Abstract
Background
Infantile neuroaxonal dystrophy (INAD) is a rare hereditary neurological disorder caused by mutations in PLA2G6. The disease commonly affects children below 3 years of age and presents with delay in motor skills, optic atrophy and progressive spastic tetraparesis. Studies of INAD in Africa are extremely rare, and genetic studies from Sub Saharan Africa are almost non-existent.
Case presentation
Two Sudanese siblings presented, at ages 18 and 24 months, with regression in both motor milestones and speech development and hyper-reflexia. Brain MRI showed bilateral and symmetrical T2/FLAIR hyperintense signal changes in periventricular areas and basal ganglia and mild cerebellar atrophy. Whole exome sequencing with confirmatory Sanger sequencing were performed for the two patients and healthy family members. A novel variant (NM_003560.2 c.1427 + 2 T > C) acting on a splice donor site and predicted to lead to skipping of exon 10 was found in PLA2G6. It was found in a homozygous state in the two patients and homozygous reference or heterozygous in five healthy family members.
Conclusion
This variant has one very strong (loss of function mutation) and three supporting evidences for its pathogenicity (segregation with the disease, multiple computational evidence and specific patients’ phenotype). Therefore this variant can be currently annotated as “pathogenic”. This is the first study to report mutations in PLA2G6 gene in patients from Sudan.
Similar content being viewed by others
Background
Infantile neuroaxonal dystrophy (INAD) is a rare hereditary neurological disorder caused by mutations in the PLA2G6 gene [1]. The disease commonly affects children below 3 years of age and presents with delay in motor skills, optic atrophy and progressive spastic tetraparesis. Brain MRI shows cerebellar atrophy, white matter abnormalities and hypointense globus pallidus. Infants with the disease are usually normal at birth but deteriorate rapidly and rarely live beyond their first decade [2].
The incidence and prevalence of INAD is currently unknown [3]. More than 85% of cases are caused by mutations in PLA2G6 gene, while the remainder has unknown genetic cause. The protein product of PLA2G6 gene has important roles in the metabolism of membrane phospholipids. Sequencing analysis of PLA2G6 gene has identified missense, nonsense, splicing disruption variants and insertion/deletions with genotype/phenotype correlation: loss of function mutations are usually associated with more severe forms of the disease, while compound heterozygous variants have usually milder presentation [4].
Studies of INAD in Africa are extremely rare, and genetic studies from Sub Saharan Africa are almost non-existent. In this first study of INAD from Sudan, we are reporting a novel homozygous splice site mutation in PLA2G6 gene in two siblings with INAD.
Case presentation
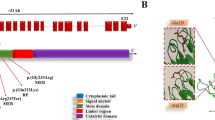
Two Sudanese siblings from a consanguineous family (F25) from White Nile area in central Sudan presented with delayed motor and intellectual development. Patient 1 presented at age 24 months with walking difficulty due to muscle weakness and poor speech. The patient was able to sit without support at age 6 months; she started walking with assistance at age 12 months but lost her ability to walk at age 2 years. She was able to say “dada, mama” at age 8 months but lost this ability by age 20 months. At age 2.5 years, the patient became non-ambulatory and completely lost her speech ability. Patient 2 presented with the same clinical features but started earlier at age 18 months, he was able to sit with support and say “dada, mama” at ages 6 and 8 months respectively. He was able to walk with support at age 12 months but at the time of examination, he was only able to roll in bed. Both patients had progressive dysphagia for both solids and liquids and behavioral changes (lack of communication in patient 1 and significant irritability in patient 2). On examination both patients had normal upper and lower limbs tone and hyperreflexia in all limbs with up-going plantar response. Cognitive status of the two patients was difficult assess but both patients appeared to be non-attentive. Both patients had marked left sided visual impairment and bilateral optic atrophy. There were no extrapyramidal signs. Brain MRI showed bilateral and symmetrical T2/FLAIR hyperintense signal changes in periventricular areas, basal ganglia, globus pallidus and to lesser degree head of caudate nucleus and putamen as well as the cerebellum. Mild generalized cerebellar atrophy was found in both patients but there was no evidence of brain iron accumulation (hypointense signals in T2 and hyperintense signals in T1 weighted images), Fig. 1.
a Pedigree and MRI of the index patient (210) of family F25 caused by splice donor mutation in PLA2G6 segregating with the disease distribution in whole family presenting with pyramidal signs and features associated with infantile neuroaxonal dystrophy (INAD). b MRI shows bilateral and symmetrical T2/FLAIR hyperintense signal changes in periventricular areas, basal ganglia, globus pallidus and to lesser degree head of caudate nucleus and putamen. c Chromotagram of Sanger sequencing showing homozygous mutation in proband, a heterozygous carrier and a control homozygous reference allele with conserved amino acid sequence. Pedigree symbols: * sampled individual; Phenotype symbols: black color: affected individuals; Genotype symbols: ++ Homozygous reference genotype; M+ Heterozygous genotype; MM Homozygous mutant genotype. Others are standard medical pedigree symbols
Whole exome sequencing (WES)
Two milliliters of saliva were collected using Oragene.Discover DNA collection kits (DNA Genotek Inc., Ottawa, ON, Canada). DNA was extracted according to prepIT.L2P manual protocol. Standard Agarose gel electrophoresis, NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and Qubit fluorometer (Promega, Madison, WI, USA) were used to qualify and quantify DNA. Sequencing was performed using the MiSeq platform (Illumina, San Diego, CA, USA) on 250 bp paired-end reads according to the manufacturer’s recommendations. Variant calling and quality control were performed using Genomics Workbench (CLC Bio, Aarhus, Denmark). SNVs and Indels were detected using probability-based and quality-based algorithms. For gene/variant prioritization, nonsense, frameshift, splice site and missense variants with a minimum depth of 30 × were selected. Minor allele frequency cutoff of 1% was used.
Sanger sequencing
To validate whole exome sequence results and segregation analysis we performed Sanger sequencing for the candidate variants for the two patients and five healthy family members using the BIGDYE chemistry on an ABI3730 sequencer (Applied Biosystems). Seqscape (Applied Biosystems) and Chromas lite software (Technelysium, South Brisbane, QLD, Australia) were used for sequence analysis and visualization. Genetic testing was done for the two patients and their parents and all available family members at the time of presentation.
Results
WES analysis identified one novel variant (NM_003560.2 c.1427 + 2 T > C) in the PLA2G6 gene as affecting a splice site at the exon 10-intron 10 junction. The mutation is predicted to disrupt splicing and skipping of exon10 with high (100%) confidence score using three different splice prediction tools: MaxEnt (http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html), NNSPLICE (http://www.fruitfly.org/seq_tools/splice.html) and HSF (http://www.umd.be/HSF3/). The mutation co-segregated with the disease in the family. It was found in a homozygous state in the two patients and homozygous reference or heterozygous in five healthy related individuals (Fig. 1).
Discussion and conclusion
INAD is a typical example of a rare hereditary neurological disease with high mortality and morbidity [2]. Mutations in PLA2G6 gene causing INAD have been described previously in many studies [4,5,6,7,8,9,10,11,12,13]. These were more commonly homozygous missense variants or deletions which lead to loss of protein function. Splice site variants have been rarely reported although they are considered also loss of function mutations [14]. The spectrum of causative mutations is obscure and possibly unknown in patients from Sub Saharan Africa despite the well-known genetic heterogeneity in this region [15]. This is the first study of INAD in Sudan and it showed a novel homozygous splicing mutation (c.1427 + 2 T > C) in exon 10 of PLA2G6 gene of two siblings with typical clinical features. The variant segregated with the disease; both parents were heterozygous and all unaffected family members had either the wild type genotype or were heterozygous. Mutations in exon 10 have been previously reported in association with INAD in dogs [16]. This comports with its role in the pathogenesis of the disease in this family. The effect on mRNA could not be checked however as no cells were available from the patients.
According to the latest ACMG guidelines [17], this variant has one very strong (loss of function mutation) and three supporting evidence for its pathogenicity (segregation with the disease, multiple computational evidence and specific patients’ phenotype). In addition, this variant was absent in Exac and 1000 genome browser databases and has a total allele frequency of 0.4065 × 10− 5 in genomAd browser with no reported homozygotes. Furthermore, the (c.1427 + 2 T > C) variant is highly predicted to cause skipping of exon 10 during mRNA maturation. Splicing alteration leads to skipping of an exon or retention of an intron both significantly altering protein structure and function [18]. Therefore this variant can be currently annotated as “pathogenic”.
Mutations in PLA2G6 gene continue to be discovered from throughout the world, but our study is the first to report cases of INAD with associated genetic changes from Sudan. However functional studies are still needed to verify the effect of the reported mutation on the expression and function of PLA2G6 protein.
Abbreviations
- ACMG:
-
American College of Medical Genetics
- DNA:
-
Deoxyribonucleic acid
- INAD:
-
Infantile neuroaxonal dystrophy
References
Khateeb S, Flusser H, Ofir R, Shelef I, Narkis G, Vardi G, et al. PLA2G6 mutation underlies infantile neuroaxonal dystrophy. Am J Hum Genet [Internet]. 2006;79(5):942–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/17033970. Cited 14 Sept 2017
Gregory A, Kurian MA, Maher ER, Hogarth P, Hayflick SJ. PLA2G6-associated neurodegeneration [Internet]. GeneReviews(®). Seattle: University of Washington; 1993. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20301718. Cited 14 Sept 2017
Genetics Home Reference. Infantile neuroaxonal dystrophy [Internet]. 2017. Available from: https://ghr.nlm.nih.gov/condition/infantile-neuroaxonal-dystrophy. Cited 14 Sept 2017.
Gebril O, Uebe S, Reuter M, Schumacher J, Jamra RA, Reis A. A new missense mutation in PLA2G6 gene among a family with infantile neuroaxonal dystrophy INAD. Egypt Pediatr Assoc Gaz [Internet]. 2016;64(4):171–6. Available from: http://www.sciencedirect.com/science/article/pii/S1110663816300520. Cited 14 Sept 2017
Kurian MA, Morgan NV, MacPherson L, Foster K, Peake D, Gupta R, et al. Phenotypic spectrum of neurodegeneration associated with mutations in the PLA2G6 gene (PLAN). Neurol Int. 2008;70(18):1623–9. Available from: http://www.neurology.org/cgi/doi/10.1212/01.wnl.0000310986.48286.8e. Cited 14 Sept 2017
Malik I, Turk J, Mancuso DJ, Montier L, Wohltmann M, Wozniak DF, et al. Disrupted membrane homeostasis and accumulation of ubiquitinated proteins in a mouse model of infantile neuroaxonal dystrophy caused by PLA2G6 mutations. Am J Pathol [Internet]. 2008;172(2):406–16. Available from: http://linkinghub.elsevier.com/retrieve/pii/S000294401061807X. Cited 14 Sept 2017
Salih MA, Mundwiller E, Khan AO, AlDrees A, Elmalik SA, Hassan HH, et al. New findings in a global approach to dissect the whole phenotype of PLA2G6 gene mutations. Palau F, editor. PLoS One [Internet]. 2013;8(10):e76831. Available from: http://dx.plos.org/10.1371/journal.pone.0076831. Cited 14 Sept 2017
Wu Y, Jiang Y, Gao Z, Wang J, Yuan Y, Xiong H, et al. Clinical study and PLA2G6 mutation screening analysis in Chinese patients with infantile neuroaxonal dystrophy. Eur J Neurol [Internet]. 2009;16(2):240–5. Available from: http://doi.wiley.com/10.1111/j.1468-1331.2008.02397.x. Cited 14 Sept 2017
Grazia I, Claudio G, Giovanna C, Maria CD, Roberta Z, Valentina P, et al. A new PLA2G6 mutation in a family with infantile neuroaxonal dystrophy. J Neurol Sci [Internet]. 2017;381:209–12. Available from: http://linkinghub.elsevier.com/retrieve/pii/S0022510X17337528. Cited 14 Sept 2017
Gregory A, Westaway SK, Holm IE, Kotzbauer PT, Hogarth P, Sonek S, et al. Neurodegeneration associated with genetic defects in phospholipase A2. Neurol Int. 2008;71(18):1402–9. Available from: http://www.neurology.org/cgi/doi/10.1212/01.wnl.0000327094.67726.28. Cited 14 Sept 2017
Romani M, Kraoua I, Micalizzi A, Klaa H, Benrhouma H, Drissi C, et al. Infantile and childhood onset PLA2G6 -associated neurodegeneration in a large North African cohort. Eur J Neurol [Internet]. 2015;22(1):178–86. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25164370. Cited 14 Sept 2017
Kapoor S, Shah MH, Singh N, Rather MI, Bhat V, Gopinath S, et al. Genetic analysis of PLA2G6 in 22 Indian families with infantile neuroaxonal dystrophy, atypical late-onset neuroaxonal dystrophy and dystonia parkinsonism complex. PLoS One [Internet]. 2016;11(5):e0155605. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27196560. Cited 14 Sept 2017
Veerapandiyan A, Chaudhari A, Aravindhan A, Hayes-Rosen C. Novel mutation in PLA2G6 gene in a patient with infantile neuroaxonal dystrophy. J Pediatr Neurol [Internet]. 2016;15(02):073–5. Available from: http://www.thieme-connect.de/DOI/DOI?10.1055/s-0036-1593885. Cited 10 Jan 2018
Ward LD, Kellis M. Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol [Internet]. 2012;30(11):1095–106. Available from: http://www.nature.com/doifinder/10.1038/nbt.2422. Cited 14 Sept 2017
Bekada A, Arauna LR, Deba T, Calafell F, Benhamamouch S, Comas D, et al. Genetic heterogeneity in Algerian human populations. Kayser M, editor. PLoS One [Internet]. 2015;10(9):e0138453. Available from: http://dx.plos.org/10.1371/journal.pone.0138453. Cited 14 Sept 2017
Tsuboi M, Watanabe M, Nibe K, Yoshimi N, Kato A, Sakaguchi M, et al. Identification of the PLA2G6 c.1579G>a missense mutation in Papillon dog neuroaxonal dystrophy using whole exome sequencing analysis. Bandapalli OR, editor. PLoS One [Internet]. 2017;12(1):e0169002. Available from: http://dx.plos.org/10.1371/journal.pone.0169002. Cited 15 Sept 2017
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med [Internet]. 2015;17(5):405–23. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25741868. Cited 23 June 2017
Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol [Internet]. 2010;220(2):152–63. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19918805. Cited 14 Sept 2017
Acknowledgements
The authors would like to send their gratitude to the patients and family members who participated in the study. We also thank the members of the DNA and cell bank of the ICM, Emeline Mundwiller, Khalid El Hachimi and Delphine Bouteiller for their valuable contribution as well as the IGenSeq platform of the Institut du Cerveau et de la Moelle épinière for genetic studies.
Funding
This study was supported financially by funded by the Agence Nationale de la Recherche (SPATAX-QUEST project, to Giovanni Stevanin), the Verum Foundation and Roger de Spoelberg Foundation (to Alexis Brice), the European Union (Neuromics projects, OMICS call, to Giovanni Stevanin) and benefited from the Programme d’Investissement d’Avenir IHU-A-ICM. MAMS was supported by the Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia via research group project number RGP-VPP-301. The funding bodies had no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
LEOE, GS, and AEA made substantial contributions to the design, administration and conceptualization of the study, interpretation of data and critically revising and drafting the manuscript. AB, GS, MEI, and MIE provided funding, acquisition of data, conception and design of the study, manuscript review and editing. INM, AAAH, MAE, MAMS, MK, MA, AY, and RAS participated in project conceptualization, data analysis and curation, review and editing. All authors approved the submission of the final manuscript and agreed to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Paris Necker Ethics Committee (France) and the Ethical Committee of the University of Khartoum Medical Campus (Sudan). Written informed consent was obtained from each family member (or parents/legal guardians in case of minors) before participation in the study.
Consent for publication
All participants (or parents/legal guardians in case of minors) provided consent for the publication of their clinical details.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Elsayed, L.E.O., Mohammed, I.N., Hamed, A.A.A. et al. Case report of a novel homozygous splice site mutation in PLA2G6 gene causing infantile neuroaxonal dystrophy in a Sudanese family. BMC Med Genet 19, 72 (2018). https://doi.org/10.1186/s12881-018-0592-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-018-0592-y