Abstract
Background
Ischemic stroke (IS) is a serious cardiovascular disease and is associated with several single nucleotide polymorphisms (SNPs). However, the role of Cytochrome P450 family 4 subfamily F member 2 (CYP4F2) gene in IS remains unknown. Our study aimed to explore whether CYP4F2 polymorphisms influenced IS risk in the Han Chinese population.
Methods
We selected 477 patients and 495 controls to do a case-control study, and five SNPs in CYP4F2 gene were successfully genotyped. And we evaluated the associations using the Chi-squared test, independent sample t test, and genetic models analyses. Logistic regression analysis was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs).
Results
In this study, rs12459936 and rs3093144 were associated with IS risk in the overall. After stratified analysis by age (> 61 years), rs3093193 and rs3093144 were related to an increased risk of IS, whereas rs12459936 was related to a decreased risk of IS. In addition, we found that three SNPs (rs3093193, rs3093144 and rs12459936) were associated with the susceptibility to IS in males. We also found five SNPs in the CYP4F2 gene had strong linkage.
Conclusions
Three SNPs (rs3093193, rs3093144 and rs12459936) in the CYP4F2 were associated with IS risk in a Chinese Han population. And, CYP4F2 gene may be involved in the development of IS.
Similar content being viewed by others
Background
Stroke is the leading neurological cause of death worldwide [1]. It is also called brain attack, typically characterized by neurological dysfunction due to acute focal injury of the central nervous system [2]. At a given age, the incidence and mortality rates of stroke are higher in men, however, women are more likely to have stroke because of longer life, and stroke events increase rapidly with aging [3]. Moreover, stroke is already becoming a global issue and it is threatening the health of human beings. Additionally, epidemiologic studies reported that the incidence of IS in China is significantly higher than that in the developed countries [4].
Hence, it is urgent to clarify the etiology of stroke. The majority of stroke are ischemic (approximately 80%), whereas 20% is due to primary hemorrhage [5]. Several studies demonstrated that some risk factors are involved in the development of ischemic stroke (IS), such as a family history of cardiovascular disease, older age, sex [6, 7], hypertension [8], diabetes [9]. However, a large proportion of IS remains unclear. Recently, the role of genetic factors in development of IS was determined in large-scale, collaborative, genome-wide association studies (GWAS) [10]. Some susceptibility genes of IS have been found, including PITX2 [11], ABO [12], and HABP2 [13]. It demonstrated that genetic factors may regulate the pathophysiological process of IS.
Cytochrome P450 (CYP450) plays a vital role in the metabolism of exogenous and endogenous compounds and is widely distributed in human [14]. Cytochrome P450 family 4 subfamily F member 2 (CYP4F2) as a member of CYP450 superfamily, is located on human chromosome 19p13.11 [15]. Previous studies have demonstrated CYP4F2 gene polymorphisms are associated with the risk of IS. For example, Nakamura et al. [16] conducted a case-control study based on the Japanese population to identify whether single nucleotide polymorphisms (SNPs) in CYP4F2 gene associated with the susceptibility to IS, they found ‘G’ allele of rs2108622 was correlated with increased risk of IS in male patients. A study in Sweden [17] reported the V433 M mutation in the CYP4F2 was correlated with cerebral infarction in male patients. It provided strong evidence on the association of CYP4F2 gene and IS. In the present study, we selected five candidate SNPs (rs3093203, rs3093193, rs12459936, rs3093144, and rs3093110) in CYP4F2 and performed a case-control study in Han Chinese population.
Methods
Subjects
The 477 cases were recruited from the First and Second Affiliated Hospital of Xi’an Jiaotong University between January 2016 and October 2018. In our study, all cases were consistent with the World Health Organization’s diagnostic criteria for IS. IS was confirmed by at least two independent neurologists using computed tomography (CT) scans and/or magnetic resonance imaging (MRI). IS patients with history of transient ischemic attack, coronary artery disease, autoimmune disease, systemic inflammatory disease, malignant tumor and other chronic diseases were excluded from the study. Healthy individuals (495 controls) were identified through the annual health assessment and were randomly recruited from the same hospital physical examination center between April 2016 and October 2018. All members were Han Chinese population living in the Shaanxi Province of China and were unrelated in at least three generations.
SNP selection and genotyping
In total, we successfully select five variants (rs3093203, rs3093193, rs12459936, rs3093144, and rs3093110) in the global population of the 1000 genome (http://www.internationalgenome.org/) with minor allele frequencies (MAFs) > 5%. Blood samples were collected in the test tubes containing ethylenediamine tetraacetic acid (EDTA) for anticoagulation and stored at − 80 °C after centrifugation at 2000 rpm for 10 min. Genomic DNA was extracted from peripheral blood samples of the 477 cases and the 495 controls using genomic DNA purification kit (GoldMag, Xi’an, China). And we used NanoDrop 2000 (Thermo Scientific, Waltham, MA) to measure the DNA concentration and purity. Agena MassARRAY Assay Design 3.0 Software [18] was used to design the primers for amplification and extension reactions. The information of primers was shown in Table 1. We used Agena MassARRAY RS1000 to perform the SNP genotyping according to the manufacturer’s instruction and used Agena Typer 4.0 software to manage and analyze the data [18, 19].
Statistical analysis
Excel (Microsoft Corp., Redmond, WA, United States) and SPSS statistical software (version 17.0, SPSS, Chicago, IL) were used for statistical analyses. All statistical tests were two-sided, and p < 0.05 was considered to be statistical significance. SNP genotype frequencies among cases and controls were calculated by Chi- squared test, and the Hardy–Weinberg equilibrium (HWE) was used to compare the actual and expected frequencies of the genotypes in the controls. In order to explore the effects of the CYP4F2 variants on the risk of IS based on the various genetic models (co- dominant, dominant, recessive, and log- additive), logistic regression analysis was used to calculate the odds ratios (ORs) and 95% CIs on PLINK software (version 1.07) [20]. Finally, the Haploview software package (version 4.2) and SNPStats (https://www.snpstats.net/start.htm?q=snpstats/start.htm) were used to estimate pairwise linkage disequilibrium (LD), haplotype construction, and genetic association at polymorphism loci [21, 22]. HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) was used to predict the potential functions of variants in CYP4F2 gene.
Results
Study participants
The basic characteristics of all study participants were shown in Table 2. Four hundred seventy-seven cases (316 males and 161 females) and 495 healthy controls (326 males and 169 females) were recruited in this case-control study. The mean ages of the patients and the controls were 60.05 ± 6.56 and 64.13 ± 10.82 years old, respectively. There was no statistically significant difference (p = 0.898) on the gender distribution, while the age distribution was statistically significant difference (p = 0.000). As expected, the prevalence of the most common risk factor for IS were significantly different between cases and controls, such as HDL cholesterol and LDL cholesterol (p < 0.001).
The association between CYP4F2 gene and the risk of IS
Table 3 summarized the basic information of the SNPs including gene, SNP ID, chromosomal position, minor allele frequency (MAF) of cases and controls and HWE test results. Five SNPs in the CYP4F2 gene were successfully genotyped for further analysis. The genotype distribution of all SNPs conformed to HWE in control subjects (p > 0.05). The differences in allele frequency distributions between patients and controls were identified by two-sided Pearson chi-squared tests. The rs12459936 was associated with decreased risk of IS (OR = 0.82, 95% CI = 0.68–0.98, p = 0.028). And the rs3093144 was associated with 1.27-fold (95% CI = 1.01–1.60, p = 0.043) increase in the risk of IS. Further, multiple inheritance models were applied for analyzing the association between CYP4F2 SNPs and IS risk by unconditional logistic regression analysis adjusted for age and gender (Table 4).
Stratification analysis by age and gender
Next, we conducted stratified analysis by age. The results showed that three SNPs were associated with the risk of IS in patients (age > 61 years old), as displayed in Table 5. Rs3093193 in CYP4F2 gene increased the susceptibility of IS in allele model (G vs. C, OR = 1.34, 95% CI = 1.02–1.77, p = 0.038), dominant model (GC/GG vs. CC, OR = 1.61, 95% CI = 1.07–2.42, p = 0.022) and log-additive model (OR = 1.44, 95% CI = 1.05–1.98, p = 0.024). Similarly, rs3093144 was observed to be associated with the higher risk of IS in allele model (T vs. C, OR = 1.53, 95% CI = 1.09–2.14, p = 0.013), dominant model (TC/TT vs. CC, OR = 1.62, 95% CI = 1.05–2.49, p = 0.029) and log-additive model (OR = 1.56, 95% CI = 1.07–2.27, p = 0.020). Conversely, rs12459936 was observed to be associated with the decreased risk of IS in allele model (T vs. C, OR = 0.73, 95% CI = 0.56–0.94, p = 0.016), co-dominant model (TT vs. CC, OR = 0.48, 95% CI = 0.26–0.88, p = 0.018), dominant model (TC/TT vs. CC, OR = 0.63, 95% CI = 0.41–0.94, p = 0.036) and log-additive model (OR = 0.69, 95% CI = 0.52–0.93, p = 0.015). In addition, these was no other significant association between SNPs and IS risk (age ≤ 61 years).
Moreover, we conducted stratified analysis by gender and found three loci associated with the risk of IS in male. Two of these SNPs were correlated with the increased risk of IS in allele model (rs3093193, G vs. C, OR = 1.35, 95% CI = 1.06–1.71, p = 0.016; rs3093144, T vs. C, OR = 1.36, 95% CI = 1.01–1.82, p = 0.040), co-dominant model (rs3093193, GG vs. CC, OR = 2.29, 95% CI = 1.21–4.32, p = 0.011; rs3093144, TT vs. CC, OR = 4.5, 95% CI = 1.22–16.64, p = 0.024), recessive model (rs3093193, GG vs. CC/GC, OR = 2.19, 95% CI = 1.18–4.05, p = 0.013; rs3093144, TT vs. CC/TC, OR = 4.32, 95% CI = 1.17–15.89, p = 0.028) and log-additive model (rs3093193, OR = 1.31, 95% CI = 1.02–1.69, p = 0.036). Conversely, rs12459936 was associated with decreased risk of IS in allele model (T vs. C, OR = 0.78, 95% CI = 0.63–0.98, p = 0.031).
Associations between haplotype analyses and IS risk
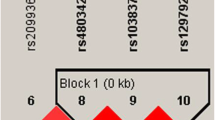
The LD and haplotype analyses of the CYP4F2 polymorphisms in the cases and controls were further studied. All SNPs in CYP4F2 gene were found to exist in LD block (Fig. 1). The distributions of different haplotypes in two groups are presented in Table 6. Although the five SNPs in the CYP4F2 gene had strong linkage, haplotype analysis did not show the significant association (p > 0.05). Then, we conducted haplotype analysis of three positive SNPs (rs3093193, rs12459936 and rs3093144) and IS risk (Table 7). The result demonstrated that the haplotype “GCT” was linked to an increased risk of IS (OR = 1.31, 95% CI = 1.01–1.70, p = 0.043).
In silico analysis
In silico analysis, HaploReg v4.1 was used to assess the function of the selected variants in CYP4F2 gene (Additional file 1: Table S1). The results suggested rs3093203, rs3093193, and rs12459936 were associated with Motifs changed, selected eQTL hits. And rs3093144 was associated with enhancer histone marks, motifs changed, selected eQTL hits. Rs3093110 was related to promoter histone marks, motifs changed, selected eQTL, GRASP QTL hits.
Discussion
In the present study, allele, genotype and haplotype frequencies of five SNPs in the CYP4F2 gene between IS patients and healthy controls were compared and stratified analyses by age and gender were conducted. We found that three SNPs (rs3093193, rs3093144 and rs12459936) in the CYP4F2 were associated with IS risk in a Chinese Han population. Rs3093144 and rs12459936 were associated with IS risk in the overall. For the individuals older than 61 years old, rs3093193 and rs3093144 were related to the increased risk of IS, whereas rs12459936 was related to the decreased risk of IS. In addition, we observed that three SNPs (rs3093193, rs3093144 and rs12459936) were associated with the susceptibility to IS in the male. And five SNPs in the CYP4F2 gene showed strong linkage. These findings suggested that genetic polymorphisms in CYP4F2 may play an important role in the etiology of IS.
Genetic variation is the molecular basis of human genetic diversity. IS is a complex polygenic disease resulting from the genetic factors and environmental risk factors [23]. Approximately 80% of stroke is IS, and the incidence of IS increases with aging. As a member of the CYP450 superfamily, CYP4F2 plays a pivotal role in the metabolism of exogenous and endogenous compounds [24]. It is expressed in the liver, heart, lungs, and kidneys, and is involved in leukotriene B4 (LTB4) and 20--hydroxy eicosane arachidonic acid (20-HETE) metabolism [25]. 20-HETE in vivo can depolarize vascular smooth muscle by blocking Ca2+ activation and K+ channels, resulting in strong vasoconstriction [26]. In hypertensive rat models, increased 20-HETE can lead to oxidative stress and endothelial cell damage, thus increasing the incidence of IS [27]. CYP4F2 is the main synthetase that catalyzes the generation of 20-HETE from arachidonic acid. Recent studies revealed that CYP4F2 was associated with the IS risk. H.-Q. Yan et al. [28] conducted a case-control study to identify the association between the selected SNPs in CYP4F2 gene and the risk of IS. The results showed that the ‘GG’ genotype of CYP4F2 rs2108622 was correlated with an increased risk of IS. In addition, Colàs-Campàs L et al. [29] found rs2108622 ‘AA’ genotype in CYP4F2 gene was significantly associated with a risk of early IS in non-valvular atrial fibrillation patients under vitamin K antagonists treatment. However, study based on this CYP4F2 gene is rare. Hence, our study discussed the relationship between CYP4F2 and IS risk. We found that three SNPs (rs3093193, rs3093144 and rs12459936) in the CYP4F2 were associated with IS risk in a Chinese Han population, suggesting CYP4F2 gene may play an important function in affecting IS.
In addition, it is worth noting that in our results, there were two SNPs (rs3093193 and rs3093144) increased the IS risk and one (rs12459936) decreased the IS risk. However, the haplotype analysis among the three SNPs mentioned above found it was significantly associated with an increased risk of IS (Table 7). For rs12459936, the minor allele ‘T’ when compared to the wild-type allele ‘C’ was protective factor. In turn, the wild-type allele ‘C’ was associated with an increased risk of stroke when compared with the minor allele ‘T’ as reference allele. Thus, based on the positive loci, the haplotype “GCT” was related to elevating risk of IS.
Furthermore, we have compared results with other stroke studies performed in the same population. Lee et al. [30] conducted that a genome-wide association study links small-vessel ischemic stroke to autophagy. And their study focused on Han Chinese in Taiwan. In addition, the study was replicated in an independent Han Chinese population. Imputation analysis also supported the association between three SNPs (rs2594966, rs2594973, rs4684776) in ATG7 gene and stroke-small-vessel occlusion (SVO). When compared to their study, our sample size was not large enough because of strict recruitment criteria. And the study failed to replicate the results within an independent sample. So that’s a part of what we’re going to do next.
Our study also has some potential limitations. The ethnicity of study subjects was limited to the Han Chinese population. Hence, whether our results could apply to other ethnicities is unclear. Furthermore, our current research is fundamental, functional studies are required to understand function of genetic variants and mechanisms underlying this association.
Conclusions
To sum up, we firstly provide new evidence for the association between CYP4F2 variants and IS risk in Han Chinese population, which may support for screening of IS in Han Chinese population and shed light on the mechanism of IS.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional file.
Abbreviations
- 20-HETE:
-
20--hydroxy eicosane arachidonic acid
- CI:
-
Confidence interval
- CT:
-
Computed tomography scans
- CYP4F2 :
-
Cytochrome P450 family 4 subfamily F member 2
- EDTA:
-
Ethylenediamine tetraacetic acid
- GWAS:
-
Genome-wide association studies
- HWE:
-
Hardy–Weinberg equilibrium
- IS:
-
Ischemic stroke
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MRI:
-
Magnetic resonance imaging
- OR:
-
Odds ratio
- SNP:
-
Single nucleotide polymorphism
References
Spence JD. Secondary stroke prevention. Nat Rev Neurol. 2010;6(9):477–86.
Sacco RL, Kasner SE, Broderick JP, Caplan LR, Connors JJ, Culebras A, Elkind MS, George MG, Hamdan AD, Higashida RT, et al. An updated definition of stroke for the 21st century: a statement for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2013;44(7):2064–89.
Reeves MJ, Bushnell CD, Howard G, Gargano JW, Duncan PW, Lynch G, Khatiwoda A, Lisabeth L. Sex differences in stroke: epidemiology, clinical presentation, medical care, and outcomes. Lancet Neurol. 2008;7(10):915–26.
Bejot Y, Daubail B, Giroud M. Epidemiology of stroke and transient ischemic attacks: current knowledge and perspectives. Rev Neurol. 2016;172(1):59–68.
Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, Farrall M, Rothwell PM, Sudlow C, Dichgans M, et al. Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke. 2012;43(12):3161–7.
Allen CL, Bayraktutan U. Risk factors for ischaemic stroke. Int J Stroke. 2008;3(2):105–16.
Hankey GJ. Potential new risk factors for ischemic stroke: what is their potential? Stroke. 2006;37(8):2181–8.
Lawes CM, Bennett DA, Feigin VL, Rodgers A. Blood pressure and stroke: an overview of published reviews. Stroke. 2004;35(4):1024.
Lawes CM, Parag V, Bennett DA, Suh I, Lam TH, Whitlock G, Barzi F, Woodward M. Blood glucose and risk of cardiovascular disease in the Asia Pacific region. Diabetes Care. 2004;27(12):2836–42.
Falcone GJ, Malik R, Dichgans M, Rosand J. Current concepts and clinical applications of stroke genetics. Lancet Neurol. 2014;13(4):405–18.
Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. The Lancet Neurology. 2012;11(11):951–62.
Malik R, Traylor M, Pulit SL, Bevan S, Hopewell JC, Holliday EG, Zhao W, Abrantes P, Amouyel P, Attia JR, et al. Low-frequency and common genetic variation in ischemic stroke: the METASTROKE collaboration. Neurology. 2016;86(13):1217–26.
Cheng YC, Stanne TM, Giese AK, Ho WK, Traylor M, Amouyel P, Holliday EG, Malik R, Xu H, Kittner SJ, et al. Genome-wide association analysis of young-onset stroke identifies a locus on chromosome 10q25 near HABP2. Stroke. 2016;47(2):307–16.
Ward NC, Croft KD, Puddey IB, Phillips M, van Bockxmeer F, Beilin LJ, Barden AE. The effect of a single nucleotide polymorphism of the CYP4F2 gene on blood pressure and 20-hydroxyeicosatetraenoic acid excretion after weight loss. J Hypertens. 2014;32(7):1495–502 discussion 1502.
Jin Y, Zollinger M, Borell H, Zimmerlin A, Patten CJ. CYP4F enzymes are responsible for the elimination of fingolimod (FTY720), a novel treatment of relapsing multiple sclerosis. Drug Metab Dispos. 2011;39(2):191–8.
Fu Z, Nakayama T, Sato N, Izumi Y, Kasamaki Y, Shindo A, Ohta M, Soma M, Aoi N, Sato M, et al. A haplotype of the CYP4F2 gene associated with myocardial infarction in Japanese men. Mol Genet Metab. 2009;96(3):145–7.
Fava C, Montagnana M, Almgren P, Rosberg L, Lippi G, Hedblad B, Engstrom G, Berglund G, Minuz P, Melander O. The V433M variant of the CYP4F2 is associated with ischemic stroke in male Swedes beyond its effect on blood pressure. Hypertension (Dallas, Tex : 1979). 2008;52(2):373–80.
Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;Chapter 2:Unit 2.12.
Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–51.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75.
Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–8.
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics (Oxford, England). 2005;21(2):263–5.
Criado-Garcia J, Fuentes F, Cruz-Teno C, Garcia-Rios A, Jimenez-Morales A, Delgado-Lista J, Mata P, Alonso R, Lopez-Miranda J, Perez-Jimenez F. R353Q polymorphism in the factor VII gene and cardiovascular risk in heterozygous familial hypercholesterolemia: a case-control study. Lipids Health Dis. 2011;10:50.
Tatarunas V, Kupstyte N, Giedraitiene A, Skipskis V, Jakstas V, Zvikas V, Lesauskaite V. The impact of CYP2C19*2, CYP4F2*3, and clinical factors on platelet aggregation, CYP4F2 enzyme activity, and 20-hydroxyeicosatetraenoic acid concentration in patients treated with dual antiplatelet therapy. Blood Coagul Fibrinolysis. 2017;28(8):658–64.
Fava C, Ricci M, Melander O, Minuz P. Hypertension, cardiovascular risk and polymorphisms in genes controlling the cytochrome P450 pathway of arachidonic acid: a sex-specific relation? Prostaglandins Other Lipid Mediat. 2012;98(3–4):75–85.
Wu CC, Gupta T, Garcia V, Ding Y, Schwartzman ML. 20-HETE and blood pressure regulation: clinical implications. Cardiol Rev. 2014;22(1):1–12.
Williams JM, Fan F, Murphy S, Schreck C, Lazar J, Jacob HJ, Roman RJ. Role of 20-HETE in the antihypertensive effect of transfer of chromosome 5 from Brown Norway to dahl salt-sensitive rats. Am J Physiol Regul Integr Comp Physiol. 2012;302(10):R1209–18.
Yan HQ, Yuan Y, Zhang P, Huang Z, Chang L, Gui YK. CYP4F2 gene single nucleotide polymorphism is associated with ischemic stroke. Genet Mol Res. 2015;14(1):659–64.
Colas-Campas L, Royo JL, Montserrat MV, Marzo C, Molina-Seguin J, Benabdelhak I, Cambray S, Purroy F. The rs2108622 polymorphism is related to the early risk of ischemic stroke in non-valvular atrial fibrillation subjects under oral anticoagulation. Pharmacogenomics J. 2018;18(5):652–6.
Lee TH, Ko TM, Chen CH, Chang YJ, Lu LS, Chang CH, Huang KL, Chang TY, Lee JD, Chang KC. A genome-wide association study links small-vessel ischemic stroke to autophagy. Sci Rep. 2017;7(1):15229.
Acknowledgments
We are grateful to all the subjects for their participation. We would also like to thank all the clinicians and hospital staffs who contributed to data collection for this study.
Funding
This study was funded by the Fundamental Research Funds for the Central Universities [grant number xjj2015018]; the special research fund for personnel training of the Second Affiliated Hospital of Xi’an Jiaotong University [grant number RC (XM) 201603]; and a special research fund for the Youth Sciences Foundation of the Second Affiliated Hospital of Xi’an Jiaotong University [grant number YJ (QN) 201402]; the Key Research and Development Project of Shaanxi Province [grant number 2019SF-193]. The funders had no role in the design of the study nor in the collection, analysis and interpretation of data or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: M Z, Y W. Performed the experiments: JJ Z, YL Z. Analyzed the data: TQ H. Contributed reagents/materials/analysis tools: XD M. Wrote the paper: HG P. Revised:M Z. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with the ethical standards of the Declaration of Helsinki and approved by the ethics committee of the Second Affiliated Hospital of Xi’an Jiaotong University. Written informed consents were obtained from all subjects before this study. The procedures were in accordance with the institutional guidelines.
Consent for publication
All subjects whose data is described in this manuscript signed informed consent in written giving permission for their data to be published anonymously.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Functional prediction results of selected loci in the database. (DOCX 13 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, Y., Zhao, J., Zhao, Y. et al. Genetic variants in CYP4F2 were significantly correlated with susceptibility to ischemic stroke. BMC Med Genet 20, 155 (2019). https://doi.org/10.1186/s12881-019-0888-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-019-0888-6