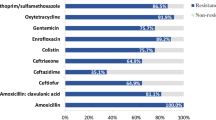
Abstract
Background
Colistin is one of the last resort therapeutic options for treating carbapenemase-producing Enterobacterales, which are resistant to a broad range of beta-lactam antibiotics. However, the increased use of colistin in clinical and livestock farming settings in Thailand and China, has led to the inevitable emergence of colistin resistance. To better understand the rise of colistin-resistant strains in each of these settings, we characterized colistin-resistant Enterobacterales isolated from farmers, swine, and hospitalized patients in Thailand.
Methods
Enterobacterales were isolated from 149 stool samples or rectal swabs collected from farmers, pigs, and hospitalized patients in Thailand between November 2014–December 2017. Confirmed colistin-resistant isolates were sequenced. Genomic analyses included species identification, multilocus sequence typing, and detection of antimicrobial resistance determinants and plasmids.
Results
The overall colistin-resistant Enterobacterales colonization rate was 26.2% (n = 39/149). The plasmid-mediated colistin-resistance gene (mcr) was detected in all 25 Escherichia coli isolates and 9 of 14 (64.3%) Klebsiella spp. isolates. Five novel mcr allelic variants were also identified: mcr-2.3, mcr-3.21, mcr-3.22, mcr-3.23, and mcr-3.24, that were only detected in E. coli and Klebsiella spp. isolates from farmed pigs.
Conclusion
Our data confirmed the presence of colistin-resistance genes in combination with extended spectrum beta-lactamase genes in bacterial isolates from farmers, swine, and patients in Thailand. Differences between the colistin-resistance mechanisms of Escherichia coli and Klebsiella pneumoniae in hospitalized patients were observed, as expected. Additionally, we identified mobile colistin-resistance mcr-1.1 genes from swine and patient isolates belonging to plasmids of the same incompatibility group. This supported the possibility that horizontal transmission of bacterial strains or plasmid-mediated colistin-resistance genes occurs between humans and swine.
Similar content being viewed by others
Introduction
Antimicrobial resistance is recognized as one of the most significant global health concerns [1]. Carbapenemase-producing Enterobacterales (CPE) are resistant to a broad range of beta-lactam antibiotics, including carbapenems [2], leaving limited treatment options to treat infections caused by CPE. In general, colistin is used as a monotherapy or in combination with other antibiotics to treat CPE infections [3]. Consequently, the increase in CPE infections has resulted in the increased use of colistin, with the inescapable risk of emerging colistin resistance [4]. Notably, because some countries, such as China and Thailand, have widely used colistin in livestock for the treatment and prevention of infection, the emergence of colistin resistance may have been additionally promoted by the inappropriate use of human antibiotics in farm animals [5, 6].
Colistin is a polypeptide antibiotic belonging to a group of polymyxins with broad-spectrum activity against Gram-negative bacteria. It binds to the lipopolysaccharide (LPS) of bacteria, resulting in the leakage of intracellular components from the cell membrane followed by bacterial death [7]. Before 2015, the most commonly reported mechanisms of colistin resistance were those mediated by chromosome-encoded mutations in the pmrAB and phoPQ genes or by mgrB gene inactivation leading to lipid A modification and subsequent interference in the colistin interaction [8]. In 2015, the first plasmid-mediated colistin resistance gene, mcr-1, was reported in China [4]. The protein encoded by mcr-1 functions by transferring a phosphoethanolamine residue to lipid A, altering its structure and thereby lowering its affinity for colistin [9]. To date, mcr-1-containing isolates have been detected for several Enterobacterales species found in food-chain production, humans, and the environment worldwide [10, 11]. According to one retrospective study in China, the mcr-1 gene was initially identified from Escherichia coli in chickens in the 1980s [12]. Recently, novel mcr family genes, mcr-2 to mcr-10, have been reported across a range of Enterobacterales [13]. The discovery of horizontal transfer of colistin resistance triggered concerns about the impact of colistin use on the spread of colistin resistance in the animal production industry, especially regarding swine [14]. In fact, a possible link between swine and farmers in terms of colistin-resistant E. coli after direct contact, i.e., horizontal transmission, was reported in Laos in 2012 [15]. These findings suggest the possible loss of colistin efficacy for the treatment of multidrug-resistant Gram-negative bacteria, such as CPE, in hospitalized patients. Thus, it is imperative to understand the genomic epidemiology between food-production animals and humans.
The aim of the present study was to determine the molecular epidemiology of mcr-mediated colistin resistance genes and their genetic environment in Escherichia coli and Klebsiella pneumoniae in farmers, livestock, and hospitalized patients in Thailand between 2014 and 2017.
Methods
Ethics statement
The study protocol was approved by the Siriraj Institutional Review Board (Si571/2015 and Si680/2016). Written informed consent was obtained from all patients. The data extracted from the medical records were de-identified to protect patients’ confidentiality. Ethical review and approval was not required for the animal study because all stool or rectal swab samples were submitted from farmers and pigs in industrial field to the diagnostic laboratory as the annual surveillance. All methods were carried out in accordance with relevant guidelines and regulations such as the Declaration of Helsinki, the Belmont Report, CIOMS guidelines, and ICH-GCP guidelines.
Population and specimen collection
Stool samples were collected from 83 hospitalized patients who were admitted for treatment in the internal medicine wards of Siriraj Hospital, Bangkok between December 2015 and June 2017, and from 28 healthy food-production animal farm workers in a northern province. One rectal swab was taken from each of 38 randomly-chosen healthy pigs raised on the same farm at which the farmers worked. The stool or rectal swab samplings of pigs and farmers were collected and submitted to the diagnostic laboratory by the farmers working at their industrial farms as a routine annual surveillance. These samples were collected between 1 November 2014 and 31 December 2017. Stool samples and rectal swabs from patients, farmers and swine were inoculated onto MacConkey agar (BD, USA) and any bacterial colonies observed after incubation at 35 °C for 24 h were subcultured for colony purification. Bacteria species were identified using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) on a Bruker Microflex LT/SH instrument (Bruker, USA) in accordance with the manufacturer’s protocol.
Antimicrobial susceptibility testing
Antibiotic susceptibility was determined by disk diffusion method. The tested antibiotics were cefoxitin, ceftriaxone, ertapenem, imipenem, meropenem, gentamicin, amikacin, nalidixic acid, ciprofloxacin, tetracycline, erythromycin, piperacillin/tazobactam, cefoperazone/sulbactam and colistin (Thermo Fisher Scientific, USA). Colistin resistance was further confirmed by the broth microdilution method. The minimum inhibitory concentrations (MICs) of colistin were determined by the broth microdilution in cation-adjusted Mueller–Hinton II broth according to Clinical and Laboratory Standards Institute (CLSI) 2017 guidelines [16]. E. coli ATCC 25922 was used as a control and a range of colistin dilutions (Chem-Impex Int’l Inc., USA) between 0.25 mg/L and 128 mg/L were performed. Breakpoints of colistin susceptibility defined by European Committee on Antimicrobial Susceptibility Testing (EUCAST) [17] were used as follows: isolates with a colistin MIC ≤ 2 mg/L were categorised as susceptible, and those with a colistin MIC > 2 mg/L were categorised as resistant.
DNA extraction and whole-genome sequencing
DNA was prepared using the Wizard® Genomic DNA Purification Kit (Promega, USA). Paired-end 150-basepair Nextera XT libraries of whole genomic DNA were sequenced on the Illumina NextSeq® 500 platform (Illumina, USA) with a target average coverage of 100-fold was performed on the 39 colistin-resistant Thailand isolates. Twelve mcr-positive isolates were selected for long-read sequencing using high molecular-weight genomic DNA and the rapid barcoding kit (SQK-RBK004) to prepare libraries, which then were sequenced on a MinION R9.4 flow cell (Oxford Nanopore, UK) with a target average coverage of 20-fold to improve the respective draft genome assemblies.
Bioinformatics analyses
All raw reads were de novo assembled using SPAdes v3.1.1 [18]. A subset of 12 genomes that also had long reads were assembled combining Illumina and Oxford Nanopore sequencing reads using Unicycler v0.4.7 [19]. Hybrid assemblies were iteratively polished using Illumina reads with Pilon v1.22 [20]. All assembled genomes were assigned bacterial species based on an average nucleotide identity (ANI) by comparing assembled genomes to all-type strain genome assemblies in GenBank using MASH v.1.1.1 [21].
A whole genome alignment was inferred from single nucleotide polymorphisms (SNPs) identified by the Northern Arizona SNP Pipeline (NASP) v1.0.2 [22]. NASP output files were run through snpEff [23], which provided variant annotations and predicted the effect of the SNP on the gene’s protein product. Additionally, the resulting NASP alignment was run through Gubbins v.2.2.1 [24] to filter out the effects of recombination before building maximum-likelihood phylogenetic tree using RAxML v8.2.12 [25] under the GTRCAT model with 100 bootstrap replicates. The resulting tree was rendered with metadata annotated using the Interactive Tree of Life v4 (iTOL) [26].
To determine phylogenetic relationship between identified novel variants of MCR proteins and previously published MCR protein variants, we followed the proposal for assignment of allele numbers for mcr-genes [27] and selected representative protein sequences of MCR-1.1 through MCR-7.1 from GenBank; additionally including sequences of MCR-8.1 (NG_061399, [28]) and MCR-8.2 (AXU00196, [29]). MEGA7 [30] was used to generate a MCR protein phylogeny tree.
To identify chromosomal colistin resistance variants, the following method was used: colistin-susceptible K. pneumoniae MGH 78578 (RefSeq accession no. CP000647-CP000652) was used as a reference for variant identification. The amino acid sequences of designated targets were aligned using Clustal Omega [31]. Jalview [32] was used to visualise the alignments and identify substitutions. Subsequently, substitutions identified among all colistin-resistant isolates were filtered against those found in colistin-susceptible isolates to sort alterations unique to colistin-resistant isolates.
Multi locus sequence typing (MLST) for K. pneumoniae and E. coli genomes was determined using LOCUST [33] and the following MLST databases: for K. pneumoniae - Pasteur Institut (http://bigsdb.pasteur.fr/klebsiella/) and for E. coli - Achtman MLST schemes (https://enterobase.warwick.ac.uk/species/ecoli/download_7_gene). Beta-lactamase and mcr genes were identified using The Resistance Gene Identifier (RGI) [34], with default parameters from the Comprehensive Antibiotic Resistance Database (CARD) [35]. LOCUST was used to identify mcr gene variants, and novel allele variants were appropriately named [27]. Identification of plasmid replicons was done using PlasmidFinder v2.0.1 [36].
Nucleotide sequence accession numbers
All sequencing reads were deposited in the SRA database under the project accession number PRJNA389557, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA389557. Relevant assemblies and mcr-genes have been deposited in GenBank under accession numbers as shown in Suppl. Tables S1 and S2).
Results
Description of colistin-resistant isolates
A total of 149 stool samples or rectal swab samples were collected from hospitalized patients (n = 83), healthy farm workers (n = 28) and swine (n = 38) between November 2014 and December 2017. The most prevalent bacteria found in these samples were E. coli (50.3%; n = 75/149) followed by K. pneumoniae (46.3%; n = 69/149) and K. quasipneumoniae subsp. similipneumoniae (3.4%; n = 5/149). A total of 39 (26.2%) bacterial isolates were identified as colistin-resistant and were further analyzed in detail in this study (Table 1). The prevalence of colistin resistant isolates was 12.1% (n = 10/83) in hospitalized patients, 10.7% (n = 3/28) in farmers and 68.4% (n = 26/38) in swine.
Seventy-five E. coli isolates were isolated from 29 swine, 23 patients and 23 farmers. Twenty five out of 75 E. coli isolates were colistin resistant (17 were from swine, 5 from patients and 3 from farmers) (Table 1). The prevalence of colistin-resistant E. coli was 58.6% (n = 17/29) in swine, 21.7% (n = 5/23) in patients and 13% (n = 3/23) in farmers, respectively. The 25 colistin-resistant E. coli isolates could be differentiated into 18 distinct sequence types (STs): ST10 (n = 3), ST34 (n = 2), and ST48 (n = 2), all of which belong to clonal complex 10 (CC10) and are frequently obtained from farmed animals, along with ST165 (n = 2), ST206 (n = 2), ST3054 (n = 2), and 12 other STs represented by single isolates (Table 1). No single E. coli genotype was shared between isolates collected from farmers, swine, and patients. Among the 25 colistin-resistant E. coli isolates, the colistin MIC range was 4–16 mg/L (Table 1). None of the E. coli isolates were resistant to ertapenem (Table 1).
The remaining 14 colistin-resistant isolates were Klebsiella spp., 11 were K. pneumoniae and three were Klebsiella quasipneumoniae subsp. similipneumoniae (Table 1). Five of the 11 colistin-resistant K. pneumoniae isolates were obtained from hospitalized patients, while the rest of the K. pneumoniae and K. quasipneumoniae subsp. similipneumoniae isolates were obtained from healthy swine; no colistin-resistant Klebsiella spp. were isolated from farmers (Table 1). Four out of five K. pneumoniae isolates from patients were ST16, which is known to be a widely-distributed disease-causing K. pneumoniae clone [37]. Among the 14 colistin-resistant Klebsiella spp. isolates, the colistin MIC range was 4–64 mg/L (Table 1). Notably, all five K. pneumoniae isolates from patients were also resistant to ertapenem (Table 1).
Mechanisms of colistin resistance
The mcr gene was detected in 87.2% (n = 34/39) of the isolates resistant to colistin. All of the E. coli isolates (n = 25) and 64.3% (n = 9/14) of the Klebsiella spp. isolates had mcr gene(s) present. Among the E. coli isolates, mcr-3 was more common in swine (94.1%, n = 16/17) than in humans (37.5%, n = 3/8), whereas mcr-1 was more common in humans (87.5%, n = 7/8) than in swine (52.9%, n = 9/17). Interestingly, 50% of E. coli isolates (n = 13/25) were found to contain two mcr genes, with 12 isolates (9 from swine, 2 from farmers and 1 from patients) coharboring mcr-1 and mcr-3 and one isolate from swine coharboring mcr-2 and mcr-3 (Table 1). All nine Klebsiella spp. isolates carried the mcr-3 allelic variant were from swine (Table 1).
Five novel allelic variants of mcr were identified among the mcr-positive E. coli and Klebsiella spp. swine isolates: mcr-2.3 (Accession: NG_065452), mcr-3.21(Accession: QDJ85872), mcr-3.22 (Accession: QDJ80325), mcr-3.23 (Accession: NG_060583), and mcr-3.24 (Accession: NG_060580) (Fig. 1). A novel mcr-2 variant, mcr-2.3 found in E. coli ECCTRSRTH05, was 98.3% similar to a reported mcr-2 gene [38] with only nine amino acid changes in the encoded protein (Suppl. Table S3). Other four novel variants of mcr-3 were identified in both E. coli and Klebsiella spp., with an average 99.7% identity to a reported mcr-3 gene [39] with only one to two amino acid differences in the encoded protein (Suppl. Table S3). Four Klebsiella spp. isolates that had the same novel variant mcr-3.22 were isolated from swine, but they belonged to different genotypes (Table 1), excluding the possibility of clonal expansion. E. coli isolates carrying mcr-1.1 and/or variants of mcr-3 did not exhibit higher levels of MIC to colistin (MIC ranged from 4 to 16 µg/ml) when compared to isolates that carried only one copy of the mentioned mcr genes. This finding suggests that the co-occurrence of mcr-1.1 and variants of mcr-3 might not provide a synergistic or an additive effect on the colistin-resistant phenotype. Additionally, the coharboring of mcr-1.1 or variants of mcr-3 and beta-lactamase (bla) genes was observed in the E. coli isolates from farmers and swine (Table 1). The most common bla genes observed in these isolates were blaCTX−M−55 and blaTEM−1 (Table 1).
Relationship between the sequences of publicly available and novel MCR proteins. The UPGMA tree was constructed, using MUSCLE, from a full-length protein alignment of 47 publicly available and 5 novel mcr variants identified in this study. The scale represents the sum of mismatches over the total length of the aligned sequence
Protein comparisons of the MCR protein (Fig. 1) showed the clustering of alleles based on their root gene; on average, the mcr-3 and mcr-2 alleles differed by an overall mean distance of 0.0345 and 0.01498, respectively, as computed using a Poisson model implemented in MEGA7 [30]. All other alleles had an average mean distance of 0.00529 within their clusters, indicating that mcr-3 is currently the most diverse mcr allele (Fig. 1). Notably, a second copy of mcr-3.21 was identified in KPCTRSRTH04 on a potential chromosomally encoded 2.8-Mbp contig, and KPCTRSRTH06 was found to contain a second copy of mcr-3 that is pseudogenized via a frameshift mutation, likely due to relaxed selection via the acquisition of a second mcr-3 copy (Table 1).
A diverse set of 12 plasmids containing mcr-1- and mcr-3 genes was identified in the E. coli and K. pneumoniae isolates (Table 1). The mcr-1.1 genes were predominantly found in isolates with IncX4 and IncI2 plasmids, while mcr-3 alleles were predominantly detected on IncFIB, IncFII, and IncR plasmids in swine isolates and the IncFII plasmid in E. coli isolated from hospitalized patients. Interestingly, the mcr-2.3 gene of ECCTRSRTH05 was identified on a potential chromosomally-encoded 2.7-Mbp contig. The IncFII (pMET) plasmid (EU383016) harboring mcr-3.21 was identified in four Klebsiella spp. isolates (Table 1). The mcr-3.22-containing plasmid in isolates KPCTRSRTH01 and KPCTRSRTH05 belonged to an unknown incompatibility group, with KPCTRSRTH01 being most similar to the mcr-3.11-containing IncR plasmid (MH341574.1) reported from China. Notably, two copies of mcr-3.21 were identified in KPCTRSRTH04. The first copy was located on the IncFII (pMET) plasmid (EU383016) and the second copy was located on a 2.8 Mbp contig, indicating that the second copy was chromosomally encoded (Table 1). The mcr-3 pseudogene was identified on the first plasmid belonging to IncFIB (JN420336) in KPCTRSRTH06 (Table 1).
In our study, none of the five K. pneumoniae isolates from patients had mcr genes, even though these isolates were highly resistant to colistin (≥ 32 mg/L) (Table 1). A detailed genomic interrogation of these genomes identified multiple mutations in the chromosomal genes, including phoPQ, mgrB, pmrCAB, pmrE and arnBCADTEF genes and intergenic regions that could potentially be associated with colistin resistance (Suppl. Table S4). Previously reported disruptions and mutations in mgrB [8, 40] and pmrB (D150Y) [41] conferring colistin resistance were observed in four of the five isolates: KPCTRPRTH01 (mgrB: Q30*), KPCTRPRTH02 (pmrB: D150Y), KPCTRPRTH03 (mgrB: W20*), and KPCTRPRTH04 (pmrB: D150Y) (Suppl. Table S4). In the two isolates with the pmrB (D150Y) mutation, KPCTRPRTH02 and KPCTRPRTH04, a pmrA pseudogene, affecting the transcriptional regulatory domain reported to increase colistin resistance [42], was also observed (Suppl. Table S4). In isolate KPCTRPRTH05, no previously identified mutations were detected. Instead, a novel S60L mutation in uncharacterized protein YcaR was identified. Located within the KDO2-lipid A biosynthesis gene cluster and co-transcribed with kdsB [43], ycaR could serve as a novel candidate gene that potentially influences colistin resistance. Additionally, an interrogation of 84 publicly available Klebsiella spp. genomes that share the same ST16 (n = 54) and ST231 (n = 30) profile as the Thailand isolates revealed only five genomes not isolated in Thailand with a similar mgrB disruption (ST16, n = 2; ST231, n = 3). All other identified mutations were unique to these STs, suggesting that new resistant mechanisms potentially have emerged in these lineages.
Phylogenetic relationship of colistin-resistant K. pneumoniae isolates
Phylogenetic analysis of the eleven colistin-resistant K. pneumoniae isolates from this study revealed high genomic diversity, with only the ST16 isolates (n = 4) clustering together and the pairwise SNP distance between any two of the ST16 isolates averaging 0.018% or 918 SNP sites (range: 0.006% or 306 SNP sites to 0.032% or 1,633 SNP sites) (Fig. 2a). None of the K. pneumoniae isolates with mcr-3 gene variants from swine clustered together (Fig. 2a), confirming that they were not the result of clonal expansion. We further compared the 11 K. pneumoniae genomes from our dataset with 117 publicly available colistin-resistant K. pneumoniae genomes to investigate the geographical spread of mcr-positive isolates (Fig. 2b). Isolates from this study were more likely to cluster together with isolates previously reported from Thailand and China, as well as a few isolates from the USA (Fig. 2b). Phylogenetic analyses revealed that mcr-positive K. pneumoniae isolates were widely dispersed across the global mcr-positive K. pneumoniae isolate tree, indicating geographical spread across countries (Fig. 2b). Interestingly, we only detected the mcr-3 gene in K. pneumoniae isolates from swine, whereas the mcr-3 gene was detected in E. coli isolates from swine and humans (Table 1).
Phylogenetic analysis ofK. pneumoniaeisolates from hospitalized patients and healthy swine in Thailand. Maximum-likelihood phylogeny tree inferred from (a) 69,435 recombinant-filtered core SNPs of K. pneumoniae genomes from Thailand analyzed in this study (n = 11), using K. pneumoniae KPCTRSRTH02 as the reference. (b) 109,250 recombinant-filtered core SNPs of 117 colistin-resistant K. pneumoniae genomes identified in the GenBank database, using K. pneumoniae MLST-15 (CP022125-CP022128) as the reference. Midpoint rooted and branch lengths were ignored. The ST (only for isolates from Thailand), host, isolation year, and geographical origin of the isolates are displayed, where information was not available, or the gene was not present, it was left blank. The presence of colistin-resistance mcr genes is indicated by a star symbol. Thailand isolates from this study are indicated in bold. Numbers at nodes represent < 90% bootstrap support
Phylogenetic relationship of colistin-resistant E. coli isolates
Phylogenetic analysis of the 25 E. coli isolates showed our isolates formed two major clades (Fig. 3). Interestingly, we found two isolates that were closely related, an isolate from a farmer (ECH + 04) and an isolate from swine (ECCTRSRTH09); both coharbored mcr-1.1 and mcr-3.1, even though these isolates were collected two years apart. Overall, two- to five-SNP differences were observed within these two genomes, which both coharbored mcr-1.1 and mcr-3.1 despite both isolates being collected 2 years apart. Our 25 E. coli genomes were compared with 408 publicly available colistin-resistant E. coli genomes to investigate the dissemination of colistin-resistant and mcr-positive isolates in Asia (Fig. 4). E. coli genomes from our study were distributed throughout and clustering with isolates from other countries, confirming that the other colistin-resistant strains similar to the ones described in our study were previously reported and suggesting spread across Asia (Fig. 4).
Phylogenetic analysis ofE. coliisolates from farmers, swine, and hospitalized patients in Thailand. Maximum-likelihood phylogeny tree inferred from 160,485 recombinant-filtered core SNPs of E. coli genomes isolated in Thailand analyzed in this study, using E. coli ECSW + 09 as the reference. The ST, host, and isolation year are shown. The presence of colistin-resistance mcr genes is indicated by a star symbol, if the gene was not present the area was left blank. The scale bar represents the number of nucleotide substitutions per site. Numbers at nodes represent <90% bootstrap support
Phylogenetic analysis ofE. coliisolates from Asia. Maximum-likelihood phylogeny inferred from 107,712 recombinant filtered core SNPs of 408 E. coli genomes isolated from Asia that shared the same ST as the isolates from Thailand (labelled with a black triangle) and 25 E. coli genomes from this study. E. coli strain K-12 substrain MG1655 (CP012868) was used as the reference. Midpoint rooted and branch lengths were ignored. The collapsed node represents 47 ST10 genomes from China and India. The host, isolation year, and geographical origin are displayed. Where information was not available it was left blank. Numbers at nodes represent < 90% bootstrap support
Discussion
We observed a colonization rate of 26.2% for colistin-resistant Enterobacterales (39/149) in our study, with E. coli and K. pneumoniae being the primary bacterial species identified. The colonization rates of colistin-resistant E. coli isolates found in the present study were relatively high, especially among the swine isolates, where the rate of colonization was at 58.6%, compared to only ~ 3% colonization rate of colistin-resistant E. coli found in 400 swine from a northern and an eastern province of Thailand in 2012 [44]. However, the high rate in our study could be explained by a small number of swine (n = 38) sampled. The prevalence of colistin-resistant isolates among farmers was 10.7%, though previous study has found no colistin-resistant isolates carriage among 30 farmers in 2012 study [44]. In the international literature, only few studies identified carriage in farmers, and reported that the prevalence of colistin-resistant isolates among farmers was < 5% [45, 46]. In contrast, the prevalence of colistin-resistant Enterobacterales among our hospitalized patients (12.1%) was not significantly different from the 17.1% prevalence reported in the most recent study conducted in 2018 in Thailand [47].
We identified E. coli with a new mcr-2 gene variant, mcr-2.3, among the swine isolates. Typically, mcr genes are found on plasmids; however, co-occurrence of genes on both plasmid and chromosome has been rarely discovered [48,49,50]. In our study, E. coli harbored mcr-2.3 on the chromosome rather than plasmid, suggesting that mcr-2.3 could have alternative pathways to get into bacterial strains. Additionally, one K. quasipneumoniae subsp. similipneumoniae isolate from swine was found to harbor two copies mcr-3.21, one on its plasmid and one on the chromosome, while the other three isolates carried mcr-3.21 on plasmids only. The discovery of an isolate with mcr on both its plasmid and chromosome suggests that the process for stabilizing the mcr-3.21 gene within the bacterial genome is still in progress. Although several studies of E. coli have shown the chromosomal location of mcr-1 and mcr-2 [50, 51], the finding of mcr-3.21 on both a plasmid and chromosome in a single isolate has recently been reported in healthy individuals, farmers and swine in Thailand [48, 49]. This finding was possible due to opportunity to do long- and short- read sequencing on some of the isolates, and hybrid assembly of these reads allowed a more closed genomes to be analyzed. The presence of these mcr variants on chromosome is concerning because it has high potential to spread mcr genes due to vertical and horizontal gene transfer. Notably, E. coli and K. quasipneumoniae are highly prevalent in swine and other livestock, where they could interact with organisms from different hosts [52], which facilitates the exchange of antimicrobial resistance carrying plasmids such as blaCTX−M on IncF plasmid as has been shown in study in China [53, 54], suggesting that this could happen with mcr-carrying plasmids as well.
The present study detected plasmids harboring mcr-1.1 and variants of mcr-3 separately or in combination in E. coli isolated from swine and farmers in Thailand. The co-occurrence of two mcr genes in a single bacterial isolate is intriguing. A few reports have described the co-occurrence of mcr-1 and mcr-3 on a single plasmid of E. coli [55, 56]. Interestingly, genes mcr-4 to mcr-10 were not identified in our study. According to previous reports, mcr-4 and mcr-5 have been detected in E. coli from Asia and European countries, whereas mcr-7.1 and mcr-8 have been detected in K. pneumoniae from China [57,58,59]. The finding of mcr and bla genes in swine isolates demonstrates the effects of amoxicillin and colistin use in pig farms in northern Thailand. Additionally, a recent study revealed that amoxicillin and colistin were also used in some small-to-medium scale pig farms in northeastern Thailand [6].
In a recent study on pig farms in Thailand, 4.5% (n = 31/696) of pathogenic E. coli harbored mcr-1, and these mcr genes were located on IncF-type [60]. Such findings relating to plasmid incompatibility groups were similar to those of our study. Regarding plasmid replicon typing, the host range of the IncF plasmids is limited to Enterobacterales, and IncF plasmids have been detected in Enterobacterales globally, associated with various antimicrobial-resistant genes [61]. IncX4-type plasmids are now considered vehicles responsible for the dissemination of the mcr-1 gene among Enterobacterales worldwide [62,63,64]. The IncX4 plasmid architecture has been reported to be highly conserved, with several studies having shown similar IncX4 plasmids carrying mcr-1 from different Enterobacterales [62, 63]. In the present study, we report E. coli isolates carrying mcr-1.1 on IncX4 plasmids from both hospitalized patients and swine. This is a significant finding because this rare plasmid has been identified in a small dataset. However, further studies involving larger datasets are now required to confirm the prevalence of this plasmid. The presence of the IncX4 plasmid was verified by long-read sequencing using MinION, which provides confidence in the results. Among the Klebsiella spp. identified in this study, IncFII (pMET) was the most common replicon type identified among isolates from swine. IncFII is a plasmid with a narrow host range that is frequently identified among E. coli strains carrying the mcr-1 gene isolated from swine worldwide [65]. Recently, IncFII was identified in Thailand and Laos along with the emergence of mcr-3-mediated plasmids such as IncP1 and IncI1 [66, 67]. The finding of IncFII (pMET) in Klebsiella spp. supports the notion that mcr-3 can potentially spread among different Enterobacterales and disseminate to neighboring countries.
We did not detect mobile colistin resistance genes in any of the patient isolates. However, the mutations in mgrB or pmrAB observed in four of five patient isolates were consistent with previous studies, suggesting that the spread of colistin-resistant K. pneumoniae in hospitals is most likely due to chromosomal mutation [68,69,70]. Interestingly, the colistin-resistant isolates with chromosomal mutations had higher MICs than the mcr-positive isolates, making clinical treatment more difficult. Moreover, a pmrA pseudogene was found in the isolates containing the mutated pmrB gene. Although the pmrA pseudogene causes an increase in the level of polymyxin B susceptibility by reducing cationic groups on the LPS [19], the isolates in our study containing both the pmrA pseudogene and the altered pmrB gene had a high level of colistin resistance, suggesting that the loss of pmrA gene function may have promoted a different lipid A modification pathway. We also identified a novel mutation in protein YcaR, which is part of the KDO2–lipid A biosynthetic pathway [43], in the K. pneumoniae isolates from hospitalized patients. However, further genomic analyses of this point mutation are required to determine whether this mutation is present in other colistin-resistant strains without the mcr gene.
There are several limitations to our study. First, direct horizontal transmission between swine and farmers could not be demonstrated because of the limited number of colistin-resistant isolates and the lack of longitudinal sampling. However, despite the study’s relatively small sample size, we were still able to demonstrate the presence of bla and colistin-resistance genes related to the antibiotics used in swine production in Thailand. Second, the fecal samples from farmers and hospital patients were collected from individuals in different provinces; consequently, it might be difficult to demonstrate linkage among bacterial clones between two provinces. However, there was similarity in the mcr gene types detected among the E. coli isolates from both locations.
Conclusion
Our study demonstrates that bacterial isolates from farmers and swine in Thailand contained multiple antimicrobial-resistance genes, such as extended spectrum beta-lactamases genes and colistin-resistance genes. Even though the present study cannot directly demonstrate horizontal transmission of bacterial strains or plasmid-mediated colistin-resistance genes between swine, farmers, and hospitalized patients, plasmid analysis revealed that the mobile colistin-resistance mcr-1.1 genes of isolates from swine and hospitalized patients belonged to the same incompatibility group. Our findings confirmed that mcr-positive E. coli isolates may be endemic in Thailand, additionally, the highly conserved IncX4 plasmids might be becoming endemic too. Notably, when we compared colistin-resistant Enterobacterales isolates from animals and farmers to the colistin-resistant Enterobacterales isolates from hospitalized patients at Siriraj Hospital, the main mechanism of colistin resistance was a chromosomal point mutation. While our study focused on plasmid-mediated mcr genes, our findings also highlight the emergence of colistin resistance mediated by specific mutations in mgrB or pmrAB present on the chromosome. The latter is important with respect to hospitalized patients because the chromosomal mutations usually confer a high level of colistin resistance (MIC ≥ 32).
Data Availability
Individual level data cannot be shared publicly because of patient confidentiality under current Thai legislation. The data that support the findings of this study are available from Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand, but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from Dr. Adhiratha Boonyasiri (Email: adhiratha.bon@mahidol.ac.th) upon reasonable request and with permission of Faculty of Medicine Siriraj Hospital, Mahidol University, Thailand and the appropriate ethics committee. All sequencing reads were deposited in the SRA database under the project accession number PRJNA389557, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA389557. Relevant assemblies and mcr-genes have been deposited in GenBank under accession numbers as shown in Suppl. Tables S1 and S2).
References
WHO. Antimicrobial resistance: global report on surveillance 2014. 2016.
Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602.
Falagas ME, Karageorgopoulos DE, Nordmann P. Therapeutic options for infections with Enterobacteriaceae producing carbapenem-hydrolyzing enzymes. Future Microbiol. 2011;6(6):653–66.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8.
Huang X, Yu L, Chen X, Zhi C, Yao X, Liu Y, et al. High prevalence of Colistin Resistance and mcr-1 gene in Escherichia coli isolated from Food Animals in China. Front Microbiol. 2017;8:562.
Strom G, Halje M, Karlsson D, Jiwakanon J, Pringle M, Fernstrom LL, et al. Antimicrobial use and antimicrobial susceptibility in Escherichia coli on small- and medium-scale pig farms in north-eastern Thailand. Antimicrob Resist Infect control. 2017;6:75.
Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. Colistin: an update on the antibiotic of the 21st century. Expert Rev anti-infective therapy. 2012;10(8):917–34.
Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643.
Erfanimanesh S, Hashemi A. Global dissemination of the MCR-1 gene. Archives of Pediatric Infectious Diseases. 2016;4(3):e38581.
Feng Y. Transferability of MCR-1/2 Polymyxin Resistance: Complex Dissemination and genetic mechanism. ACS Infect Dis. 2018;4(3):291–300.
Liu Y, Liu JH. Monitoring colistin resistance in Food Animals, an urgent threat. Expert Rev anti-infective therapy. 2018;16(6):443–6.
Schwarz S, Johnson AP. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother. 2016;71(8):2066–70.
Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg microbes infections. 2020;9(1):508–16.
Rhouma M, Beaudry F, Theriault W, Letellier A. Colistin in Pig Production: Chemistry, mechanism of Antibacterial Action, Microbial Resistance Emergence, and one health perspectives. Front Microbiol. 2016;7:1789.
Olaitan AO, Thongmalayvong B, Akkhavong K, Somphavong S, Paboriboune P, Khounsy S, et al. Clonal transmission of a colistin-resistant Escherichia coli from a domesticated pig to a human in Laos. J Antimicrob Chemother. 2015;70(12):3402–4.
Institute CaLS. CLSI Supplement M100. 27th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. Performance Standards for Antimicrobial Susceptibility Testing. 2017.
EUCAST. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1. European Committee on Antimicrobial Susceptibility Testing. 2017.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a New Genome Assembly Algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Wick RR, Judd LM, Gorrie CL, Holt KE, Unicycler. Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE. 2014;9(11):e112963.
Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17(1):132.
Sahl JW, Lemmer D, Travis J, Schupp JM, Gillece JD, Aziz M, et al. NASP: an accurate, rapid method for the identification of SNPs in WGS datasets that supports flexible input and output formats. Microb genomics. 2016;2(8):e000074.
Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92.
Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Letunic I, Bork P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–w9.
Partridge SR, Di Pilato V, Doi Y, Feldgarden M, Haft DH, Klimke W, et al. Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J Antimicrob Chemother. 2018;73(10):2625–30.
Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg microbes infections. 2018;7(1):122.
Ma K, Feng Y, Liu L, Yao Z, Zong Z. A cluster of colistin-and carbapenem-resistant Klebsiella pneumoniae carrying bla NDM-1 and mcr-8.2. J Infect Dis. 2020;221(Supplement2):237–S42.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4.
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W41.
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–91.
Brinkac LM. LOCUST: a custom sequence locus typer for classifying microbial isolates. 2017;33(11):1725–6.
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348–57.
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45(D1):D566–d73.
Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–903.
Bathoorn E, Rossen JW, Lokate M, Friedrich AW, Hammerum AM. Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a dutch patient without travel history abroad, August 2015. Euro Surveill. 2015;20(41).
Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27).
Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z et al. Novel plasmid-mediated Colistin Resistance Gene mcr-3 in Escherichia coli. MBio. 2017;8(3).
Nordmann P, Jayol A, Poirel L. Rapid Detection of Polymyxin Resistance in Enterobacteriaceae. Emerg Infect Dis. 2016;22(6):1038–43.
Wand ME, Bock LJ, Bonney LC, Sutton JM. Mechanisms of increased resistance to Chlorhexidine and Cross-Resistance to Colistin following exposure of Klebsiella pneumoniae clinical isolates to Chlorhexidine. Antimicrob Agents Chemother. 2017;61(1).
Lee H, Hsu FF, Turk J, Groisman EA. The PmrA-regulated pmrC gene mediates phosphoethanolamine modification of lipid A and polymyxin resistance in Salmonella enterica. J Bacteriol. 2004;186(13):4124–33.
Strohmaier H, Remler P, Renner W, Hogenauer G. Expression of genes kdsA and kdsB involved in 3-deoxy-D-manno-octulosonic acid metabolism and biosynthesis of enterobacterial lipopolysaccharide is growth phase regulated primarily at the transcriptional level in Escherichia coli K-12. J Bacteriol. 1995;177(15):4488–500.
Boonyasiri A, Tangkoskul T, Seenama C, Saiyarin J, Tiengrim S, Thamlikitkul V. Prevalence of antibiotic resistant bacteria in healthy adults, foods, food animals, and the environment in selected areas in Thailand. Pathogens and Global Health. 2014;108(5):235–45.
Nakano A, Nakano R, Nishisouzu R, Suzuki Y, Horiuchi S, Kikuchi-Ueda T, et al. Prevalence and relatedness of mcr-1-mediated colistin-resistant Escherichia coli isolated from livestock and farmers in Japan. Front Microbiol. 2021;12:664931.
Effelsberg N, Kobusch I, Linnemann S, Hofmann F, Schollenbruch H, Mellmann A, et al. Prevalence and zoonotic transmission of colistin-resistant and carbapenemase-producing Enterobacterales on german pig farms. One Health. 2021;13:100354.
Wangchinda W, Pati N, Maknakhon N, Seenama C, Tiengrim S, Thamlikitkul V. Collateral damage of using colistin in hospitalized patients on emergence of colistin-resistant Escherichia coli and Klebsiella pneumoniae colonization and infection. Antimicrob Resist Infect control. 2018;7:84.
Leangapichart T, Stosic MS, Hickman RA, Lunha K, Jiwakanon J, Angkititrakul S, et al. Exploring the epidemiology of mcr genes, genetic context and plasmids in Enterobacteriaceae originating from pigs and humans on farms in Thailand. J Antimicrob Chemother. 2023;78(6):1395–405.
Phuadraksa T, Wichit S, Arikit S, Songtawee N, Yainoy S. Co-occurrence of mcr-2 and mcr-3 genes on chromosome of multidrug-resistant Escherichia coli isolated from healthy individuals in Thailand. Int J Antimicrob Agents. 2022;60(4):106662.
Sun J, Li XP, Fang LX, Sun RY, He YZ, Lin J, et al. Co-occurrence of mcr-1 in the chromosome and on an IncHI2 plasmid: persistence of colistin resistance in Escherichia coli. Int J Antimicrob Agents. 2018;51(6):842–7.
AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, et al. mcr-1 and mcr-2 variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. 2017;72(10):2745–9.
Ling Z, Yin W, Li H, Zhang Q, Wang X, Wang Z et al. Chromosome-mediated mcr-3 variants in Aeromonas veronii from Chicken meat. Antimicrob Agents Chemother. 2017;61(11).
Yang Q, Sun J, Li L, Deng H, Liu B, Fang L, et al. IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front Microbiol. 2015;6:964.
Zhang Y, Sun Y-H, Wang J-Y, Chang M-X, Zhao Q-Y, Jiang H-X. A novel structure harboring bla CTX-M-27 on IncF plasmids in Escherichia coli isolated from swine in China. Antibiotics. 2021;10(4):387.
Hernandez M, Iglesias MR, Rodriguez-Lazaro D, Gallardo A, Quijada N, Miguela-Villoldo P et al. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill. 2017;22(31).
Li R, Zhang P, Yang X, Wang Z, Fanning S, Wang J, et al. Identification of a novel hybrid plasmid coproducing MCR-1 and MCR-3 variant from an Escherichia coli strain. J Antimicrob Chemother. 2019;74(6):1517–20.
Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A et al. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill. 2017;22(31).
Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018.
Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother. 2017;72(12):3317–24.
Khine NO, Lugsomya K, Kaewgun B, Honhanrob L, Pairojrit P, Jermprasert S, et al. Multidrug Resistance and Virulence factors of Escherichia coli Harboring plasmid-mediated Colistin Resistance: mcr-1 and mcr-3 genes in contracted Pig Farms in Thailand. Front Vet Sci. 2020;7:582899.
Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol. 2007;73(6):1976–83.
Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179.
Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, et al. Genetic analysis of the IncX4 plasmids: implications for a Unique Pattern in the mcr-1 Acquisition. Sci Rep. 2017;7(1):424.
Bai F, Li X, Niu B, Zhang Z, Malakar PK, Liu H et al. A mcr-1-Carrying conjugative IncX4 plasmid in colistin-resistant Escherichia coli ST278 strain isolated from dairy cow feces in Shanghai, China. Front Microbiol. 2018;9(2833).
Deng Y, Zeng Z, Chen S, He L, Liu Y, Wu C, et al. Dissemination of IncFII plasmids carrying rmtB and qepA in Escherichia coli from pigs, farm workers and the environment. Clin Microbiol Infect. 2011;17(11):1740–5.
Tansawai U, Yu Y, Kiddee A, Assawatheptawee K, Sands K, Hassan B, et al. Emergence of mcr-3-mediated IncP and IncFII plasmids in Thailand. J Global Antimicrob Resist. 2021;24:446–7.
Hadjadj L, Baron SA, Olaitan AO, Morand S, Rolain J-M. Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front Microbiol. 2019;10:2720.
Otter JA, Doumith M, Davies F, Mookerjee S, Dyakova E, Gilchrist M, et al. Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci Rep. 2017;7(1):12711.
Guducuoglu H, Gursoy NC, Yakupogullari Y, Parlak M, Karasin G, Sunnetcioglu M, et al. Hospital Outbreak of a Colistin-Resistant, NDM-1- and OXA-48-Producing Klebsiella pneumoniae: high mortality from Pandrug Resistance. Microb drug Resist (Larchmont NY). 2018;24(7):966–72.
Giani T, Arena F, Vaggelli G, Conte V, Chiarelli A, De Henrici L, et al. Large nosocomial outbreak of Colistin-Resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrB deletion mutant. J Clin Microbiol. 2015;53(10):3341–4.
Acknowledgements
We greatly appreciate Professor Xavier Didelot, School of Life Sciences, University of Warwick for his valuable advice. We thank Katie Oakley, PhD, from Edanz Group (https://en-author-services.edanzgroup.com/) for editing a draft of this manuscript and helping to draft the abstract. EJ acknowledges the generous support of The Rosetree Trust and The Stoneygate Trust as part of her Imperial College Research Fellowship. We also acknowledge the National Institute for Health Research Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance at Imperial College London in partnership with the UK Health Security Agency, in collaboration with, Imperial Healthcare Partners, University of Cambridge and University of Warwick. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or UK Health Security Agency.
Funding
This work has been funded in whole or part with federal funds from the National Institute of Allergy and Infectious Diseases; National Institutes of Health; Department of Health and Human Services [U19AI110819].
Author information
Authors and Affiliations
Contributions
LB, AB, VT, and DF conceived the study. LB, AB, RW, CG, and KN undertook data analysis with input from CS and TT. EJ provided additional input into the framing of the results. LB, AB, EJ, VT, and DF produced the first draft of the manuscript. All authors contributed to the final draft.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Siriraj Institutional Review Board (Si571/2015 and Si680/2016). Written informed consent was obtained from all patients. The data extracted from the medical record were de-identified to protect patients’ confidentiality. Ethical review and approval was not required for the animal study because all stool or rectal swab samples were submitted from farmers and pigs in industrial field to the diagnostic laboratory as the annual surveillance. All methods were performed in accordance with relevant guidelines and regulations such as the Declaration of Helsinki, the Belmont Report, CIOMS guidelines, and ICH-GCP guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Boonyasiri, A., Brinkac, L.M., Jauneikaite, E. et al. Characteristics and genomic epidemiology of colistin-resistant Enterobacterales from farmers, swine, and hospitalized patients in Thailand, 2014–2017. BMC Infect Dis 23, 556 (2023). https://doi.org/10.1186/s12879-023-08539-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-023-08539-8