Abstract
Background
Caudal regression syndrome (CRS) or sacral agenesis is a rare congenital disorder characterized by a constellation of congenital caudal anomalies affecting the caudal spine and spinal cord, the hindgut, the urogenital system, and the lower limbs. CRS is a complex condition, attributed to an abnormal development of the caudal mesoderm, likely caused by the effect of interacting genetic and environmental factors. A well-known risk factor is maternal type 1 diabetes.
Method
Whole exome sequencing and copy number variation (CNV) analyses were conducted on 4 Caucasian trios to identify de novo and inherited rare mutations.
Results
In this pilot study, exome sequencing and copy number variation (CNV) analyses implicate a number of candidate genes, including SPTBN5, MORN1, ZNF330, CLTCL1 and PDZD2. De novo mutations were found in SPTBN5, MORN1 and ZNF330 and inherited predicted damaging mutations in PDZD2 (homozygous) and CLTCL1 (compound heterozygous). Importantly, predicted damaging mutations in PTEN (heterozygous), in its direct regulator GLTSCR2 (compound heterozygous) and in VANGL1 (heterozygous) were identified. These genes had previously been linked with the CRS phenotype.
Two CNV deletions, one de novo (chr3q13.13) and one homozygous (chr8p23.2), were detected in one of our CRS patients. These deletions overlapped with CNVs previously reported in patients with similar phenotype.
Conclusion
Despite the genetic diversity and the complexity of the phenotype, this pilot study identified genetic features common across CRS patients.
Similar content being viewed by others
Background
Caudal Regression Syndrome (CRS; Caudal Dysgenesis Syndrome, Caudal Dysplasia Sequence, Congenital Sacral Agenesis; OMIM 600145) is a rare (1 in 7,500–100,000 births [1, 2]) congenital disorder characterized by varying degrees of spinal column agenesis. Associated with it are anomalies of central nervous, genito-urinary, cardiac, respiratory and gastro-intestinal systems [3] with anorectal malformations (ARMs) being the most common.
CRS has been attributed to abnormal fetal development of the caudal mesoderm before the fourth week of gestation [4]. During the abnormal gastrulation, prospective notochordal cells, that are wrongly specified in terms of their rostrocaudal positional encoding, are eliminated. Eventually, fewer or even no cells will be available to form the notochord at a given abnormal segmental level. The consequences of such segmental notochordal paucity are manifold and affect the development of the spinal column and spinal cord as well as other organs that rely on the notochord as their inductor. If the prospective notochord is depleted a wide array of segmental vertebral malformations may develop including segmentation defects, indeterminate or block vertebrae, or absence of several vertebrae. Because of lack of neural induction and absence of a floor plate, fewer prospective neuroectodermal cells will be induced to form the neural tube. The resulting malformation essentially depends on the segmental level and the extent of the abnormality along the longitudinal embryonic axis, with subsequent interference on the processes of primary and/or secondary neurulation [5]. However, what triggers such abnormal events is not known.
Caudal spinal abnormalities are the defining characteristics of CRS. Cama et al. [6] and Pang et al. [7] classified the disorder into 5 categories according to the degree of caudal spine involvement: Type I) total sacral agenesis with normal or short transverse pelvic diameter and some lumbar vertebrae possibly missing; Type II) total sacral agenesis without involvement of lumbar vertebrae; Type III) subtotal sacral agenesis or sacral hypodevelopment; Type IV hemisacrum and Type V) coccygeal agenesis.
Maternal type 1 diabetes is a risk factor for CRS, as it is for many other congenital disorders [8]. Maternal type 1 diabetes confers a higher relative risk (252) for CRS than for any congenital diosorder [9]. The exact mechanism by which maternal diabetes affects fetal development in humans remains unclear [10]. While animal studies have shown that embryos exposed to higher levels of glucose develop growth anomalies, hyperglycemia has not been associated with abnormal fetal development in humans [11]. During normal pregnancies insulin sensitivity is reduced at the start of the third trimester in order to provide metabolic fuel for both mother and the developing fetus. However, since insulin is unable to cross the placenta, the fetus starts producing its own insulin in order to metabolize nutrition. It has been suggested that insulin, antibodies to insulin, or some other abnormality of carbohydrate metabolism could affect the development of a genetically susceptible fetus [12, 13].
Evidence for a genetic cause is provided by the existence of familial segregation as well as animal models. While the most severe forms of CRS present sporadically, milder CRS forms can be transmitted within families in a dominant manner with reduced penetrance and phenotypic variability [4]. Currarino syndrome (CS) is characterized by sacral agenesis type IV, presacral mass, and ARM. CS has been associated with mutations in the MNX1 gene [14–19]. Yet MNX1 mutations account for only 50% of sporadic and 90% of familial cases [19]. Although private mutations in genes such as VANGL1 [20], HOXD13 [21] and PTEN [22] have been described in sporadic cases with caudal dysgenesis and/or vertebrae anomalies, no firm genetic association has been established.
A CRS-like phenotype can be induced by administration in animals of retinoic acid (RA), lithium, cadmium, sulphamide, or organic solvents [23, 24]. Several mutated genes including Cyp26a1, Hoxd13 [25], Wnt-3a [26], Acd, Ptf1a, and Pcsk5 underlie a CRS-like phenotype in mice [10, 27], yet mutations in the human orthologs have never been identified in CRS patients. Interestingly, the reverse is also true; Mnx1 (formerly Hxlb9) mutant mice do not present Currarino syndrome features [28]. These exceptions to human-mouse phenotypic correlation suggest differences in genetic etiology between humans and experimental organisms [10].
In order to search for genetic risk factors for CRS the exomes of five sporadic CRS cases and their respective healthy parents were sequenced. Due to the sporadic nature of the disease we have focused on de novo or recessive inherited damaging genetic variants. In addition we also used a SNP chip assay in order to identify rare and de novo CNVs.
Methods
Subjects
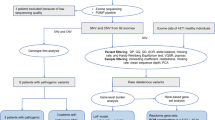
The records of patients treated between 1995–2010 at the Neurosurgery Department of Giannina (Genoa, Italy) and at the AbaCid-Genética, Grupo HM Hospitales (Madrid, Spain) for congenital anomalies of the spine were reviewed. For all patients, family history, cardiac, respiratory and endocrine data were collected. This included family history for diabetes. Neurological, neurophysiological (Somatosensory evoked potential, SEP), radiological, neuroradiologic (MRI), orthopaedic, physical, urological (urodynamic, cystography) and surgical assessments were performed for each case. For this pilot study, we selected four Italian trios (CR5, CR17, CR41, CR46) as well as one trio from Spain (CURR20). We only selected cases within this period who had sporadic lower spine agenesis. Patients were also affected with additional anomalies of axial skeleton and internal organs. One child had a mother with diabetes type I. The local ethical committees approved the study and written informed consent was obtained from all patients and parents. Subjects are identified by the trio ID suffixed by either A, B or C indicating father, mother or child respectively.
Bioinformatics processing
Capture, alignment and base-calling
Whole exome sequencing (WES) was performed at the Centre of Genomic Sciences of the University of Hong Kong, Hong Kong. The exomes of all five trios were sequenced using Illumina HiSeq PE100 and captured with TruSeq Exome Enrichment kit (FC–121–1024, Illumina Inc.). The exome sequences were alignment against Human Genome HG-19 using BWA MEM [29]. Duplicated reads were flagged with Picard-tools [30]. The GATK tool set was used to realign indels, perform base recalibration, remove duplicates, perform indel and SNP calling, and for genotype refinement to improve accuracy of genotype calls [31]. Data quality for each variant was scored and a hard threshold used to remove low data quality variants. We used the GATK recommended criteria for this (see Additional file 1). Relatedness of our participants was investigated using PLINK [32]. We then assessed variants for their potential pathogenicity and frequency, retaining for further analysis only variants that were rare. We considered a variant to be rare if its minor allele frequency was ≤1% in each of several public databases (see Additional file 1). We considered a variant to be potentially deleterious according to a score obtained from KGGSeq [33]. KGGSeq’s prediction algorithm makes use of available biological information (the mutation’s effect on the gene, i.e. stop gain or loss, frameshift, splice site, missense), as well as scoring from other publicly available prediction algorithms (PolyPhen-2, SIFT and others). KGGSeq scores were only used as an informative instrument, variants were not removed from the list of possible disease relevant candidates based on KGGSeq score alone.
De novo, homozygous and compound heterozygous mutations
Single nucleotide variants (SNVs) and small indels
Subsequent analysis of de novo and compound heterozygous, as well as homozygous, mutations was performed using KGGSeq [33]. For recessive disease models (homozygous and compound heterozygous) we only considered variants with a minor allele frequency (MAF) of less than 1%. We defined a de novo mutation as a first time genetic alteration of a specific locus in a proband. Compound heterozygous mutations were defined as the co-occurrence of two nonsynonymous alleles, one paternal, the other maternal, within a gene. The probability of de novo mutations in each gene was estimated using the framework of Samocha et al. [34].
These probabilities were used to guide the assessment of de novo mutations and not to filter variants. Since a similar framework was not available for compound heterozygous mutations we made use of the only large control trio dataset publicly available, the Genome of the Netherlands (GoNL) [35]. The GoNL is a population dataset containing 250 unaffected parents-offspring trios. We estimated the background compound heterozygous mutation rate per gene from the GoNL dataset. We prioritized compound heterozygous mutations found in our CRS cases in genes where the background rate was low. Thus we classed as a candidate risk locus any gene for which a recessive or de novo model could be constructed in any of our trios using the set of rare potentially deleterious variants we had identified. Detected de novo, compound heterozygous and homozygous mutations were validated via Sanger sequencing of trio DNA (i. e. genotypes were validated in both parents and child).
Analysis of kinship revealed misattributed paternity within one family (CR46). Hence the family CR46 was excluded from all family based analyses (de novo, compound heterozygous and homozygous mutation analysis).
Copy number variation
We investigated copy number variation (CNV) in the families CR5, CR17 and CR41 with Illumina’s HumanCoreExome-24 beadchip. Quality control of the assayed genotypes was performed using GenomeStudio (Illumina Inc.) using the default settings. CURR20 was also genotyped using CytoScan® HD Array, but failed initially quality control and was therefore excluded from the analysis. CNV calling and de novo CNV detection was performed using PennCNV [36]. We identified potentially disease associating CNVs as follows. We retained for further analysis, CNVs which allowed construction of a recessive disease model for any gene in any of our trios. We also retained de novo CNVs and rare CNVs. We deemed a CNV to be rare if it did not overlap with any CNV detected in the 1000 Genome Project. De novo CNVs were validated by quantitative real-time PCR (ABI Prism 7900 Sequence Detection System; Applied Biosystems) using TaqMan® Copy Number Assay (Catalog #: 4400291). Ensembl’s genome browser was used to determine genes or regulatory elements affected by the CNVs [37]. Initially we attempted to call CNVs from the exome sequencing data on our trios using three programs (EXCAVATOR [38], CoNIFER [39], and CONTRA [40]), but found no consistency between used tools. Similar previous studies have demonstrated limited power for CNV detection from exome sequencing data tools [41] (Table 1).
Results
After extensive quality control and MAF (MAF ≤ 1%) filtering we retained 229,849 variants of which 92.4% were known in dbSNP137. Hence we only retained variants below the frequency of 1% and those which are novel. Out of these, 7,442 missense, 184 frameshift, 227 nonframe-shift, 150 splicing, 173 stop-gain and 5 stop-loss variants in 3,872 different genes were analyzed in respect to de novo, compound heterozygous and homozygous mutations. Of these variants 236 were either heterozygogous or homozygous in all analyzed cases (see Additional file 2). Out of these, 226 were missense and 10 stop-gain variants distributed across 35 different genes. A further 65 of these variants were predicted to be damaging by KGGSeq. We identified two rare mutations in the known CRS related genes PTEN (rs202004587, missense, p.A79T) and VANGL1 (rs74117015, stop-gain, p.Ser338Ter) in CR5C inherited from the mother and father respectively (Table 2). Both were predicted to be damaging.
De novo variants
In total we identified three de novo mutations, two missense and one frameshift mutations in three different genes: MORN1 (p.Gly107Arg), SPTBN5 (p.Glu25Lys), and ZNF330 (p.Lys3fs) in patients CR41C, CR5C, and CURR20C respectively (Table 2). MORN1 encodes MORN (membrane occupation and recognition nexus) repeats [42]. The exact function of this gene is not known, however, in Toxoplasma gondii it is known to be involved in nuclear cell division [43]. Furthermore MORN repeats are known to be part of a number of genes, including junctophilins [44] which are involved in cardiomyopathy [45]. Notably, MORN1 was reported to be produced by insulin producing cells (IPCs) derived from pancreatic stem cells [46]. The estimated probability for a de novo mutation to occur in MORN1 is 0.8%, but 59% of all analyzed genes have a lower probability [34]. Pathogenicity analysis by KGGSeq suggests that the de novo mutation is damaging.
SPTBN5 (OMIM: 605916) is a beta-spectrin encoding protein. It plays an important role in linking proteins, lipids, and cytosolic factors of the cell membrane to the cytoskeletal filament systems of the cell [47]. SPTBN5 is expressed in the cerebellum, pancreas, kidney, and bladder, as well as in a number of other systems. The gene has not been associated with any disease or disorder. The estimated gene-specific probability of de novo mutation is 1.8% and 99% of genes have a lower probability of having a de novo mutation making this this gene less likely to be causally related. Further, KGGSeq’s pathogenic prediction algorithm suggests that this variant is benign.
ZNF330 (OMIM: 609550) is a zinc finger protein with no known disease association and is mainly present in the nucleus during interphase as well as at the centromeres during mitosis [48]. Interestingly, this gene is differentially expressed in pancreatic Islets of Langerhans and in peripheral blood mononuclear cells [49]. The estimated gene-based de novo mutation probability is 0.6%, relatively low but still within the 28th percentile of all genes. KGGSeq was unable to predict a pathogenic score for this variant.
Thus, given the pathogenic nature of the two de novo variants and their expression pattern in pancreatic cells, MORN1 and ZNF330 are candidate CRS genes.
We detected one de novo CNV deletion on 3q13.13 in CR5C (Table 3). The deletion does not seem to encompass any gene or functional element, yet it overlaps with CNVs previously reported in patients with a similar phenotype. In particular, a documented de novo deletion in a Japanese patient with OEIS (omphalocele, exstrophy of the cloaca, imperforate anus, spinal defects) complex who also had a sacrum malformation (DECIPHER: 971) overlaps with the de novo CNV identified in CR5C.
Homozygous and compound heterozygous mutations
In total we identified 8 compound missense heterozygous and one homozygous missense mutations (PDZD2) which passed the described filtering criteria (detailed in Table 2). Strikingly, mutations in genes related to diabetes were detected in two patients. None of the affected genes were recurrent. The two genes associated with diabetes were PDZD2 and CLTCL1 and were found mutated in CR5C and CR17C respectively.
PDZD2 (p.Ser1106Phe) has been shown to be an important promoter of fetal pancreatic progenitor cell proliferation [50, 51]. Ma et al. [52] showed that expression of PDZD2 is specific to pancreatic beta cells. Furthermore, higher concentrations of secreted PDZD2 in rat insulinoma cell lines were correlated with higher rates of cell proliferation and inhibited transcription of INS, an insulin promoter.
CLTCL1 (p.Arg1620His, p.Val44Phe) is involved in the intracellular trafficking of glucose transporter GLUT4. Intracellular trafficking of the glucose transporter GLUT4 from storage compartments to the plasma membrane is triggered in muscle and fat during the body’s response to insulin [53].
A compound heterozygous mutation in GLTSCR2 (Glioma Tumor Suppressor Candidate Region Gene 2) was identified in patient CR5C (p.Arg190Trp, p.Thr284Met). GLTSCR2, is expressed at high levels in pancreas and heart, is a tumor suppressor gene and a direct regulator of PTEN. Mutations of PTEN have been previously identified in a patient affected with VACTERL (Vertebral anomalies, Anal atresia, Cardiac defects, Tracheoesophageal fistula and/or Esophageal atresia, Renal & Radial anomalies and Limb defects) [22] which has commonalities with CRS [27]. This is especially interesting considering that patient CR5C is also harbouring a rare inherited mutation in PTEN itself.
A compound heterozygous mutation (p.Ala1616Thr, p.Thr3620Leu) in DNAH10, an inner arm dynein heavy chain, was identified in CR41C. Dynein proteins are implicated in many disorders such as motor neuropathies, cortical development diseases, as well as congenital malformations such as heterotaxia and situs inversus. Moreover, cytoplasmic Dyneins have been reported to interact with Kinesin (KIF1A, mutated in patient CR17C) for interkinetic nuclear migration in neural stem cells [54].
We detected a homozygous CNV deletion encompassing part of chromosome 8p23.2 in patient CR5C (Table 3). The CNV does not overlap with known genes but is contained within a duplication found in a patient with abnormal sacrum (DECIPHER: 271204). This documented patient, while also harboring another deletion (7q34–7q36.3), displayed a great variety of phenotypes including central hypertonia, hypermetropia, long thorax, narrow mouth, seizures, strabismus, and deep set eyes. Additional detected rare CNVs overlapped with a number of other genes, however, none were known to be related to CRS (see Additional file 1).
Discussion
Within this pilot study we have identified a number of novel risk loci potentially connected to CRS. We have also found a number of mutations for already known genetic risk factors [10]. Here we highlight these preliminary findings and discuss their relevance for future studies.
Foremost, all four patients were affected by a homozygous, compound heterozygous mutations or de novo variant in a diabetes-relevant (CLTCL1 and PDZD2) or pancreatic expressed (MORN1 and ZNF330) gene. While these results are not definitive it is in line with the increased CRS risk for children born to diabetic mothers. In addition, one de novo (chr3q13.13) and one homozygous CNV (chr8p23.2) overlap with CNVs reported in patients with similar phenotype. Identification of overlapping CNVs in patients with similar phenotype is the central aim of DECIPHER [55]. Since many patients with rare diseases harbor novel or extremely rare variants, it is crucial to accumulate evidence across patients in order to foster understanding of the disease. Furthermore, we identified a heterozygous mutation in GLTSCR2, a direct regulator of PTEN. PTEN has been previously associated with CRS-like phenotypes (VACTER) [10]. Interestingly, the same subject (CR5C) has also an inherited mutation within PTEN and the CRS-related gene VANGL1. These results further strengthen the role of PTEN and VANGL1 in CRS. Likewise, the identification of compound heterozygous mutations in DNAH10 and KIF1A could suggest an involvement of ciliary proteins.
There are, a number of limitations to our study. Our sample size is small, but to be expected given the disease prevalence and the costs of the genetic assays used. We were not able to identify recurrent affected genes across different patients. We did not look for mutations assuming other than recessive inheritance because the yield of true to false positives would be poor. Many types of genetic variation were not assayed, for instance we only assayed exomic SNVs. Lastly, models involving environment, such as gene-environment interactions within the utero, could not be investigated.
The diversity of identified potential disease mechanisms matches that of previous studies [10, 56–58] and also reflects the phenotypic diversity associated with CRS [56]. We previously showed that if a disorder is genetically complex, one should expect large genetic heterogeneity across patients [59]. Thus the number of candidate genes identified is not surprising and is similar to that reported for other complex rare genetic disorders [60]. While the presence of several possible disease mechanisms does not necessary suggest a multigenic model, the large number of candidate genes identified within this study, as well as those reported by others [23, 27, 61–64] suggests that CRS might be caused by a multitude of private genetic risk factors. This makes identification of a common underlying genetic architecture challenging. Furthermore, differences in the genetic etiology between humans and experimental organisms makes it difficult to investigate the exact causal mechanism. Many aspects of the disease are still poorly characterized, for example, disease prevalence. While some studies have estimated that 1 in 7,700 children might be affected [1], others suggest it might be as rare as 1 in 100,000 births [2]. This further complicates estimation of the number of disease causing mechanisms [59].
Conclusions
Despite the complexity of the phenotype, we were able to identify common genetic characteristics across patients, potentially causally related to the known risk factors and supposed disease etiology. Our data, although limited to a small group of patients, support a multigenic model for CRS. Future studies should consider larger accumulated samples across multiple centers in order to identify common genetic characteristics via whole genome or whole exome sequencing.
Abbreviations
- ARM:
-
Anorectal malformation
- CNV:
-
Copy number variation
- CRS:
-
Caudal regression
- Hh:
-
Hedgehog protein
- IPC:
-
Insulin producing cells
- MAF:
-
Minor allele frequency
- QC:
-
Quality control
- SEP:
-
Somatosensory evoked potential
- SNP:
-
Single nucleotide polymorphism
- SNV:
-
Single nucleotide variation
- WES:
-
Whole exome sequencing
References
Unsinn KM, Geley T, Freund MC, Gassner I. US of the spinal cord in newborns: spectrum of normal findings, variants, congenital anomalies, and acquired diseases. Radiographics. 2000;20:923–38.
Entezami M: Ultrasound Diagnosis of Fetal Anomalies. Thieme; 2003. https://www.amazon.com/gp/product/1588902129?ie=UTF8&tag=radiopaediaor-20&linkCode=as2&camp=1789&creative=9325&creativeASIN=1588902129.
Duhamel B. From the mermaid to anal imperforation: the syndrome of caudal regression*. Arch Dis Child. 1961;36:152–5.
Barkovich AJ, Raghavan N, Chuang S, Peck WW. The wedge-shaped cord terminus: a radiographic sign of caudal regression. AJNR Am J Neuroradiol. 1989;10:1223–31.
Tortori-Donati P, Fondelli MP, Rossi A, Raybaud CA, Cama A, Capra V. Segmental spinal dysgenesis: neuroradiologic findings with clinical and embryologic correlation. AJNR Am J Neuroradiol. 1999;20:445–56.
Cama A, Palmieri A, Capra V, Piatelli GL, Ravegnani M, Fondelli P. Multidisciplinary management of caudal regression syndrome (26 cases). Eur J Pediatr Surg. 1996;6 Suppl 1:44–5.
Pang D, Hoffman HJ, Menezes AH. Sacral agenesis and caudal spinal cord malformations. Neurosurgery. 1993;32:755–79.
Passarge E, Lenz W. Syndrome of caudal regression in infants of diabetic mothers: observations of further cases. Pediatrics. 1966;37:672–5.
Reece EA, Hobbins JC. Diabetic embryopathy: pathogenesis, prenatal diagnosis and prevention. Obstet Gynecol Surv. 1986;41:325–35.
Semba K. Etiology of Caudal Regression Syndrome. Hum Genet Embryol. 2013;3:1–7.
Persaud O. Maternal diabetes and the consequences for her offspring. J Devel Disabil. 2007;13:101–33.
Boskovic R, Feig DS, Derewlany L, Knie B, Portnoi G, Koren G. Transfer of insulin lispro across the human placenta: In vitro perfusion studies. Diabetes Care. 2003;26:1390–4.
Challier JC, Hauguel S, Desmaizieres V. Effect of insulin on glucose uptake and metabolism in the human placenta. J Clin Endocrinol Metab. 1986;62:803–7.
Hagan DM, Ross AJ, Strachan T, Lynch SA, Ruiz-Perez V, Wang YM, Scambler P, Custard E, Reardon W, Hassan S, Nixon P, Papapetrou C, Winter RM, Edwards Y, Morrison K, Barrow M, Cordier-Alex MP, Correia P, Galvin-Parton PA, Gaskill S, Gaskin KJ, Garcia-Minaur S, Gereige R, Hayward R, Homfray T. Mutation analysis and embryonic expression of the HLXB9 currarino syndrome gene. Am J Hum Genet. 2000;66:1504–15.
Garcia-Barceló M, So M-T, Lau DK-C, Leon TY-Y, Yuan Z-W, Cai W-S, Lui VC-H, Fu M, Herbrick J-A, Gutter E, Proud V, Li L, Pierre-Louis J, Aleck K, van Heurn E, Belloni E. Population differences in the polyalanine domain and 6 new mutations in HLXB9 in patients with currarino syndrome. Clin Chem. 2006;52:46–52.
Wang Y, Wu Y. A novel HLXB9 mutation in a chinese family with currarino syndrome. Eur J Pediatr Surg. 2012;22:243–5.
Ross AJ, Ruiz-Perez V, Wang Y, Hagan DM, Scherer S, Lynch SA, Lindsay S, Custard E, Belloni E, Wilson DI, Wadey R, Goodman F, Orstavik KH, Monclair T, Robson S, Reardon W, Burn J, Scambler P, Strachan T. A homeobox gene, HLXB9, is the major locus for dominantly inherited sacral agenesis. Nat Genet. 1998;20:358–61.
Kim AY, Yoo S-Y, Kim JH, Eo H, Jeon TY. Currarino syndrome: variable imaging features in three siblings with HLXB9 gene mutation. Clin Imaging. 2013;37:398–402.
Belloni E, Martucciello G, Verderio D, Ponti E, Seri M, Jasonni V, Torre M, Ferrari M, Tsui LC, Scherer SW. Involvement of the HLXB9 homeobox gene in currarino syndrome. Am J Hum Genet. 2000;66:312–9.
Kibar Z, Torban E, McDearmid JR, Reynolds A, Berghout J, Mathieu M, Kirillova I, De Marco P, Merello E, Hayes JM, Wallingford JB, Drapeau P, Capra V, Gros P. Mutations in VANGL1 associated with neural-tube defects. N Engl J Med. 2007;356:1432–7.
Garcia-Barceló M-M, Wong KK, Lui VC, Yuan Z, So M, Ngan ES, Miao X, Chung PH, Khong P, Tam PK. Identification of a HOXD13 mutation in a VACTERL patient. Am J Med Genet A. 2008;146A:3181–5.
Reardon W, Zhou XP, Eng C. A novel germline mutation of the PTEN gene in a patient with macrocephaly, ventricular dilatation, and features of VATER association. J Med Genet. 2001;38:820–3.
Padmanabhan R. Retinoic acid-induced caudal regression syndrome in the mouse fetus. Reprod Toxicol. 1998;12:139–51.
Rojansky N, Fasouliotis SJ, Ariel I, Nadjari M. Extreme caudal agenesis. Possible drug-related etiology? J Reprod Med. 2002;47:241–5.
Young T, Rowland JE, van de Ven C, Bialecka M, Novoa A, Carapuco M, van Nes J, de Graaff W, Duluc I, Freund J-N, Beck F, Mallo M, Deschamps J. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev Cell. 2009;17:516–26.
Greco TL, Takada S, Newhouse MM, McMahon JA, McMahon AP, Camper SA. Analysis of the vestigial tail mutation demonstrates that Wnt-3a gene dosage regulates mouse axial development. Genes Dev. 1996;10:313–24.
Szumska D, Pieles G, Essalmani R, Bilski M, Mesnard D, Kaur K, Franklyn A, El Omari K, Jefferis J, Bentham J, Taylor JM, Schneider JE, Arnold SJ, Johnson P, Tymowska-Lalanne Z, Stammers D, Clarke K, Neubauer S, Morris A, Brown SD, Shaw-Smith C, Cama A, Capra V, Ragoussis J, Constam D, Seidah NG, Prat A, Bhattacharya S. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 2008;22:1465–77.
Catala M. Genetic control of caudal development. Clin Genet. 2002;61:89–96.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013. p. 0:3.
Picard [http://broadinstitute.github.io/picard/]. Accessed 22 Nov 2015.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Li M-X, Gui H-S, Kwan JSH, Bao S-Y, Sham PC. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 2012;40:e53.
Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki J a, Rehnström K, Mallick S, Kirby A, Wall DP, MacArthur DG, Gabriel SB, DePristo M, Purcell SM, Palotie A, Boerwinkle E, Buxbaum JD, Cook EH, Gibbs R a, Schellenberg GD, Sutcliffe JS, Devlin B, Roeder K, Neale BM, Daly MJ. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–50.
The Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46(8):818–25.
Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF A, Hakonarson H, Bucan M. PennCNV: an integrated hidden markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–74.
Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, Gil L, Girón CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Kähäri AK, Keenan S, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Overduin B, Parker A, Patricio M, Perry E, Pignatelli M, et al. Ensembl 2015. Nucleic Acids Res. 2014;43:D662–669.
Magi A, Tattini L, Cifola I, D’Aurizio R, Benelli M, Mangano E, Battaglia C, Bonora E, Kurg A, Seri M, Magini P, Giusti B, Romeo G, Pippucci T, De Bellis G, Abbate R, Gensini GF. EXCAVATOR: detecting copy number variants from whole-exome sequencing data. Genome Biol. 2013;14:R120.
Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–32.
Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, Tothill RW, Halgamuge SK, Campbell IG, Gorringe KL. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–13.
Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, Guo H, Jiang Q, Allen AS, Zhu M. An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat. 2014;35:899–907.
Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P, Guyer M, Peck AM, Derge JG, Lipman D, Collins FS, Jang W, Sherry S, Feolo M, Misquitta L, Lee E, Rotmistrovsky K, Greenhut SF, Schaefer CF, Buetow K, Bonner TI, Haussler D, Kent J, Kiekhaus M, Furey T, Brent M, et al. The status, quality, and expansion of the NIH full-length cDNA project: the mammalian gene collection (MGC). Genome Res. 2004;14:2121–7.
Ferguson DJP, Sahoo N, Pinches RA, Bumstead JM, Tomley FM, Gubbels M-J. MORN1 has a conserved role in asexual and sexual development across the apicomplexa. Eukaryot Cell. 2008;7:698–711.
Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22.
Matsushita Y, Furukawa T, Kasanuki H, Nishibatake M, Kurihara Y, Ikeda A, Kamatani N, Takeshima H, Matsuoka R. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet. 2007;52:543–8.
Ebrahimie M, Esmaeili F, Cheraghi S, Houshmand F, Shabani L, Ebrahimie E. Efficient and simple production of insulin-producing cells from embryonal carcinoma stem cells using mouse neonate pancreas extract, as a natural inducer. PLoS One. 2014;9:e90885.
Stabach PR, Morrow JS. Identification and characterization of beta V spectrin, a mammalian ortholog of Drosophila beta H spectrin. J Biol Chem. 2000;275:21385–95.
Bolívar J, Díaz I, Iglesias C, Valdivia MM. Molecular cloning of a zinc finger autoantigen transiently associated with interphase nucleolus and mitotic centromeres and midbodies. Orthologous proteins with nine CXXC motifs highly conserved from nematodes to humans. J Biol Chem. 1999;274:36456–64.
Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H, Degenhardt J, Mayba O, Gnad F, Liu J, Pau G, Reeder J, Cao Y, Mukhyala K, Selvaraj SK, Yu M, Zynda GJ, Brauer MJ, Wu TD, Gentleman RC, Manning G, Yauch RL, Bourgon R, Stokoe D, Modrusan Z, Neve RM, de Sauvage FJ, Settleman J, Seshagiri S, Zhang Z. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol. 2014;33:306–12.
Tsang SW, Shao D, Cheah KSE, Okuse K, Leung PS, Yao K-M. Increased basal insulin secretion in Pdzd2-deficient mice. Mol Cell Endocrinol. 2010;315:263–70.
Suen PM, Zou C, Zhang YA, Lau TK, Chan J, Yao KM, Leung PS. PDZ-domain containing-2 (PDZD2) is a novel factor that affects the growth and differentiation of human fetal pancreatic progenitor cells. Int J Biochem Cell Biol. 2008;40:789–803.
Ma RYM, Tam TSM, Suen APM, Yeung PML, Tsang SW, Chung SK, Thomas MK, Leung PS, Yao K-MM. Secreted PDZD2 exerts concentration-dependent effects on the proliferation of INS-1E cells. Int J Biochem Cell Biol. 2006;38:1015–22.
Vassilopoulos S, Esk C, Hoshino S, Funke BH, Chen C-Y, Plocik AM, Wright WE, Kucherlapati R, Brodsky FM. A role for the CHC22 clathrin heavy-chain isoform in human glucose metabolism. Science. 2009;324:1192–6.
Tsai J-W, Lian W-N, Kemal S, Kriegstein AR, Vallee RB. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci. 2010;13:1463–71.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP, Lu X, Shaw CA, Patel A, Li J, Cooper ML, Wells WR, Sullivan CM, Sahoo T, Yatsenko SA, Bacino CA, Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, Fiegler H, Firth H, Sanlaville D, Winter R, et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet. 2009;84:524–33.
Singh SK, Singh RD, Sharma A. Caudal regression syndrome--case report and review of literature. Pediatr Surg Int. 2005;21:578–81.
Kaciński M, Jaworek M, Skowronek-Bała B. Caudal regression syndrome associated with the white matter lesions and chromosome 18p11.2 deletion. Brain Dev. 2007;29:164–6.
Chan BWH, Chan K-S, Koide T, Yeung S-M, Leung MBW, Copp AJ, Loeken MR, Shiroishi T, Shum ASW. Maternal diabetes increases the risk of caudal regression caused by retinoic acid. Diabetes. 2002;51:2811–6.
Campbell DD, Porsch RM, Cherny SS, Capra V, Merello E, De Marco P, Sham PC, Garcia-Barceló M-M. Cost effective assay choice for rare disease study designs. Orphanet J Rare Dis. 2015;10:10.
Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14:681–91.
Adra A, Cordero D, Mejides A, Yasin S, Salman F, O’Sullivan MJ. Caudal regression syndrome: etiopathogenesis, prenatal diagnosis, and perinatal management. Obstet Gynecol Surv. 1994;49:508–16.
Vlangos C, O’Connor B, Morley M. Caudal regression in adrenocortical dysplasia (acd) mice is caused by telomere dysfunction with subsequent p53-dependent apoptosis. Dev. 2009;334:418–28.
Knight B. Caudal regression syndrome: a case report. AANA J. 2011;79:281–2.
Evans JA, Vitez M, Czeizel A. Congenital abnormalities associated with limb deficiency defects: a population study based on cases from the hungarian congenital malformation registry (1975–1984). Am J Med Genet. 1994;49:52–66.
Hogan MC, Griffin MD, Rossetti S, Torres VE, Ward CJ, Harris PC. PKHDL1, a homolog of the autosomal recessive polycystic kidney disease gene, encodes a receptor with inducible T lymphocyte expression. Hum Mol Genet. 2003;12:685–98.
Yim J-H, Kim Y-J, Ko J-H, Cho Y-E, Kim S-M, Kim J-Y, Lee S, Park J-H. The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death. Cell Death Differ. 2007;14:1872–9.
Rivire JB, Ramalingam S, Lavastre V, Shekarabi M, Holbert S, Lafontaine J, Srour M, Merner N, Rochefort D, Hince P, Gaudet R, Mes-Masson AM, Baets J, Houlden H, Brais B, Nicholson GA, Van Esch H, Nafissi S, De Jonghe P, Reilly MM, Timmerman V, Dion PA, Rouleau GA. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet. 2011;89:219–301.
Nahorski MS, Al-Gazali L, Hertecant J, Owen DJ, Borner GHH, Chen Y-C, Benn CL, Carvalho OP, Shaikh SS, Phelan A, Robinson MS, Royle SJ, Woods CG. A novel disorder reveals clathrin heavy chain-22 is essential for human pain and touch development. Brain. 2015;138:2147–60.
Landstrom AP, Weisleder N, Batalden KB, Bos JM, Tester DJ, Ommen SR, Wehrens XHT, Claycomb WC, Ko J-K, Hwang M, Pan Z, Ma J, Ackerman MJ. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42:1026–35.
Maiti AK, Mattéi MG, Jorissen M, Volz A, Zeigler A, Bouvagnet P. Identification, tissue specific expression, and chromosomal localisation of several human dynein heavy chain genes. Eur J Hum Genet. 2000;8:923–32.
Huizing M, Didier A, Walenta J, Anikster Y, Gahl WA, Krämer H. Molecular cloning and characterization of human VPS18, VPS 11, VPS16, and VPS33. Gene. 2001;264:241–7.
Acknowledgements
We thank all participants who made this study possible. We also thank the A.S.B.I. (Associazione Spina Bifida Italia).
Funding
Seed Funding Programme for Basic Research, University of Hong Kong, Project Code:201410159002 to MMGB. Small Project Funding, University of Hong Kong, Project Code 201209176012 to DC. EM; PDM, VC would like to acknowledge Ricerca Corrente Ministero della Salute Italia 5× Mille, Aletti- Volpati Trust Onlus and private funding resources.
Availability of data and materials
All called variants are included in the suplementary material.
Authors’ contributions
RMP, DDC, SSC, GC, MS and MMGB analyzed the data and drafted the paper. VC, EM, PDM and LR played a major role in collecting the samples and phenotyping the patients. MS performed most of the laboratory work. PCS and PKT reviewed the study proposal, provided feedback on the study progress and mansucript. MMGB proposed the study idea. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Ethical Commitee Istituto Giannina Gaslini (IGG-VACA 213/2013), the CEIm HM Hospitales, and the institutional review board of The University of Hong Kong together with the Hong Kong Hospital Authority (Institutional Review Board: UW 10–399). Written informed consent was obtained from all patients and parents.
Author information
Authors and Affiliations
Corresponding authors
Additional files
Additional file 1:
Supplementary Methods and Material. (DOCX 51 kb)
Additional file 2:
All called variants after QC and MAF. (XLSX 1329 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Porsch, R.M., Merello, E., De Marco, P. et al. Sacral agenesis: a pilot whole exome sequencing and copy number study. BMC Med Genet 17, 98 (2016). https://doi.org/10.1186/s12881-016-0359-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12881-016-0359-2