Abstract
Background
X-linked spondyloepiphyseal dysplasia tarda (SEDT-XL) is a skeletal disorder characterized by defective structures of vertebral bodies and/or of epiphyses of the long bones, resulting in moderately short stature and early joint degeneration. TRAPPC2 gene, which is important for collagen secretion, has been reported as causative for SEDT-XL.
Case presentation
Here, we report two variants of TRAPPC2 gene of SEDT-XL patients, a missense variant of start codon, c.1A > T, and a deletion variant, c.40delG. To understand molecular consequence of the variants, we establish an in vitro gene expression assay system and demonstrate that both mutated genes are transcribed, but are not properly translated, indicative of the pathogenic nature of those TRAPPC2 variants.
Conclusions
In the current study, we provide additional experimental data showing that loss-of-function TRAPPC2 variants are probably causative for SEDT-XL phenotype. These findings further contribute to the understanding the clinical picture related to TRAPPC2 gene.
Similar content being viewed by others
Background
X-linked spondyloepiphyseal dysplasia tarda (SEDT-XL) is a skeletal dysplasia affecting male subjects, characterized by defective structures of vertebral bodies and/or of epiphyses of the long bones, leading to short stature and premature joint degeneration [1,2,3]. SEDT-XL has been linked with variants in transport protein particle complex subunit 2 (TRAPPC2) [1, 4, 5]. TRAPPC2 protein consists of 140 amino acids without described enzymatic domains. To date, 32 insertion or deletion, 10 splicing, 9 nonsense, and 6 missense variants of TRAPPC2 gene in SEDT-XL patients have been listed in the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) [6]. Based on the reported pathogenic variants in TRAPPC2 in different ethnic groups, it was suggested that there is no specific population with increased risks for inherited SEDT-XL [7]. Most variants are predicted to cause premature truncation. However, only a few of the variants were studied for its functional defect of protein such as c. 93 + 5G > A or p.D47Y [2, 3, 8, 9]. Interestingly, locations of TRAPPC2 variants are not necessarily related to the phenotype of affected SEDT-XL individuals. In one study, a phenotype of a patient with a deletion of 19 amino acids was not significantly distinguishable from that of another patient with a deletion of 71 amino acids [3, 4]. Although it still remains elusive how the phenotype of SEDT-XL patients are linked to loss of TRAPPC2 functions, it was proposed that loss of TRAPPC2 might affect either the Golgi integrity [3] or the collagen secretion level [2]. We previously reported a missense variant at the start codon (c.1A > T) [10], and detected a novel single nucleotide deletion (c.40delG) in patients presented with SEDT-XL phenotype. In this study, we investigated and found that both variants are transcribed, but they are not expressed at all, as confirmed by immunoblotting, implying that loss of TRAPPC2 function is causative for SEDT-XL phenotypes of these patients.
Case presentation
Approval to perform this study was obtained from the institutional review board of Seoul National University Hospital, Seoul, Republic of Korea. Two cases of Korean SEDT-XL patients with East Asian ethinicity, were found to harbor sequence variation of TRAPPC2 gene. The first patient, a 49-year-old man was referred for right hip pain starting from mid-thirties with abnormal radiographic findings. His height was 138 cm (z-score − 6.2) and arm span 152 cm. Short neck and increased anteroposterior diameter of the thoracic cage were noted. The height and body proportion of the parents and three siblings of the proband were in normal range. Radiographs revealed generalized platyspondyly with hump-like protruding at the endplates of the lumbar spine on the lateral view, a diagnostic finding of SEDT-XL (Fig. 1a-b). Hip radiograph showed degenerative osteoarthritis of the hip joint resulting in profound hip joint space narrowing. Target sequencing including TRAPPC2 and subsequent Sanger sequencing revealed a hemizygous sequence variation of c.1A > T, predicting p.Met1Lys. His mother could not be examined for mutation as she was not alive. The second patient was a 15-year-old boy referred for short stature and back pain. His clinical finding was previously reported [11]. His height was 146.8 cm (z-score − 4.3). Lateral spine radiographs showed humps at the posterior half of the endplates, similar to the first patient and informative finding for the diagnosis of of SEDT-XL(Fig. 1c-d). Sanger sequencing for TRAPPC2 revealed a hemizygous sequence variation, c.40delG, predicting p.Asp14IlefsX27, which was inherited from the mother. She was short (146 cm, z-score − 3.12), but did not show any radiographic abnormality, back pain or joint pain [11]. Both variants were not annotated in dbSNP, 1000 Genomes database, the Genome Aggregation Database (gnomAD), NHLBI Exome Sequencing Project Exome Variant Server (EVS), or Exome Aggregation Consortium (ExAC) Browser.
Lateral spine and hip radiographs of two SEDT-XL patients. a, b Radiographs taken from the proband with the c.1A > T, TRAPPC2 variant at age of 45 years. a Thoracolumbar lateral spine shows uniform platyspondyly and hump-like bony projections at the centroposterior portions of upper and lower endplates, and narrowing or almost obliteration of the intervertebral disc spaces. b Pelvis radiograph shows narrow iliac wings with relatively long ischium and pubic bones, deep acetabulum and degenerative osteoarthtis with narrow hip joints. c, d Radiographs from the proband with the c.40delG, TRAPPC2 variant at age 15 years. c Spine lateral radiograph shows platyspondyly with elongated vertebral bodies and hump-like bony protrusion at the upper and lower endplates, dominantly noted in the lumbar spine. d Hip shows relatively long ischium and pubic bones. Deep acetabulum and dysplastic femoral heads with premature degenerative hip joint narrowing are noted
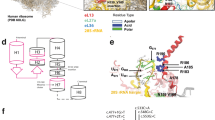
The first variant, c.1A > T, misses its translational start codon, and consequently, it would not be translated. However, it is possible to use the in-frame cryptic initiating ATG codon, located 57 nucleotides downstream from the start codon, which might lead to express the N-terminal truncated TRAPPC2 mutant if translated (Fig. 2a). The predicted protein from the second mutation, c.40delG, is C-terminal truncated mutant, p.Asp14IlefsX27 (Fig. 2a). In an attempt to understand molecular consequences of the identified TRAPPC2 variants, we established the in vitro experimental system, which enabled us to determine expression of both TRAPPC2 variants at the level of transcription and translation. To this end, we cloned wild-type (WT) TRAPPC2 cDNA from the cDNA library prepared from U2OS cells. Each mutation was introduced using QuikChange II XL Site-Directed Mutagenesis Kit (Agilent). To study expression levels of mutants, WT and two TRAPPC2 variants were constructed to the pcDNA3.1 plasmid of which the HA tag was added to 3′ end to each cDNA to prevent translation of HA from causing expression of mutants that would otherwise be not translated (Fig. 2b). After obtaining each plasmid, we confirmed that the variants were correctly reproduced in the vector by Sanger sequencing (Fig. 2c). Next, to determine expression levels, 293 T cells were transfected with control, wild-type or mutant TRAPPC2 expression vector individually, and 24 h later, the prepared cell lysates were fractionated with immunoblot analysis to characterize expression patterns of those two mutants. As shown in Fig. 2d, only WT was detected; the missense mutation, c.1A > T, and the frameshift mutation, c.40delG, failed to express the TRAPPC2 mutant. No proteins expression could be due to either no translation or no transcription. To test if mutants were transcribed or not, we performed reverse transcription polymerase chain reaction (RT-PCR). We designed a set of primers to detect TRAPPC2 transcripts produced by the vectors only and to exclude the endogenous TRAPPC2 transcripts (arrows in the Fig. 2b). Cell lysates for RT-PCR were prepared in the same way as for immunoblotting. After harvesting and lysing cells, lysates were first used to isolate total mRNA with RNeasy Mini Kit (Qiagen) according to a given instruction. Then, the purified RNA was subjected to produce cDNA library with reverse transcriptase reaction (SuperScript III First-Strand Synthesis Kit, Thermo). Next, using the cDNA library, we performed PCR reaction with primers that specifically cover the TRAPPC2 cDNA produced by the transformed vectors. When the PCR products were run on an agarose gel, both WT and two mutants revealed a band at size of 700 bp (Fig. 2e), suggesting two possible scenarios: the first is that two TRAPPC2 variants are transcribed, but not translated. The other is that these variants are transcribed and translated, but the translated proteins are degraded.
Functional analysis of TRAPPC2 variants found in the SEDT-XL patients (a) Predicted amino acid sequences of the TRAPPC2 variants found in the SEDT-XL individuals. The c.1A > T missense variant would change the starting codon Methionine to Leucine. The deletion variant, c.40delG, would only produce the first 13 amino acids correctly and then abruptly terminate the translation. The gene accession number for human TRAPPC2 mRNA is indicated. b DNA constructs used in this study. Wild-type (WT) or the TRAPPC2 variants are expressed under the CMV promoter. HA tag is introduced at the C-terminus of the cDNAs and thus the HA tag does not affect translation of the TRAPPC2 variants. The arrows indicate primers for RT-PCR. These primers are specific to the vector used in this experiment to exclude any endogenous TRAPPC2 genes. c Chromatogram displaying the mutations in the TRAPPC2 expression vectors. A missense mutation at the position 1 showed a change of base from adenine to thymine in the left pair. A deletion of guanine at the position 40 is demonstrated in the right pair. d-e Analyses of the TRAPPC2 variant expression at protein and transcript level. 293 T HEK cells were transfected with individual pCDNA3.1, WT, or TRAPPC2 variant expressing vector, and cell lysates were subjected to Western blot analysis with HA antibody (d) and used to prepare total RNA for RT-PCR (e). The size markers for DNA fragments and proteins are indicated
Discussion and conclusions
We anticipated that both mutant TRAPPC2 proteins would be expressed at a smaller size if they were translated at all. In c.1A > T mutant, the initial starting codon is missing, but there is another in-frame ATG codon 57 nucleotides downstream of the start codon; it may lead to the N-terminal truncated TRAPPC2 transcript at a smaller size. Another variant, c.40delG, causes premature termination at amino acid 37, and thus if expressed, it would produce a protein that is about a third of WT in size. However, our data demonstrated that both mutants were transcribed, but the translation products of these mRNA are not properly translated as it either lost the start codon or is likely degraded by nonsense-mediated RNA decay. Absence of any protein would likely cause significant changes in cellular processes, which would be followed by developmental problems or disorder. In the current study, we provide experimental evidences showing that two TRAPPC2 variants identified from SEDT-XL patients lead to no TRAPPC2 protein production. Although here we showed that loss-of-function TRAPPC2 variants is causative for the SEDT-XL, further studies will be required to understand molecular mechanisms of pathophysiology of the disease.
Both SEDT-XL individuals showed defects in skeletal development and have been through difficulties due to the abnormalities as described above. Complete absence of a single protein, especially when it is involved in a crucial pathway, should take a toll on a cell, and on an individual, possibly lethally. However, our data shows that two SEDT-XL individuals are survived without the TRAPPC2 protein, although they both had abnormal bone structures and related pain, suggesting existence of an alternate protein or a pathway that takes over a role of TRAPPC2. TRAPPC2 is associated with numerous proteins, either directly or indirectly [12]. If it is not expressed, and yet affected individuals can be survived, then there is highly likely another protein that compensates for the absence of this multi-interacting protein, and identifying the substituting protein would be helpful in gene therapies of SEDT-XL. One possible set of candidates is pseudogenes of TRAPPC2. There are nine pseudogenes, and the most likely compensator is TRAPPC2B (also known as SEDLP1); it is located on chromosome 19q13.4 and has potential to produce a protein identical to that coded by TRAPPC2 [13]. Since TRAPPC2B is not on X chromosome, and yet can encode an identical protein, it is highly possible that individuals with TRAPPC2 variants who survived absence of the protein may compensate by utilizing TRAPPC2B. Individuals with TRAPPC2 variants can restore the pathway of protein transport by a means of TRAPPC2B if they inherited both normal copies of chromosome 19. However, if either one or both copies of TRAPPC2B are damaged, then, the normal pathway would be only partially overcome, or not at all, resulting in different degrees of phenotypes in patients.
Abbreviations
- cDNA:
-
complimentary deoxyribonucleic acid
- HA:
-
Hemagglutinin
- mRNA:
-
messenger ribonucleic acid
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SEDT-XL:
-
X-linked spondyloepiphyseal dysplasia tarda
- TRAPPC2 :
-
Transport protein particle complex subunit 2
- WT:
-
Wild-type
References
Gedeon AK, Tiller GE, Le Merrer M, Heuertz S, Tranebjaerg L, Chitayat D, Robertson S, Glass IA, Savarirayan R, Cole WG, et al. The molecular basis of X-linked spondyloepiphyseal dysplasia tarda. Am J Hum Genet. 2001;68(6):1386–97.
Brunet S, Sacher M. In sickness and in health: the role of TRAPP and associated proteins in disease. Traffic (Copenhagen, Denmark). 2014;15(8):803–18.
Savarirayan R, Thompson E, Gecz J. Spondyloepiphyseal dysplasia tarda (SEDL, MIM #313400). Eur J Hum Genet. 2003;11(9):639–42.
Christie PT, Curley A, Nesbit MA, Chapman C, Genet S, Harper PS, Keeling SL, Wilkie AO, Winter RM, Thakker RV. Mutational analysis in X-linked spondyloepiphyseal dysplasia tarda. J Clin Endocrinol Metab. 2001;86(7):3233–6.
Shaw MA, Brunetti-Pierri N, Kadasi L, Kovacova V, Van Maldergem L, De Brasi D, Salerno M, Gecz J. Identification of three novel SEDL mutations, including mutation in the rare, non-canonical splice site of exon 4. Clin Genet. 2003;64(3):235–42.
Stenson PD, Mort M, Ball EV, Evans K, Hayden M, Heywood S, Hussain M, Phillips AD, Cooper DN. The human gene mutation database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum Genet. 2017;136(6):665–77.
Tiller GE, Hannig VL: X-Linked Spondyloepiphyseal Dysplasia Tarda. In: GeneReviews((R)). edn. Edited by Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A. Seattle WA: University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle; 1993.
Ryu H, Park J, Chae H, Kim M, Kim Y, Ok IY. X-linked spondyloepiphyseal dysplasia tarda: identification of a TRAPPC2 mutation in a Korean pedigree. Ann Lab Med. 2012;32(3):234–7.
Zong M, Wu XG, Chan CW, Choi MY, Chan HC, Tanner JA, Yu S. The adaptor function of TRAPPC2 in mammalian TRAPPs explains TRAPPC2-associated SEDT and TRAPPC9-associated congenital intellectual disability. PLoS One. 2011;6(8):e23350.
Bae JS, Kim NK, Lee C, Kim SC, Lee HR, Song HR, Park KB, Kim HW, Lee SH, Kim HY, et al. Comprehensive genetic exploration of skeletal dysplasia using targeted exome sequencing. Genet Med. 2016;18(6):563–9.
Kim HJ, Lee BH, Kim YM, Kim GH, Kim OH, Yoo HW. A case with spondyloepiphyseal dysplasia tarda with TRAPPC2 mutation. J Genet Med. 2012;9(1):31–4.
Sacher M, Shahrzad N, Kamel H, Milev MP. TRAPPopathies: an emerging set of disorders linked to variations in the genes encoding transport protein particle (TRAPP)-associated proteins. Traffic (Copenhagen, Denmark). 2019;20(1):5–26.
Gecz J, Hillman MA, Gedeon AK, Cox TC, Baker E, Mulley JC. Gene structure and expression study of the SEDL gene for spondyloepiphyseal dysplasia tarda. Genomics. 2000;69(2):242–51.
Acknowledgements
We are grateful to the affected individuals and their families for their invaluable contribution.
Funding
TJC is supported by National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning of the Korean government (NRF-2014M3C9A2064684: Genome Technology to Business Translation Program), which has been used for the patient recruitment and care, and determining genetic variants. YK is supported by NRF funded by the Ministry of Science, ICT & Future Planning of the Korean government (NRF-2014M3C9A2064688: Genome Technology to Business Translation Program and NRF-2016R1A5A1011974), which have been used for validation of the pathogenicity of identified variants and in vitro functional studies. All the decisions regarding to the current studies are made by authors, not by funders. The funders are not involved in the study design, data collection and analysis, performing experiments and in writing the manuscript.
Availability of data and materials
All the data and materials used in this studies are fully available upon request.
Author information
Authors and Affiliations
Contributions
YK and TJC conceived the whole study design. TJC, MHS, HRS, OHK conducted clinical evaluation of the patients and obtained genetic materials. JYW, DK, SYP, HRL, JSL, JHP designed and performed experiments. YK and TJC wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The institutional Review Boards of the Seoul National University Hospital, Seoul, Republic of Korea approved the studies, and the patients consented to participate this study.
Consent for publication
The affected individuals provided written informed consent for the personal and clinical information to be published.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Won, J.Y., Kim, D., Park, S.Y. et al. Novel loss-of-function variants of TRAPPC2 manifesting X-linked spondyloepiphyseal dysplasia tarda: report of two cases. BMC Med Genet 20, 70 (2019). https://doi.org/10.1186/s12881-019-0802-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-019-0802-2