Abstract
Background
Previous studies evaluated the association of IL-4 C33T polymorphism and risk of bronchial asthma but failed to establish a consistent conclusive association. In the present meta-analysis, we intend to define a more reliable estimate of the association in the presence of filling published literature.
Methods
An exhaustive search in Web of Science, Scopus, and PubMed databases was performed to identify all relevant publications before September 2020, and 24 publications (28 studies) with 6587 cases and 8408 controls were included in final analysis. The association between polymorphism and risk of asthma were measured by Odd ratios (ORs) and 95% confidence intervals (CIs). Moreover, Cochran’s Q and the I2 statistics were used to evaluate the degree of heterogeneity between studies.
Results
In the overall study populations, a significant positive association was detected under all genotype models and announced the IL-4 C33T polymorphism as a potential risk factor in the pathogenesis of asthma. In the subgroup analysis by age, a significant association between IL-4 C33T polymorphism and risk of asthma in different age groups was identified in allelic model, which highlighted the predisposing role of the T allele for the asthma risk in all three age groups. Furthermore, the results of subgroup analysis by continent were heterogenous. Accordingly, IL-4 C33T polymorphism was a risk factor in Europeans (all models except heterozygote comparison), Americans (all models except recessive and homozygote comparison) and Asians (just recessive and allelic model). Finally, the ethnicity-specific analysis disclosed a significant association between IL-4 C33T polymorphism and asthma risk in Caucasians (all genotype models except heterozygote comparison), while this association was not significant in African-Americans.
Conclusions
This study suggests that IL-4 C33T polymorphism potentially acts as a risk factor for asthma in different ethnicities and age groups.
Similar content being viewed by others
Background
Asthma is a chronic, complex respiratory disorder in which allergen-triggered inflammatory reactions in the airways contribute to the development of symptoms, including breathlessness, cough, wheezing, and dyspnea. It has been estimated that asthma affect about 300 million people in the world [1]. Prognostic markers to detect high-risk individuals are urgently required for early identification and preventive attention. In the scientific community, genetic vulnerability to asthma is one of the main research interests [2]. In the recent decade, many studies have been focused to elucidate the susceptibility genes of asthma and several single nucleotide polymorphisms (SNPs) in these genes have been described to be related with asthma risk in different populations [3, 4]. Among different genes, interleukin 4 (IL-4) gene has been comprehensively investigated [5, 6]. IL-4 plays a major function in isotype class switching of B cells to IgE production, type 2 immune responses, and it is involved in recruitment of mast cell [7, 8]. It has thus been proposed that IL-4 may have an imperative role in the development and persistent of asthma [7]. IL-4 gene is located on long arm of chromosome 5 (5q31), a region that has been associated with asthma or related disorders such as bronchial hyper responsiveness (BHR) and atopy [9]. The IL-4 C33T single nucleotide polymorphism (rs2070874) which is located on the untranslated region (UTR) has been represented to be linked with elevated serum IgE levels and risk of asthma [10,11,12]. There are several studies in which association between IL-4 C33T polymorphism and asthma risk have been evaluated [5, 6, 10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31]. Nevertheless, this association remains inconsistent and inconclusive in several studies. Probably, this could be because of the small samples size examined in these studies and the small effect size of the polymorphism that failed to provide sufficient statistical power to identify statically significant associations. Accordingly, we conducted a meta-analysis to conclude a more exact estimation of the relation between the IL-4 C33T polymorphism and risk of asthma.
Methods
We carried out this meta-analysis by following the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement [32]. Since our study did not contain any experimental procedure on humans and animals, thus no ethics committee confirmation was applicable.
Search strategy
A comprehensive systematic search was applied through three major databases (MEDLINE, web of science, and Scopus) to find all potential publications considering the association between IL-4 C33T polymorphism and asthma risk released before September 2020. We searched (“asthma” [Mesh] OR “asthmatic”) AND (“interleukin-4” OR “IL-4” OR “interleukin 4”) AND (“single nucleotide polymorphism” OR “SNP” OR “polymorphisms” OR “mutation” OR “variation”), as main key words. Besides, cross reference check of review studies were screened for additional relevant papers.
Inclusion and exclusion criteria
Initial search strategy yield 1873 studies that exported to Endnote X8. The duplicate studies which were common among databases were removed and title and abstract of remain studies were reviewed by two investigators. In cases that we could not categorize retrieved studies by title and abstract, full-text verification was performed. Eventually, studies considered eligible if met the following criteria: 1) publications that evaluate the association between IL-4 C33T polymorphism and the risk of asthma; 2) publications with extractable data to estimate odds ratios (ORs) and 95% confidence intervals; 3) publications that report genotype or allele distributions of case and controls. Duplicate articles, review articles, editorials, case reports, book chapters, republished data, comments, and studies with insufficient data after contacting with authors were all excluded. The usage of these criteria results in 24 eligible paper for the meta-analysis.
Data extraction and quality assessment
Two researchers independently and according to a standard checklist extracted requisite data including: the first author family, country of origin, ethnicity, number of subjects in the case and the control groups for each gender, mean or range of age, applied genotyping method, distribution of alleles in cases and controls, journal and year of publication. The methodological quality of included study was scored using the Newcastle-Ottawa Scale (NOS) criteria [33]. Accordingly, publications with scores 0–3, 4–6 or 7–9 were low, moderate or high-quality, respectively.
Statistical analysis
All data analysis was accomplished using SPSS (version 23; Chicago, IL, USA) and Stata (version 14; Stata Corporation, College Station, TX) softwares. The strength of association between SNP and the risk of asthma was measured via Odd ratios (ORs) and 95% confidence intervals (CIs). Moreover, the degree of heterogeneity between studies was assessed by the Q test (Q-statistic P value> 0.10, no heterogeneity vs. Q-statistic P value< 0.10, significant heterogeneity) and the I2 test (I2 < 25%, no heterogeneity; I2 = 25–50%, moderate heterogeneity; I2 = 50–75%, large heterogeneity, I2 > 75%, extreme heterogeneity) [34]. In the presence of heterogeneity random effect model (REM) should be used. Otherwise, fixed effect model (FEM) should be applied [35]. To estimate whether the results were substantially changed by any individual study, sensitivity analysis by sequential omitting of each included study was performed. Furthermore, publication bias was investigated, using Begg’s and Egger’s tests along with visual examination of the funnel plot (p value< 0.05 considered statistically significant) [36].
Results
Study characteristics
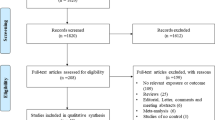
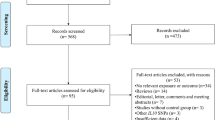
A total of 1873 articles were identified through the systematic literature search of databases. After excluding 219 duplicate studies and removing 1496 irrelevant publications based on titles/abstracts, 158 studies went under full-text screening. Of which, 134 paper were excluded. Finally, twenty-four studies qualified for quantitative analysis Fig. 1. All included studies were performed between 2000 to 2016 and had good methodological score ranging 5 to 8. Case-control design was common between eligible studies and different genotyping method were used by included studies. Tables 1 and 2 summarized the characteristics and allele distribution, genotype frequency of the eligible studies.
Meta-analysis of IL-4 C33T polymorphism and the risk of asthma
Twenty-four studies with 6587 cases and 8408 healthy controls were included in final meta-analysis of overall population. Of them, 15 publications were carried out in Asian countries, 5 publications were in American countries and 4 publications were in Europe. The pooled OR indicated that IL-4 C33T polymorphism increase risk of asthma across all genotype models including dominant model (OR = 1.15, 95% CI = 1.04–1.26, P = ≤0.001, FEM), recessive model (OR = 1.16, 95% CI = 1.06–1.28, P = ≤0.001, FEM), allelic model (OR = 1.14, 95% CI = 1.07–1.21, P = ≤0.001, FEM), CC vs. TT model (OR = 1.21, 95% CI = 1.02–1.43, P = 0.02, FEM) and CT vs. TT model (OR = 1.10, 95% CI = 1–1.22, P = 0.05, FEM) Fig.2. The detailed findings for different analysis models are shown in Table 3.
Subgroup analysis
We categorized studies into different subgroups on the basis of age, continent and ethnicity. The results of pooled ORs, heterogeneity tests and publication bias tests for different analysis models are reported in Table 3.
Subgroup analysis by age
In this group, we stratified included publications into three groups including: adult (8 articles), children (7 articles) and mixed (cover both ranges; 9 articles). Overall, the results rejected significant association between IL-4 C33T polymorphism and risk of asthma in different age group except for allelic model [adults (OR = 1.14, 95% CI = 1.02–1.26, P = 0.02, FEM), mixed (OR = 1.14, 95% CI = 1.01–1.29, P = 0.03, REM), children (OR = 1.13, 95% CI = 1.04–1.24, P = ≤0.001, FEM)] and recessive model (just in children (OR = 1.18, 95% CI = 1.03–1.35, P = 0.01, REM)) Fig. 3.
Subgroup analysis by continent
Our included studies performed in Asia (15 articles), Europe (4 articles), America (4 articles), and Oceania (1 article). Since there was only one study for Oceania, we exclude it. The final findings indicated strong significant association between IL-4 C33T polymorphism and asthma risk in European population across dominant model (OR = 1.23, 95% CI = 1.01–1.50, P = 0.03, FEM), recessive model (OR = 2.94, 95% CI = 1.54–5.62, P = ≤0.001, FEM), allelic model (OR = 1.30, 95% CI = 1.10–1.54, P = ≤0.001, FEM) and CC vs. TT (OR = 3, 95% CI = 1.56–5.76, P = ≤0.001, FEM). Moreover, there was a significant association between IL-4 C33T polymorphism and risk of asthma in American population under dominant model (OR = 1.26, 95% CI = 1.05–1.51, P = ≤0.001, FEM), allelic model (OR = 1.17, 95% CI = 1.03–1.33, P = 0.01, FEM), and CT vs. TT model (OR = 1.23, 95% CI = 1.02–1.49, P = 0.03, FEM). Eventually, Significant positive association was revealed in Asians just in recessive model (OR = 1.14, 95% CI = 1.02–1.26, P = 0.01, FEM), and allelic model (OR = 1.12, 95% CI = 1.03–1.21, P = ≤0.001, FEM) Fig. 3.
Subgroup analysis by ethnicity
Finally, we stratified eligible articles according ethnicity including Caucasians (20 articles), and African-Americans (4 articles). The results showed significant association between IL-4 SNP (C33T) and asthma risk in Caucasians under dominant model (OR = 1.15, 95% CI = 1.04–1.28, P = 0.008, FEM), recessive model (OR = 1.17, 95% CI = 1.06–1.30, P = 0.002, FEM), allelic model (OR = 1.14, 95% CI = 1.07–1.22, P = ≤0.001, FEM),and CC vs. TT model (OR = 1.23, 95% CI = 1.01–1.49, P = 0.03, FEM) but not CT vs. TT model (OR = 1.1, 95% CI = 0.98–1.22, P = 0.09, FEM). However, there was no significant association between IL-4 C33T polymorphism and risk of asthma in American-African population across all genotype models Fig. 3.
Evaluation of heterogeneity
No significant heterogeneity was detected for IL-4 C33T polymorphism neither in overall population nor subgroup analysis, therefore we did not perform mete-regression analysis for possible parameters (Table 3).
Sensitivity analysis and publication bias
Begg’s and Egger’s tests were performed to estimate the publication biases of studies. As showed in Table 3 no evidence of publication bias was detected in overall populations and subgroup analysis. Also, symmetric shape of Begg’s funnel plot confirm this finding Fig. 4. Moreover, the impact of individual study on pooled OR was evaluated by sensitivity analysis, which confirmed stability of our results Fig. 5.
Discussion
The cytokine IL-4 act as a key player in the development and pathogenesis of allergic inflammation [37] and atopy [38] through the induction of the heavy chain isotype switching, secretion of IgE antibody (IgE synthesis) by B cells, functioning as a growth factor for Th2 cells [37]. The IL-4 promotes IgE-dependent immune responses as it induces overexpression of IgE receptors on the surface of various immune cells: FcεRI on basophils and mast cells; and FcεRII (CD23) on mononuclear phagocytic cells and B lymphocytes [39]. The IL-4 tilts the immune response to anti-inflammatory, inhibiting macrophages pro-inflammatory effect and downregulating secretion of pro-inflammatory cytokines [40]. The IL-4 critically, initiate immediate allergic responses by triggering IgE-mediated mast cell activation [41]. The IL-4 plays a pivotal role in the priming of naïve T cell towards Th2 differentiation as well as exacerbate allergic inflammation through induction of vascular adhesion molecule 1 (VCAM-1) that recruit leukocytes and promote their survival [39]. The IL-4 induce airway remodeling encountered in asthma by its role in the proliferation of bronchial fibroblasts, myofibroblasts, and airway smooth muscles [38].
At the turn of the millennium genetic polymorphisms of the IL-4 gene in the development and maintenance of asthma have drawn increasing consideration. Modulation of the immune system is the common denominator in IL-4 polymorphisms [40]. Suzuki and coworkers found a single nucleotide polymorphism of C replacement of T at position 33 bp of exon 1 (C33T) of the IL-4 proximal promoter region [42]. Asthmatic patients with C33T have higher serum level of IL-4 and IgE [43]. Anovazzi and colleagues studied IL-4 haplotypes and reported that the studied haplotypes induce an opposing immune response, as well they recorded minimal functional activity in polymorphisms involving the promoter region [40]. An increasing body of evidence has demonstrated that C33T of the IL-4 gene untranslated region (UTR) of chromosome 5q was associated with elevated serum IgE levels and the risk of asthma [44, 45]. However, this association remains inconclusive. If this is indeed the case, a meta-analysis with big sample size, sufficient statistical power, and subgroup analysis was needed.
Our current meta-analysis composed of 24 publications (28 studies) involving 6587 cases and 8408 controls, we systematically assessed the relationship between IL-4 C33T polymorphism and asthma susceptibility. Cumulatively, the result illustrated IL-4 C33T polymorphism as a risk factor in the pathogenesis of asthma. The result indicated that the presence of T allele across different genetic models increased asthma risk by 10 to 21%. In the subgroup analysis by age, the results rejected the significant association between IL-4 C33T polymorphism and risk of asthma in different age groups except for allelic model, which highlighted the predisposing role of the T allele for the asthma risk in all three age groups. Subgroup analysis by continent revealed a significant association between IL-4 C33T polymorphism and asthma risk in the European population. In the Asian population, there was a significant association between IL-4 C33T polymorphism and the risk of asthma under recessive and allelic models. In contrast, the American population showed a significant association under dominant and allelic models.
Additionally, subgroup analysis was conducted according to ethnicity; the results showed a significant association between IL-4 C33T polymorphism and asthma risk in Caucasians under all models except CT vs TT model. While, no significant association between IL-4 C33T polymorphism and the risk of asthma in the American-African population were detected.
It should be noted that our results are not in agreement with those of Liu et al. [46] meta-analysis on the role of IL-4 C33T polymorphism and asthma. They suggested a significant association between whites and Asians. While they reported a significant association between the IL-4 C33T polymorphism and asthma risk in the overall population, they did not find a significant association among atopic and non-atopic asthma patients in subgroup analysis. Furthermore, in contrast to our meta-analysis, in the subgroup analysis by age, they reported an increased risk of asthma among children but not in the adult. Finally, while they reported evidence of publication bias, we identified no evidence of publication bias for the overall population and subgroup analysis under all genetic models.
The main reason for these discrepancies raised could be from the fact that Liu and colleagues included 18 studies with 5523 cases and 5618 controls. However, our meta-analysis encompasses 28 studies including 6587 cases and 8408 controls from different ethnicities and continents.
The C33T single nucleotide polymorphism is detected on the 5′ untranslated regions (UTR) of the IL-4 gene [42]. The 5′ UTRs region of mRNA may contain many gene regulatory elements (GRE) that regulate the localization, translation and degradation of transcripts [47]. In the eukaryotic mRNAs, the 5′ UTRs regulate both cap-dependent and cap-independent translation initiation of mRNA [48]. Researchers revealed a relationship between IL-4 C33T polymorphism and elevated serum IgE levels in a group of the Japanese population [49]. While the exact mechanisms by which the IL-4 C33T allele modulates the gene expression of the IL-4 remain elusive, it has been suggested that this variation may influence the stability of mRNA, as well as transcriptional or translational efficiency of the IL-4 gene, highlighting that the 5′ UTR may involve many cis-acting elements [47, 50,51,52].
Heterogeneity and publication bias, which may affect the results of meta-analyses, should always be considered. The result of this study did not show significant heterogeneity. Moreover, there was no significant publication bias in the overall population and subgroup analysis under all genetic models. Consequently, heterogeneity and publication bias did not appear to have inclined the results. Sensitivity analyses were also performed. There was a little variation of the estimates after exclusion of a single study and the significance of the pooled ORs was not affected proposing the consistency of this result.
The current study had some limitations. First, most included articles were from the Asia continent with Caucasian race and there was no study from Africans; accordingly, the results of this meta-analysis may not be appropriate to Africans. Second, in some studies, the diagnostic criteria and asthma phenotype were not clearly determined; while the asthma diagnostic criteria were primarily based on physical examination, clinical history, and pulmonary function tests (PFT), there did exist a little dissimilarity among studies. Third, the overall results were based on unadjusted estimates; a more precise evaluation should be accompanied when all singular raw data are accessible, which would facilitate the adjustment by other potential co-variants such as; age, gender, obesity, environmental factors, smoking status, and other lifestyles. Fourth, due to a lack of extractable data, we failed to address gene-environment and gene-gene interactions. In contrast to these limitations, two main strengths of our meta-analysis include; Firstly, a large number of patients and the healthy individuals were pooled from various studies, which considerably augment the statistical power of the meta-analysis. Secondly, no evidence of publication biases was identified, representing that the whole collected data may be unbiased.
Conclusion
Taken together, this study suggests that IL-4 C33T polymorphism potentially acts as a risk factor for asthma in different ethnicities and age groups. Nevertheless, large sample studies from different continents and races with homogeneous asthmatic patients and well-matched healthy subjects are still needed. Furthermore, gene-environment and gene-gene interactions should also be regarded in future studies. With taking these factors into account in future studies, it would ultimately lead to our comprehensive and better understanding of the association between the IL-4 C33T polymorphism and asthma susceptibility.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Abbreviations
- IL-4:
-
interleukin 4
- SNP:
-
Single nucleotide polymorphisms
- PRISMA:
-
Preferred Reporting Items for Systematic reviews and Meta-Analyses
- NOS:
-
Newcastle-Ottawa Scale
References
Subbarao P, Mandhane PJ, Sears MR. Asthma: epidemiology, etiology and risk factors. CMAJ. 2009;181(9):E181–90.
Ferreira MA, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49(12):1752.
Makoui MH, et al. Vitamin D receptor gene polymorphism and susceptibility to asthma: meta-analysis based on 17 case-control studies. Asthma & Immunology: Annals of Allergy; 2019.
Rogers AJ, et al. Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009;179(12):1084–90.
Beghe B, et al. Polymorphisms in the interleukin-4 and interleukin-4 receptor α chain genes confer susceptibility to asthma and atopy in a Caucasian population. Clin Exp Allergy. 2003;33(8):1111–7.
Isidoro-García M, et al. Interleukin-4 (IL4) and Interleukin-4 receptor (IL4RA) polymorphisms in asthma: a case control study. Clin Mol Allergy. 2005;3(1):15.
Jamal M, et al. Recent Progress on circular RNA research in acute myeloid leukemia. Front Oncol. 2019;9:1108.
Makoui MH, et al. Vitamin D receptor gene polymorphism and susceptibility to asthma: meta-analysis based on 17 case-control studies. Ann Allergy Asthma Immunol. 2020;124(1):57–69.
Noguchi E, et al. Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exper Allergy. 1998;28(4):449–53.
Suzuki I, et al. Association between a C+ 33T polymorphism in the IL-4 promoter region and total serum IgE levels. Clin Exp Allergy. 2000;30(12):1746–9.
Basehore MJ, et al. A comprehensive evaluation of IL4 variants in ethnically diverse populations: association of total serum IgE levels and asthma in white subjects. J Allergy Clin Immunol. 2004;114(1):80–7.
Jiang, P., et al. Several interleukin-4 and interleukin-13 gene single nucleotide polymorphisms among Chinese asthmatic patients. In Allergy and asthma proceedings. 2009. OceanSide Publications.
Park BL, et al. Interleukin 3 (IL3) polymorphisms associated with decreased risk of asthma and atopy. J Hum Genet. 2004;49(10):517.
Donfack J, et al. Variation in conserved non-coding sequences on chromosome 5q and susceptibility to asthma and atopy. Respir Res. 2005;6(1):145.
Battle NC, et al. Ethnicity-specific gene–gene interaction between IL-13 and IL-4Rα among African Americans with asthma. Am J Respir Crit Care Med. 2007;175(9):881–7.
Amirzargar A, et al. 2 polymorphisms in IL4 and IL4RA confer susceptibility to asthma. J Investig Allergol Clin Immunol. 2009;19(6):433.
Daley D, et al. Analyses of associations with asthma in four asthma population samples from Canada and Australia. Hum Genet. 2009;125(4):445–59.
Haller G, et al. Sequencing the IL4 locus in African Americans implicates rare noncoding variants in asthma susceptibility. J Allergy Clin Immunol. 2009;124(6):1204–1209. e9.
Wang J-Y, et al. An association study of 13 SNPs from seven candidate genes with pediatric asthma and a preliminary study for genetic testing by multiple variants in Taiwanese population. J Clin Immunol. 2009;29(2):205–9.
Berce V, Potoˇnik U. Association of Q551R polymorphism in the interleukin 4 receptor gene with nonatopic asthma in Slovenian children. Wien Klin Wochenschr. 2010;122(2):11–8.
Michel S, et al. Unifying candidate gene and GWAS approaches in asthma. PLoS One. 2010;5(11):e13894.
Undarmaa S, et al. Replication of genetic association studies in asthma and related phenotypes. J Hum Genet. 2010;55(6):342.
Wu X, et al. Association and gene-gene interactions of eight common single-nucleotide polymorphisms with pediatric asthma in middle China. J Asthma. 2010;47(3):238–44.
Xue-Xi Y, et al. Association of TGF-[beta] 1, IL-4 and IL-13 gene polymerphisms with asthma in a Chinese population. Asian Pac J Allergy Immunol. 2011;29(3):273.
Chen Y, Yang X, Huang Y, Liu E, Wang L. Association of the interactions between IL-4 and Mina gene with children asthma. Immun J. 2011:416–9.
Huang H-R, Zhong Y-Q, Wu J-F. The association between IFN-γ and IL-4 genetic polymorphisms and childhood susceptibility to bronchial asthma. Gene. 2012;494(1):96–101.
Davoodi P, et al. A preliminary study on the association of single nucleotide polymorphisms of interleukin 4 (IL4), IL13, IL4 receptor alpha (IL4Rα) & toll-like receptor 4 (TLR4) genes with asthma in Indian adults. Indian J Med Res. 2015;142(6):675–80.
Micheal S, et al. IL4 gene polymorphisms and their association with atopic asthma and allergic rhinitis in Pakistani patients; 2013.
Miyake Y, Tanaka K, Arakawa M. Relationship between polymorphisms in IL4 and asthma in Japanese women: the Kyushu Okinawa maternal and child health study. J Investig Allergol Clin Immunol. 2013;23(4):242–7.
Wang RS, Jin HX, Shang SQ, Liu XY, Chen SJ, Jin ZB. Relacion entre la expresión de IL-2 e IL-4 y sus polimorfismos y losriesgos de padecer infección por mycoplasma pneumoniae y asmaen ninos. Arch Bronconeumol. 2015:619–26.
Li L, et al. Role of interleukin-4 genetic polymorphisms and environmental factors in the risk of asthma in children. Genet Mol Res. 2016;15(4):534–43.
Moher D, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–9.
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5.
Higgins JP, et al. Measuring inconsistency in meta-analyses. Bmj. 2003;327(7414):557–60.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48.
Egger M, et al. Bias in meta-analysis detected by a simple, graphical test. Bmj. 1997;315(7109):629–34.
Gour N, Wills-Karp M. IL-4 and IL-13 signaling in allergic airway disease. Cytokine. 2015;75(1):68–78.
Bagnasco D, et al. A critical evaluation of anti-IL-13 and anti-IL-4 strategies in severe asthma. Int Arch Allergy Immunol. 2016;170(2):122–31.
Borish, L.C, et al. Efficacy of soluble IL-4 receptor for the treatment of adults with asthma. J Allergy Clin Immunol. 2001;107(6):963–970.
Anovazzi G, et al. Functionality and opposite roles of two interleukin 4 haplotypes in immune cells. Genes Immun. 2017. 18(1):33–41.
Gould HJ, Sutton BJ. IgE in allergy and asthma today. Nat Rev Immunol. 2008;8(3):205–17.
Suzuki I, et al. A new polymorphism in the 5’flanking region of the human interleukin (IL)-4 gene. Immunogenetics. 1999;49(7):738–739.
Gervaziev YV, Kaznacheev VA, Gervazieva VB. Allelic polymorphisms in the interleukin-4 promoter regions and their association with bronchial asthma among the Russian population. Int Arch Allergy Immunol. 2006;141(3):257–64.
Graves PE, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105(3):506–13.
Suzuki, I., et al., Association between a C+33T polymorphism in the IL-4 promoter region and total serum IgE levels. Clinical <html_ent glyph="@amp;" ascii="&"/> Experimental Allergy, 2000. 30(12): p. 1746–1749.
Liu S, Li T, Liu J. Interleukin-4 rs2243250 polymorphism is associated with asthma among Caucasians and related to atopic asthma. Cytokine. 2012;59(2):364–9.
Hinnebusch, A.G., I.P. Ivanov, and N.J.S. Sonenberg, Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352(6292):1413–1416.
Leppek, K., R. Das, and M.J.N.r.M.c.B. barna, Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. 2018. 19(3): p. 158.
Yang H-J. Association between the Interleukin-4 gene C-589T and C+33T polymorphisms and asthma risk: a meta-analysis. Arch Med Res. 2013;44(2):127–35.
Mandola MV, et al. A 6 bp polymorphism in the thymidylate synthase gene causes message instability and is associated with decreased intratumoral TS mRNA levels. Pharmacogenetics. 2004;14(5):319–27.
Kim M, Kogan N, Slack FJ. Cis-acting elements in its 3′ UTR mediate post-transcriptional regulation of KRAS. Oncotarget. 2016;7(11):11770–84.
Wang R-S, et al. Associations of IL-2 and IL-4 expression and polymorphisms with the risks of mycoplasma pneumoniae infection and asthma in children. Archivos de Bronconeumología (English Edition). 2015;51(11):571–8.
Acknowledgements
None.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
RR generated the idea. DI and MME analyzed and interpreted the data. GAS and BR prepared the original draft. MA, DI, and GAS critically revised the paper. RR supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Imani, D., Eslami, M.M., Anani-Sarab, G. et al. Interleukin-4 gene polymorphism (C33T) and the risk of the asthma: a meta-analysis based on 24 publications. BMC Med Genet 21, 232 (2020). https://doi.org/10.1186/s12881-020-01169-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-020-01169-w