Abstract
Background
Hearing loss is the most common sensory defect, and it affects over 6% of the population worldwide. Approximately 50–60% of hearing loss patients are attributed to genetic causes. Currently, more than 100 genes have been reported to cause non-syndromic hearing loss. It is possible and efficient to screen all potential disease-causing genes for hereditary hearing loss by whole exome sequencing (WES).
Methods
We collected 5 consanguineous pedigrees from Pakistan with hearing loss and applied WES in selected patients for each pedigree, followed by bioinformatics analysis and Sanger validation to identify the causal genes.
Results
Variants in 7 genes were identified and validated in these pedigrees. We identified single candidate variant for 3 pedigrees: GIPC3 (c.937 T > C), LOXHD1 (c.6136G > A) and TMPRSS3 (c.941 T > C). The remaining 2 pedigrees each contained two candidate variants: TECTA (c.4045G > A) and MYO15A (c.3310G > T and c.9913G > C) for one pedigree and DFNB59 (c.494G > A) and TRIOBP (c.1952C > T) for the other pedigree. The candidate variants were validated in all available samples by Sanger sequencing.
Conclusion
The candidate variants in hearing-loss genes were validated to be co-segregated in the pedigrees, and they may indicate the aetiologies of hearing loss in such patients. We also suggest that WES may be a suitable strategy for hearing-loss gene screening in clinical detection.
Similar content being viewed by others
Background
Hearing loss is the most common sensory defect, and it affects ~ 1/500 newborns [1] and 466 million people worldwide (https://www.who.int/pbd/deafness/estimates/en/). Approximately 50% ~ 60% of hearing loss patients are attributed to genetic causes [1, 2]. Hereditary hearing loss is a genetically heterogeneous disorder [3] that can be divided into syndromic hearing loss and non-syndromic hearing loss, among which non-syndromic hearing loss is the predominant type, with a proportion of ~ 80% [4]. Currently, more than 100 genes have been reported to cause non-syndromic hearing loss (https://hereditaryhearingloss.org/), and the total number of genes related to hearing loss is expected to be several hundred.
There are mature gene panels for hearing-loss detection, and the genes involved range from 4 to more than 100. However, except for several genes, such as GJB2 [5,6,7] or SLC26A4 [8,9,10], most causal genes contribute a small fraction to the disorder. Therefore, in clinical detection, we may not obtain a satisfactory result by gene panel screening for many cases. As whole exome sequencing (WES) technology has rapidly developed and its cost has become less expensive, it is possible and efficient to screen all potential disease-causing genes for hereditary hearing loss by WES [11, 12].
Recessive inheritance hearing loss is worth studying because such patients usually have unaffected parents, which makes the disorder seem to be “sudden onset”, and this situation is more difficult to prevent. Consanguineous pedigrees represent a suitable natural model to study recessive disorders [13]. In Pakistan, there are numerous consanguineous pedigrees because of their customs, and these pedigrees may provide more opportunities to study and recognize such disorders [14, 15].
In this study, we collected 5 consanguineous pedigrees with hearing loss from Pakistan and applied WES to identify the causal genes. We identified several variants in hearing-loss genes that co-segregated in the pedigrees, and they may indicate the aetiologies of hearing loss in such patients.
Methods
Participants and clinical diagnosis
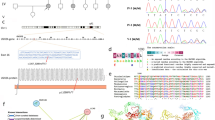
In the present study, we collected 5 consanguineous pedigrees containing 22 patients with hearing loss from rural areas in Pakistan. All the patients showed different degrees of hearing loss. The most likely inheritance mode for these pedigrees was autosomal recessive (Fig. 1). The study was approved by the ethical committee of the National Institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan, and all participants provided written informed consent (written informed consent of participants under the age of 16 were obtained from their parents or legal guardians).
Pedigree figures of the five consanguineous families with genotypes of available individuals. The squares and the circles represent males and females, respectively. The black-filled symbols indicate patients with hearing loss, and a symbol with a diagonal line indicates a deceased family member. The symbols with numbers indicate the availability of the individual’s DNA. The candidate variants are listed under each pedigree, and the genotypes of the individuals for the variants are marked. “+/+” indicates homozygous variant, “+/−” indicates heterozygous variant, and “−/−” indicates reference
DNA extraction and whole exome sequencing
According to the manufacturer’s instructions, genomic DNA was isolated from the peripheral blood leukocytes of all participants using a DNA QIAamp mini kit (Qiagen, Hilden, Germany). One patient from each pedigree was selected, and WES was performed. Exons were captured using the BGI-Exome kit V4 and sequenced by BGI-seq 500 with 100 bp paired-end sequencing.
Bioinformatics analysis
Low-quality reads were removed by SOAPnuke [16], and then the reads were mapped to the human genome reference (UCSCGRCh37/hg19) by the Burrows–Wheeler Aligner (BWA-MEM, version 0.7.10) [17]. Variants were called using the Genome Analysis Tool Kit (GATK, version 3.3) [18]. Variant Effect Predictor (VEP) [19] was used to annotate and classify all the variants. After that, all the variants were filtered based on their frequency in public databases (e.g., 1000 Genomes Project, Exome Sequencing Project and ExAC) and our in-house databases, and the variants with MAF < 0.005 were retained. Then, homozygous variants and compound heterozygous variants were selected because the most likely inheritance mode for these pedigrees was autosomal recessive. Finally, we applied several variant prediction tools including SIFT [20] (http://provean.jcvi.org/), PolyPhen2 [21] (http://genetics.bwh.harvard.edu/pph2/), MutationTaster [22] (http://www.mutationtaster.org/) and CADD [23] (https://cadd.gs.washington.edu/snv), to predict the functional impact of candidate variants [24].
Sanger validation
DNA from all available samples in the five pedigrees was Sanger sequenced to validate the variants and confirm their co-segregation in the pedigree. Forward and reverse primers were designed by Primer3. After PCR amplification, the purified product was sequenced on ABI 3730XL DNA Analyzer.
Results
Clinical features
All the patients showed different degrees of hearing loss. In the first pedigree (HL1), all patients showed severe deafness, and one patient (III1) was selected for WES. In the second pedigree (HL2), all patients showed congenital profound deafness and muteness, and one patient (IV1) was selected for WES. In the third pedigree (HL3), all patients showed moderate deafness (their hearing loss started after seizures), and one patient (V1) was selected for WES. In the fourth pedigree (HL4), all patients showed congenital profound deafness, and one patient (IV1) was selected for WES. In the fifth pedigree (HL5), all patients showed moderate deafness, and one patient (III5) was selected for WES.
Genetic analysis
WES was applied in the selected patients. The average depth of the target region was 146X with a coverage of 99.85%, and the coverage of the target region that was sequenced at least 10 times (depth > = 10 X) was 98.20% (Table 1). For each individual, more than ten thousand variants that may influence protein were identified. After frequency filtration (MAF < 0.005), approximately 15 ~ 32 exon variants were retained. Further inheritance model filtration retained 1 ~ 6 candidate variants (Table 1). All the rare variants detected in the exon region for the pedigrees are listed in the supplementary table 1. The original WES sequencing data of the samples were deposited in the CNSA (see “Availability of data and materials” section) and the samples information were listed in supplementary table 2.
We identified a stop codon lost homozygous variant, GIPC3: c.937 T > C, from the patient in the HL1 pedigree, and the variant prediction tools provided a benign prediction. For the patient from the HL2 pedigree, we identified 4 variants in 2 genes. However, one gene was reported to cause autosomal dominant hearing loss, therefore, we first analyzed the other gene. Then, a homozygous variant, LOXHD1: c.6136G > A, was regarded as a candidate variant for this pedigree. The variant prediction tools indicated a damaging prediction. For the patient from the HL3 pedigree, 6 variants in 5 genes were identified at first, and further analysis indicated that only 2 genes may cause autosomal recessive deafness. Therefore, the homozygous variant, TECTA: c.4045G > A, and two compound heterozygous variants, c.3310G > T and c.9913G > C in MYO15A, were regarded as candidate variants. The variant prediction tools indicated benign prediction for both MYO15A variants and damaging prediction for the TECTA variant. For the patient from the HL4 pedigree, 3 variants in 3 genes were identified, and two of them may cause autosomal recessive deafness. The two homozygous variants were DFNB59: c.494G > A and TRIOBP: c.1952C > T. The variant prediction tools indicated damaging prediction for the first variant and benign prediction for the last variant. For the patient from the HL5 pedigree, only one homozygous candidate variant was identified, TMPRSS3: c.941 T > C. The variant prediction tools indicated a damaging prediction. None of these variants were previously reported to cause hearing loss. The detailed information is listed in Table 2.
In summary, we identified one most likely causing variant for the HL1, HL2 and HL5 pedigrees and two most likely causing candidate variants for the HL3 and HL4 pedigrees.
Sanger validation
To validate co-segregation in the pedigree, we applied Sanger sequencing to all available samples. In total, 6 samples (II1 ~ 2 and III1 ~ 4) were sequenced for the HL1 pedigree, 6 samples (III1 ~ 2 and IV1 ~ 4) were sequenced for the HL2 pedigree, 4 samples (IV1 and V1 ~ 3) were sequenced for the HL3 pedigree, and 21 samples (I1, II1 ~ 11 and III1 ~ 9) were sequenced for the HL5 pedigree. For the HL4 pedigree, the initial samples collected were degraded, and we failed to collect additional samples. All the variants selected for Sanger sequencing were co-segregated in the pedigrees except for the HL4 pedigree (Fig. 2).
Discussion
In this study, we identified several variants in genes reported to cause hearing loss that co-segregated in the pedigrees. For the HL1 pedigree, the variant in the GIPC3 gene may cause the disorder. A protein with 312 amino acid residues is encoded by GIPC3, which contains a PDZ domain and three low-complexity regions [25]. The PDZ domain is responsible for the survival of hair cells and spiral ganglion in the ears. GIPC3 was the causal gene of autosomal recessive deafness (type 15), non-syndromic genetic deafness and audiogenic seizures. Currently, 11 pathogenic variants in this gene have been identified in ClinVar.
For the HL2 pedigree, the variant in the LOXHD1 gene was likely the causative variant. A highly conserved stereociliary protein is encoded by LOXHD1, which contains 15 PLAT domains that is responsible for protein interactions with the plasma membrane [26]. It was reported that Loxhd1 maintained the cochlear hair cells’ function in mice [27]. LOXHD1 was the causal gene of autosomal recessive deafness (type 77). Currently, 28 pathogenic variants in this gene have been identified in ClinVar.
For the HL3 pedigree, a homozygous variant in TECTA and two compound heterozygous variants in MYO15A were the likely candidate variants. A protein with 2155 amino acid residues is encoded by TECTA, which is a non-cellular matrix overlying the cochlear neuroepithelium. The function of this protein is to amplify and transmit sound [28, 29]. TECTA was the causal gene of autosomal recessive deafness (type 21), and 40 pathogenic variants in this gene have been identified in ClinVar. MYO15A encodes a protein that plays a crucial role in hair cells of the inner ear to maintain normal hearing [30]. MYO15A was the causal gene of autosomal recessive deafness (type 3), and 112 pathogenic variants in this gene have been identified in ClinVar. Take the inheritance mode of this pedigree (autosomal recessive) into consideration, we preferred that the homozygous variant in TECTA was more likely to be responsible than the compound heterozygous variants in MYO15A.
For the HL4 pedigree, homozygous variants were detected in both DFNB59 and TRIOBP. A protein with 352 amino acids is encoded by DFNB59, and is responsible for the signal transmit of auditory nerve [31]. DFNB59 was the causal gene of autosomal recessive deafness (type 59), and 9 pathogenic variants in this gene have been identified in ClinVar. A protein with 652 amino acids is encoded by TRIOBP, which regulates adherens junctions and recognizes actin cytoskeleton [32]. Currently, the function of TRIOBP remains unclear, and no pathologies except hearing loss were caused by pathogenic variants in this gene. TRIOBP was the causal gene of autosomal recessive deafness (type 28), and 26 pathogenic variants in this gene have been identified in ClinVar. It was predicted to be likely benign in the deafness variation database for the variant in TRIOBP. Therefore, we thought the variant in DFNB59 was more likely to be the causal variant than the TRIOBP variant.
For the HL5 pedigree, the variant in TMPRSS3 may cause the disorder. The protein encoded by this gene plays a crucial role in activating the ENaC sodium channel [33], and it regulates the Na+ concentration in the inner ear [34]. TMPRSS3 was the causal gene of autosomal recessive deafness (type 8), and 23 pathogenic variants in this gene have been identified in ClinVar.
We calculated the density of reported pathogenic variants in these genes, which were 11.7/kb, 4.2/kb, 6.2/kb, 10.6/kb, 8.5/kb, 3.7/kb and 16.8/kb for GIPC3, LOXHD1, TECTA, MYO15A, DFNB59, TRIOBP and TMPRSS3, respectively. The density may indicate the degree of understanding or focus for different genes. Genes with low density, such as LOXHD1 and TRIOBP, may have potential research value.
The majority of causal genes we identified for these pedigrees were not common hearing-loss genes. If we applied a common hearing-loss gene panel to screen these patients, we would obtain negative results, and the causal gene/variant for the patients would be missed. Therefore, WES may be a better strategy than panel sequencing for hearing-loss screening even in clinical detection.
Conclusion
In conclusion, we applied WES in five consanguineous pedigrees (one patient per pedigree) from Pakistan with hearing loss, followed by Sanger sequencing for all available samples among the pedigrees to identify the causal genes for them. Several variants in hearing-loss genes were validated to be co-segregated in the pedigrees, and they may indicate the aetiologies of hearing loss in such patients. Moreover, we suggest that WES may be a suitable strategy for hearing-loss screening in clinical detection.
Availability of data and materials
The data that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb. The download link is http://ftp.cngb.org/pub/CNSA/data2/CNP0000508/, and the samples information can be found in supplementary table 2.
The download links of public data used in this study are listed below.
The human genome reference (hg19):ftp://hgdownload.soe.ucsc.edu/goldenPath/hg19/chromosomes/
1000 Genomes Project:ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/
Exome Sequencing Project: http://evs.gs.washington.edu/evs_bulk_data/ESP6500SI-V2-SSA137.GRCh38-liftover.snps_indels.vcf.tar.gz
Abbreviations
- ABI:
-
Applied Biosystems
- BGI:
-
The Beijing Genomics Institute
- BWA:
-
Burrows–Wheeler Aligner
- CADD:
-
Combined Annotation Dependent Depletion
- DFNB59:
-
Autosomal Recessive Deafness Type 59
- DNA:
-
DeoxyriboNucleic Acid
- ENaC:
-
Epithelial sodium channel
- ExAC:
-
The Exome Aggregation Consortium
- GATK:
-
Genome Analysis Tool Kit
- GIPC3:
-
GIPC PDZ Domain Containing Family Member 3
- GRCh37:
-
The Genome Reference Consortium Human Genome Build 37
- LOXHD1:
-
Lipoxygenase Homology Domains 1
- MAF:
-
Minor allele frequency
- MYO15A:
-
Myosin XVA
- NIBGE:
-
National institute for Biotechnology and Genetic Engineering
- PCR:
-
Polymerase chain reaction
- PDZ:
-
PSD95/Dlg/ZO-1
- PLAT:
-
Plasminogen activator
- SIFT:
-
Sorting tolerant from intolerant
- SOAP:
-
Short Oligonucleotide Analysis Package
- TECTA:
-
Tectorin Alpha
- TMPRSS3:
-
Transmembrane Serine Protease 3
- TRIOBP:
-
TRIO And F-Actin Binding Protein
- UCSC:
-
University of California, Santa Cruz
- VEP:
-
Variant Effect Predictor
- WES:
-
Whole exome sequencing
References
Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354(20):2151–64.
Hu S, Sun F, Zhang J, Tang Y, Qiu J, Wang Z, et al. Genetic etiology study of ten Chinese families with Nonsyndromic hearing loss. Neural plasticity. 2018;2018:4920980.
Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010;68(2):293–308.
Morton NE. Genetic epidemiology of hearing impairment. Ann N Y Acad Sci. 1991;630:16–31.
Bakhchane A, Bousfiha A, Charoute H, Salime S, Detsouli M, Snoussi K, et al. Update of the spectrum of GJB2 gene mutations in 152 Moroccan families with autosomal recessive nonsyndromic hearing loss. European J Med Genetics. 2016;59(6–7):325–9.
Tlili A, Al Mutery A, Eddine K, Ahmad Mohamed W, Mahfood M, Hadj Kacem H. Prevalence of GJB2 mutations in affected individuals from United Arab Emirates with autosomal recessive Nonsyndromic hearing loss. Genetic Testing Mol Biomarkers. 2017;21(11):686–91.
Dalamon V, Lotersztein V, Beheran A, Lipovsek M, Diamante F, Pallares N, et al. GJB2 and GJB6 genes: molecular study and identification of novel GJB2 mutations in the hearing-impaired Argentinean population. Audiol Neuro-otology. 2010;15(3):194–202.
Park HJ, Shaukat S, Liu XZ, Hahn SH, Naz S, Ghosh M, et al. Origins and frequencies of SLC26A4 (PDS) mutations in east and south Asians: global implications for the epidemiology of deafness. J Med Genet. 2003;40(4):242–8.
Tsukada K, Nishio SY, Hattori M, Usami S. Ethnic-specific spectrum of GJB2 and SLC26A4 mutations: their origin and a literature review. Ann Otology Rhinology Laryngology. 2015;124(Suppl 1):61S–76S.
Albert S, Blons H, Jonard L, Feldmann D, Chauvin P, Loundon N, et al. SLC26A4 gene is frequently involved in nonsyndromic hearing impairment with enlarged vestibular aqueduct in Caucasian populations. European J Human Genetics. 2006;14(6):773–9.
Zazo Seco C, Wesdorp M, Feenstra I, Pfundt R, Hehir-Kwa JY, Lelieveld SH, et al. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in the Netherlands. European J Human Genetics. 2017;25(3):308–14.
Bademci G, Foster J 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genetics Med. 2016;18(4):364–71.
Bittles A. Consanguinity and its relevance to clinical genetics. Clin Genet. 2001;60(2):89–98.
Hamamy H, Antonarakis SE, Cavalli-Sforza LL, Temtamy S, Romeo G, Kate LP, et al. Consanguineous marriages, pearls and perils: Geneva international consanguinity workshop report. Genetics Med. 2011;13(9):841–7.
Li L, Chen Y, Jiao X, Jin C, Jiang D, Tanwar M, et al. Homozygosity mapping and genetic analysis of autosomal recessive retinal dystrophies in 144 consanguineous Pakistani families. Invest Ophthalmol Vis Sci. 2017;58(4):2218–38.
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y, Li S, et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7(1):1–6.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25(14):1754–60.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016;17(1):122.
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–2.
Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5.
Li MX, Kwan JS, Bao SY, Yang W, Ho SL, Song YQ, et al. Predicting mendelian disease-causing non-synonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 2013;9(1):e1003143.
Reed BC, Cefalu C, Bellaire BH, Cardelli JA, Louis T, Salamon J, et al. GLUT1CBP(TIP2/GIPC1) interactions with GLUT1 and myosin VI: evidence supporting an adapter function for GLUT1CBP. Mol Biol Cell. 2005;16(9):4183–201.
Bateman A, Sandford R. The PLAT domain: a new piece in the PKD1 puzzle. Current biology : CB. 1999;9(16):R588–90.
Grillet N, Schwander M, Hildebrand MS, Sczaniecka A, Kolatkar A, Velasco J, et al. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet. 2009;85(3):328–37.
Verhoeven K, Van Laer L, Kirschhofer K, Legan PK, Hughes DC, Schatteman I, et al. Mutations in the human alpha-tectorin gene cause autosomal dominant non-syndromic hearing impairment. Nat Genet. 1998;19(1):60–2.
Balciuniene J, Dahl N, Jalonen P, Verhoeven K, Van Camp G, Borg E, et al. Alpha-tectorin involvement in hearing disabilities: one gene--two phenotypes. Hum Genet. 1999;105(3):211–6.
Redowicz MJ. Myosins and deafness. J Muscle Res Cell Motil. 1999;20(3):241–8.
Delmaghani S, del Castillo FJ, Michel V, Leibovici M, Aghaie A, Ron U, et al. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat Genet. 2006;38(7):770–8.
Shahin H, Walsh T, Sobe T, Abu Sa'ed J, Abu Rayan A, Lynch ED, et al. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am J Hum Genet. 2006;78(1):144–52.
Vallet V, Chraibi A, Gaeggeler HP, Horisberger JD, Rossier BC. An epithelial serine protease activates the amiloride-sensitive sodium channel. Nature. 1997;389(6651):607–10.
Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, et al. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet. 2002;11(23):2829–36.
Acknowledgments
The authors thanked all the participating patients and the families in this study for their cooperation.
Funding
No funding was obtained for this study.
Author information
Authors and Affiliations
Contributions
YZ analyzed and interpreted the WES data and write the manuscript. MT analyzed and interpreted the patient data and modified the manuscript. SH analyzed and interpreted the WES data. UA performed Sanger sequencing. JZ designed the project and guided the analysis of WES data. SB designed the project, guided the analysis of patient data and helped the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the ethical committee of National institute for Biotechnology and Genetic Engineering (NIBGE), Faisalabad, Pakistan and all participants provided written informed consent (written informed consent of participants under the age of 16 were obtained from their parents or legal guardians).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhou, Y., Tariq, M., He, S. et al. Whole exome sequencing identified mutations causing hearing loss in five consanguineous Pakistani families. BMC Med Genet 21, 151 (2020). https://doi.org/10.1186/s12881-020-01087-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-020-01087-x