose gel stained with ethidium bromide followed by Sanger sequencing reactions.
Sanger sequencing
Sequencing products were run on an ABI 3130XL Genetic Analyzer (Macrogene Company South Korea), according to the manufacturer’s guidelines. Data analysis was performed with Chromas software version 2.6.5 (Technelysium, South Brisbane, Australia).
Results
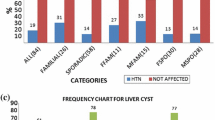
Twenty-two patients with an average age of 36.6 ± 7.3 years, suffering from ADPKD were studied. The sequencing results of the patientsare reported in Table 2 and Fig. 1. In patient 45.1, variations in rs10960 polymorphism in exon 44 led to the conversion of isoleucine to valine (p.Ile4045Val). This type of variation, considered as missense, was recorded in the PKDB database with a minor allele frequency of 0.239. Moreover, the single nucleotide variant (p.Ile4045Val), was also found in patient 410.2. In four patients, including 45.3, 410.1, 417.1, and 419.1, exon 45 had a synonymous mutation (p.Ala4092=) and was reported as rs3087632 with MAF: 0.262 in the database PKDB. Moreover, the missense mutation converting glutamine to arginine (p.Gln4005Arg) had occurred in exon 44 of the patient 421 and was recorded as uncertain significance in the PKDB database.
DNA sequencing results of Iranian patients with ADPKD. Patients including a) patient# 421 (p.Gln4005Arg); b) patient# 48.1 (p.Leu4031X); c) patient# 418.2 (p.Val4035Met); d) patient# 45.1 (p.Ile4045Val); e) patient# 410.2 (p.Ile4045Val); f) patient# 47.1 (p.Ser4013=); g) patient# 45.3 (p.Ala4092=); h) patient# 417.1 (p.Ala4092=); i) patient# 419.1(p.Ala4092=); j) patient# 410.1 (p.Ala4092=); k) patient# 422 (p.Thr4073Ala)
Novel variants
The first variant was observed in patient 45.5. This variant caused a synonymous variant in exon 44 (p.Gly4068=). Patient 47.1, a variant of rs200796474 was also synonymous, with a serine converted to serine (p.Ser4013=). A leucine to stop codon mutation, was observed in exon 44 of patient 48.1 (p.Leu4031X). In patient 411.2, a missense converting arginine to leucine was observed in exon 45 and the missense mutation converting arginine to leucine was also found in the same exon of the same patient (CGT/CTT). The missense change converting valine to methionine occurred in exon 44 of patient 418.2 (p.Val4035Met). In addition, the missense variation converting threonine to alanine was found in some part of the exon 45 in the patient 422 (p.Thr4073Ala)(Table 2).
In silico functional analysis
Nucleotide changes in the PKD1 gene was determined based on reference genomic sequemces NC_000016.10. The detected sequence variations reported in this study were checked with the list of Autosomal Dominant Polycystic Kidney Disease Mutation Database (PKDB) and PKD gene variants in the Human Gene Mutation Database (HGMD) [31].
The pathogenicity prediction of novel variations were analyzed by Mutation Taster [32]. We checked related protein products for sequence and length alteration by altered CDS (NM_001009944) using expasy translate tools. The prediction obtained of the potential effect of each variant has been shown in Table 2. In the current study, mutations were named based on CDS according to standard mutation nomenclature for molecular diagnostic aims.
The UniProt database, UniProtKB ID Q8IYM9 (http://www.uniprot.org), the NCBI dbSNP database (https://www.ncbi.nlm.nih.gov/SNP/), and 1000 Genomes (http://www.1000genomes.org/) have been used to retrieve polymorphism data. Functional effects of SNPs were predicted using Polyphen-2 (http://genetics.bwh.harvard.edu/pp2).
Discussion
To date, 2322 pathogenic meuations for PKD1 and 278 for PKD2 have been reported in the PKDB [33] but their relative frequencies are unknown. Moreover, Daoust et al., identified a family in the French-Canadian population in which a classical clinical presentation of ADPKD resulted from a mutation at a locus genetically distinct from all the previously described loci for this disease. This suggests an existence of a third genetic locus for ADPKD [5].
In the current study, 16 end exons of PKD1 gene were studied. The sequencing results have shown that exons 35, 36 and 36 were non-polymorphic, with no mutations, and the most mutations occurred in exons 44 and 45. In most of the patients, variants were mostly missense and same-sense types. Our results have shown that there is no definite hot spot in PKD1 and thus, a complete PKD1 mutation analysis is needed for genetic diagnosis of ADPKD in the Iranian patients. Our newly detected mutations in the Iranian population have made the PKD mutation database richer, a result of great importance in the genetic consultation of ADPKD patients.
Regarding the large genes involved in ADPKD, screening all of their regions would be expensive and time-consuming; hence, to overcome this issue a database could be generated for mutations of polycystic kidney disease among the Iranian population to determine the most common mutations and to characterize mutation hot spots in this population. Furthermore, considering the clinical similarity of ADPKD with other kidney cystic diseases, causing incorrect clinical diagnosis in the absence of familial history, molecular study for PKD1 with or without PKD2 in suspected patients is recommended.
Identified pathogenic mutations in the present study could be confirmed in future studies with more ADPKD families. Besides, genotype-phenotype correlation studies could be performed to determine the severity of each variant and the outcome of patients associated with a specific variant.
Conclusion
In the current study, we demonstrated nine novel missense or synonymous variants in PKD1. These data will contribute to an improved diagnostic and genetic counseling in clinical settings.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the GenBank repository, [BankIt2382235 BSeq#1 MT993938]. Any additional information related to this study is available from the author for correspondence upon reasonable request.
Abbreviations
- ADPKD:
-
Autosomal dominant polycystic kidney disease
- ESRDs:
-
End-stage renal diseases
- PCR:
-
Polymerase chain reaction
- PKDB:
-
Polycystic Kidney Disease Mutation Database
- HGMD:
-
Human Gene Mutation Database
References
Torres VE, Harris PC, Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369(9569):1287–301.
Grantham JJ. Clinical practice. Autosomal dominant polycystic kidney disease. N Engl J Med. 2008;359(14):1477–85.
Torres VE, Harris PC. Autosomal dominant polycystic kidney disease. Nefrologia. 2003;23(Suppl 1):14–22.
Srivastava A, Patel N. Autosomal dominant polycystic kidney disease. Am Fam Physician. 2014;90(5):303–7.
Daoust MC, Reynolds DM, Bichet DG, Somlo S. Evidence for a third genetic locus for autosomal dominant polycystic kidney disease. Genomics. 1995;25(3):733–6.
Yeo WS, Ng QX. Disruptive technological advances in vascular access for dialysis: an overview. Pediatr Nephrol. 2017;29:1–6.
Johnson AM, Gabow PA. Identification of patients with autosomal dominant polycystic kidney disease at highest risk for end-stage renal disease. J Am Soc Nephrol. 1997;8(10):1560–7.
Gabow PA. Autosomal dominant polycystic kidney disease—more than a renal disease. Am J Kidney Dis. 1990;16(5):403–13.
Fick GM, Johnson AM, Hammond WS, Gabow PA. Causes of death in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 1995;5(12):2048–56.
Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, San Millan JL, et al. The polycystic kidney disease1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet. 1995;10(2):151–60.
Zhou J. Polycystins and primary cilia: primers for cell cycle progression. Annual Rev Physiol. 2009;71:83–113. https://doi.org/10.1146/annurev.physiol.70.113006.100621 PMID 19572811.
Mochizuki N, Cho G, Wen B, Insel PA. Identification and cDNA cloning of a novel human mosaic protein, LGN, based on interaction with G alpha i2. Gene. 1996;181(1–2):39–43.
Chen ML, Hoshi T, Wu CF. Heteromultimeric interactions among K+ channel subunits from shaker and eag families in Xenopus oocytes. Neuron. 1996;17(3):535–42.
Hateboer N, Lazarou LP, Williams AJ, Holmans P, Ravine D. Familial phenotype differences in PKD11. Kidney Int. 1999;56(1):34–40.
The Polycystic Kidney Disease 1 Gene Encodes a 14 Kb Transcript and Lies Within a Duplicated Region on Chromosome 16. The European Polycystic Kidney Disease Consortium. Cell. 1994;77(6):881–94. https://pubmed.ncbi.nlm.nih.gov/8069919/.
Veldhuisen B, Spruit L, Dauwerse HG, Breuning MH, Peters DJ. Genes homologous to the autosomal dominant polycystic kidney disease genes (PKD1 and PKD2). Eur J Hum Genet. 1999;7(8):860–72.
Loftus BJ, Kim UJ, Sneddon VP, Kalush F, Brandon R, Fuhrmann J, et al. Genome duplications and other features in 12 Mb of DNA sequence from human chromosome 16p and 16q. Genomics. 1999;60(3):295–308.
Krawczak M, Cooper DN. The human gene mutation database. Trends Genet. 1997;13(3):121–2.
Peral B, Gamble V, Strong C, Ong AC, Sloane-Stanley J, Zerres K, et al. Identification of mutations in the duplicated region of the polycystic kidney disease 1 gene (PKD1) by a novel approach. Am J Hum Genet. 1997;60(6):1399–41.0.
Watnick T, Phakdeekitcharoen B, Johnson A, Gandolph M, Wang M, Briefel G, et al. Mutation detection of PKD1 identifies a novel mutation common to three families with aneurysms and/or very- early-onset disease. Am J Hum Genet. 1999;65(6):1561–71.
Watnick TJ, Gandolph MA, Weber H, Neumann HP, Germino GG. Gene conversion is a likely cause of mutation in PKD1. Hum Mol Genet. 1998;7(8):1239–43.
Watnick TJ, Piontek KB, Cordal TM, Weber H, Gandolph MA, Qian F, et al. An unusual pattern of mutation in the duplicated portion of PKD1 is revealed by use of a novel strategy for mutation detection. Hum Mol Genet. 1997;6(9):1473–81.
Thomas R, McConnell R, Whittacker J, Kirkpatrick P, Bradley J, Sandford R. Identification of mutations in the repeated part of the autosomal dominant polycystic kidney disease type 1 gene, PKD1, by long-range PCR. Am J Hum Genet. 1999;65(1):39–49.
Veldhuisen B, Saris JJ, de Haij S, Hayashi T, Reynolds DM, Mochizuki T, et al. A spectrum of mutations in the second gene for autosomal dominant polycystic kidney disease (PKD2). Am J Hum Genet. 1997;61(3):547–55.
Daniells C, Maheshwar M, Lazarou L, Davies F, Coles G, Ravine D. Human gene mutations. Gene symbol: PKD1. Disease: Polycystic kidney disease. Hum Genet. 1998;10:127 (1)2.
Perrichot RA, Mercier B, Simon PM, Whebe B, Cledes J, Ferec C. DGGE screening of PKD1 24.Gene reveals novel mutations in a large cohort of 146 unrelated patients. Hum Genet. 1999;105(3):231–9.
Van Raay TJ, Burn TC, Connors TD, Petry LR, Germino GG, Klinger KW, et al. A 2.5 kb polypyrimidine tract in the PKD1 gene contains at least 23 H-DNA-forming sequences. Microb Comp Genomics. 1996;1(4):317–27.
Blaszak RT, Potaman V, Sinden RR, Bissler JJ. DNA structural transitions within the PKD1 gene. Nucleic Acids Res. 1999;27(13):2610–7.
Phakdeekitcharoen B, Watnick TJ, Ahn C, Whang DY, Burkhart B, Germino GG. Thirteen novel mutations of the replicated region of PKD1 in an Asian population. Kidney Int. 2000;58(4):1400–12.
Rossetti S, Strmecki L, Gamble V, Burton S, Sneddon V, Peral B, et al. Mutation analysis of the entire PKD1 gene: genetic and diagnostic implications. Am J Hum Genet. 2001;68(1):46–63.
Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, et al. The human gene mutation database: 2008 update. Genome Med. 2009;1(1):13.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–2.
Liu B, Chen S-C, Yang Y-M, Yan K, Qian Y-Q, Zhang J-Y, et al. Identification of novel PKD1 and PKD2 mutations in a Chinese population with autosomal dominant polycystic kidney disease. Sci Rep. 2015;5:17.
Acknowledgments
We thank Dr. Hassan Boskabadi, from Mashhad University of Medical Sciences for providing human blood samples. Special thanks also go to Mr. Mohammad Bagher Eskandari at Ghaem Genetic Diagnostic Laboratory Mashhad for his technical assistance. We thank all the family members for their participation and their physicians for the clinical evaluation of the patients.
Funding
This study was financially supported by grants from Vice-Chancellor of Research, Mashhad University of Medical Sciences, Mashhad, Iran, (Grant# 901006).
The funders of this study do not have any roles in study design, data collection, analysis, result interpretation, writing and the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
FK performed the experiments. MAK and AT participated in the study design and scientific discussion of the data. MAK supervised the study. All authors contribute to the writing, read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Mashhad University of Medical Sciences. A written informed consent was obtained from each individual participating in this study.
Consent for publication
Our study is not a case report, and identifying images or other personal or clinical detail of participants that compromise anonymity are not included. Consent to publish from the patients, or in case of minors, the patients’ guardians is “Not Applicable”.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Khadangi, F., Torkamanzehi, A. & Kerachian, M.A. Identification of missense and synonymous variants in Iranian patients suffering from autosomal dominant polycystic kidney disease. BMC Nephrol 21, 408 (2020). https://doi.org/10.1186/s12882-020-02069-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12882-020-02069-0