Abstract
Background
Searching the risk factors for carbapenem-resistant Enterobacteriaceae (CRE) infection is important in clinical practice. In the present study, we aim to investigate bacterial characteristics of colonizing strains and their correlation with subsequent CRE infection.
Methods
Between May 2018 and January 2019, patients hospitalized in the department of haematology and intensive care unit (ICU) were screened for CRE by rectal swabs and monitored for the outcome of infection. We identified the species and carbapenemase-encoding genes of colonizing strains and performed antimicrobial susceptibility tests and multilocus sequence typing (MLST). Risk factors for subsequent CRE infections were ascertained by univariate and multivariable analysis.
Results
We collected a total of 219 colonizing strains from 153 patients. Klebsiella pneumoniae was the most abundant species, and MLST analysis showed rich diversity. K. pneumoniae carbapenemase (KPC) was predominant in the infection group (72.4%). In the non-infection group, 35.4% of strains were non-carbapenemase-producing CRE (NCP-CRE), and New Delhi metallo-β-lactamase (NDM) was predominant (42.2%). The rate of high-level carbapenem resistance (minimum inhibitory concentration [MIC] ≥ 64 mg/L for meropenem and ertapenem, ≥ 32 mg/L for imipenem) was remarkably higher in the infection group than in the non-infection group (P < 0.001). Univariate analysis showed that K. pneumoniae, high-level carbapenem resistance, CP-CRE and KPC-CRE were infection risk factors after CRE colonization. On multivariable analysis with different carbapenemase dichotomizations, KPC-CRE (adjusted odds ratio [aOR], 4.507; 95% confidence interval [CI], 1.339–15.171; P = 0.015) or imipenem MIC ≥ 32 mg/L (aOR, 9.515; 95% CI, 1.617–55.977; P = 0.013) were respectively identified as independent risk factors for subsequent infection.
Conclusions
Patients colonized with KPC-CRE or strains with an imipenem MIC ≥ 32 mg/L were at particularly high risk of subsequent CRE infections during their hospital stay.
Similar content being viewed by others
Background
Carbapenem-resistant Enterobacteriaceae (CRE) infections are of major concern to clinicians and public health authorities due to increasing prevalence, rapid regional dissemination, limited therapeutic options and deleterious patient outcomes (mortality rates are 40 ± 10%) [1]. CRE carriage is responsible for the incidence of clinical infection [2,3,4,5,6], and it has been reported that colonization with CRE was associated with at least a two-fold increased risk of infection by the colonizing strain [3]. Many guidelines for the prevention and control of these organisms have been developed by health organizations, including the Centers for Disease Control and Prevention (CDC), the European Centre for Disease Prevention and Control, and the World Health Organization [7,8,9]. A series of clinical reports have shown that individually or nationally directed infection control interventions can effectively reduce CRE transmission and infection rates [4,5,6]. Moreover, researchers have increasingly explored strategies to decolonize CRE to interrupt pathways between colonization and subsequent infections [10,11,12,13]. Despite remarkable effects, there are various challenges to implement these interventions or strategies [14]. Searching CRE colonization patients who are at high risk of infection is an urgent priority, as it can guide us whether additional interventions are needed and limit decolonization strategy use.
Although many studies have been performed to identify risk factors for clinical CRE infections, which highlighted the analysis of clinical data of patients [3, 15,16,17,18], few have explored bacterial characteristics of colonizing strains and their correlation with subsequent CRE infection. It was reported that patients colonized with carbapenemase-producing CRE (CP-CRE) were more likely than non-carbapenemase-producing CRE (NCP-CRE)-colonized patients to develop CRE infections during hospitalization [19]. Previous studies illustrated that some characteristics of infection strains, namely, carbapenemase-encoding genes and minimum inhibitory concentration (MIC) values of carbapenem, were closely related to the outcome of CRE-infected patients [20]. Therefore, we postulated that some microbiological parameters of CRE-colonizing isolates may be risk factors of subsequent infections in patients colonized with CRE.
The study were conducted among patients hospitalized in department of haematology and intensive care unit (ICU), which are at particularly high risk of infecting CRE during their hospital stay as compromised immune systems, lengthy unit stays, and significant rates of device and antibiotic utilization [19]. We identified the microbiological parameters of colonizing isolates, including genus and species, phenotypic carbapenem resistance profile, carbapenemase production status, carbapenemase-encoding genes and multilocus sequence typing (MLST). Subsequently, univariate and multivariable analyses were conducted to find their correlation with subsequent CRE infection. We found that imipenem MIC ≥ 32 mg/L or Klebsiella pneumoniae carbapenemase (KPC)-positive CRE-colonized patients were at high risk of subsequent CRE infections during their hospital stay.
Materials and methods
Research setting and ethics statement
From May 2018 to January 2019, we carried out this study at Tongji Hospital, the largest hospital in the region of central China. In the department of haematology and ICU, we initiated a coordinated and comprehensive intervention of CRE, which mainly included implementation of CRE screening, contact precautions, patient isolation, and antibiotic management [7,8,9, 21]. All inpatients (including transferred and re-admitted patients) were routinely screened for CRE by rectal swabs on admission and twice a week thereafter until discharge or infection. For CRE-colonized and -infected patients, isolation precautions were applied according to CDC guidelines, and removed if they developed decolonization [7]. Decolonization was defined as two consecutive negative cultures within 48 h, and the second sample was confirmed to be carbapenemase-negative by the Xpert® Carba-R Assay (Cepheid, Sunnyvale, California).
We collected all CRE-colonizing strains from adult patients (≥ 18 years old), and divided them into infection and non-infection groups, according to whether the colonized patient had a subsequent CRE infection. The infection group included all CRE colonizing isolates obtained from patients who subsequently had CRE infections. Strains isolated from colonized patients who did not develop CRE infections during their hospital stay were subsumed by the non-infected group.
This study was approved by the ethical committee of Tongji hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
Bacterial isolate collection and identification
Rectal swabs were consecutively obtained from patients and screened for CRE with selective chromogenic agar (Zhengzhou Dianshi biotechnology Co., Ltd., China). Cultured isolates were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, Bruker Daltonics Inc., Billerica, Massachusetts), and then carbapenem (meropenem and imipenem) antimicrobial susceptibility testing was performed to confirm CRE by the disk diffusion method [22]. Enterobacteriaceae that were resistant to meropenem or imipenem were classified as CRE.
Antibiotic susceptibility testing
According to the Clinical and Laboratory Standards Institute (CLSI) guidelines [22], we performed antibiotic susceptibility testing using the broth microdilution method to determine MICs of cefepime, cefoxitin, ceftazidime, aztreonam, ertapenem, imipenem, meropenem, gentamicin, amikacin, minocycline, ciprofloxacin, fosfomycin, piperacillin-tazobactam, trimethoprim-sulfamethoxazole, colistin and tigecycline. All antibiotics, except tigecycline and colistin, were interpreted according to the standard of the CLSI document. For tigecycline and colistin, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoint was used. Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC27853 were used as quality control standards.
Investigation of resistance mechanisms
For all CRE strains, the modified carbapenem inactivation method (mCIM) was conducted to identify carbapenemase production [23]. For CP-CRE strains, polymerase chain reaction (PCR) was performed to detect five common carbapenemase-encoding genes, including blaKPC, blaIMP, blaVIM, blaNDM and blaOXA-48 [24]. CP-CRE strains without common genes were further tested uncommon carbapenemase-encoding genes, including blaGES, blaVEB, blaPER, blaSME and blaIMI [25,26,27,28]. The PCR products were sequenced and analysed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST).
Multilocus sequence typing
Multilocus sequence typing (MLST) of K. pneumoniae was performed following the protocol described on the Pasteur Institute MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). The sequences of seven housekeeping genes and sequence types (STs) were assigned using online MLST databases.
Statistical analysis
Data were analysed using SPSS v.19.0 software (SPSS Inc., Chicago, IL, USA). MICs were analysed both as ordinal and as dichotomized variables. We calculated the Youden index (sensitivity + specificity – 1) at each possible cutoff value for dichotomized MICs. The significant differences between different groups were analysed using the chi-square (χ2) test or Fisher’s exact test, as appropriate. Univariate logistic regression analyses were carried out to assess the relevant risk factors of CRE infection. Only significantly different factors were subsequently included in multivariable analyses, which were constructed using stepwise model selection and manually curated. Statistical significance was determined as P < 0.05. Because carbapenemase can be distinguished by production status and different carbapenemase-encoding genes, we conducted multivariable analyses twice. Odds ratio (OR) with 95% confidence interval (CI) was presented for the logistic regression analysis.
Results
Study population and distribution of CRE colonizing isolates
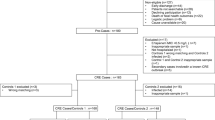
A total of 219 CRE colonizing strains were collected from 153 patients, of whom 29 individuals developed CRE infections during hospitalization (Supplementary Fig. 1). In the infection group, we obtained 23 colonizing isolates from the ICU and 35 isolates from the department of haematology. K. pneumoniae was the most abundant species (81.0%), followed by E. coli (12.1%). In the non-infection group, there were 161 strains and 70.2% of strains were from the department of haematology. Thereinto, K. pneumoniae accounted for 53.4% and E. coli accounted for 29.2%. The two groups differed remarkably in terms of bacterial species (P = 0.001), and there were no obvious differences in medical department proportions (P = 0.170) (Table 1).
Characteristics of K. pneumoniae STs
A total of 36 distinct STs were identified among 133 carbapenem-resistant K. pneumoniae (CR-KP) (Supplementary Table 1). As depicted in Table 1, ST11 was the most prevalent ST in the infection group (89.4%). In the non-infection group, a total of 33 STs were identified, among which ST11 was the most common type (30.2%), followed by ST37 (11.6%), ST15 (8.1%) and ST147 (7.0%). A univariable analysis showed a difference in the proportion of STs between the two groups (P < 0.001).
Screening for carbapenemase-encoding genes
From all strains, 155 (70.8%) were found to produce carbapenemases (Table 1). The major carbapenemase-encoding genes were KPC-type (n = 69) and New Delhi metallo-β-lactamase (NDM)-type (n = 77). All detected KPC-type genes were blaKPC-2, while NDM-type genes included blaNDM-1 (n = 34), blaNDM-4 (n = 1), blaNDM-5 (n = 40), and blaNDM-7 (n = 2). Other common carbapenemase-encoding genes, namely blaIMP-4 (n = 2), blaVIM-1 (n = 2) and blaOXA-48 (n = 1), were also found. Uncommon genes were not detected. Two strains co-harbouring blaKPC-2 and blaNDM-1 and two CP-CRE strains which didn’t harbour the tested genes were found.
In the infection group, 87.9% of strains were CP-CRE and KPC (72.4%) was the most common carbapenemase type. In the non-infection group, approximately 64.6% of strains were CP-CRE, among which NDM (42.2%) was the most abundant, followed by KPC (16.8%). There were noticeable differences in carbapenemases between the two groups (P < 0.001). The KPC production status was remarkably associated with CRE infection after intestinal CRE colonization.
Antimicrobial susceptibility testing results
The antimicrobial susceptibility of CRE colonizing isolates is shown in Table 2. In total, rectal CRE strains showed high susceptibility to colistin (92.2%), followed by tigecycline (83.1%). Compared with K. pneumonia, E. coli was more susceptible to gentamicin (59.3% vs. 21.1%), amikacin (87.0% vs. 45.1%) and fosfomycin (74.1% vs. 11.3%). NDM-positive strains were more susceptible to aztreonam (37.7% vs. 0), gentamicin (46.8% vs. 7.2%), amikacin (87.0% vs. 14.5%) and fosfomycin (63.6% vs. 0) than KPC-producing strains. NCP-CRE was more resistant to tigecycline and trimethoprim-sulfamethoxazole than CP-CRE. The infection and non-infection groups differed significantly in terms of susceptibility to gentamicin (15.5% vs. 37.9%), amikacin (31.0% vs. 71.4%), fosfomycin (6.9% vs. 41.6%), tigecycline (93.1% vs. 79.5%) and trimethoprim-sulfamethoxazole (48.3% vs. 19.3%).
Evaluation of cutoff values for dichotomized carbapenem MICs
According to carbapenem breakpoints, there were no obvious differences between the two groups except for the rate of resistance to imipenem (Table 1). The distributions of MICs of carbapenem in the two groups are shown in Fig. 1. In the infection group, there was only one peak in every carbapenem antibiotic and the peak value was high. However, the distribution in the non-infection group was relatively gentle, and the peak value was lower. Youden index was calculated to determine the most appropriate cutoff values for dichotomized MICs (Supplementary Table 2). When MICs for meropenem and ertapenem were dichotomized at < 64 mg/L vs. ≥ 64 mg/L and MICs for imipenem were dichotomized at < 32 mg/L vs. ≥ 32 mg/L, the Youden indexes were the highest. Colonizing strains with high carbapenem MICs (MIC ≥ 64 mg/L for meropenem and ertapenem, ≥ 32 mg/L for imipenem) were risk factors for subsequent infection (P < 0.001).
The distribution of minimum inhibitory concentrations (MICs) of carbapenem (meropenem, imipenem and ertapenem) in different groups of carbapenem-resistant Enterobacteriaceae colonizing strains. A Meropenem MIC distribution in the infection group (n = 58). B Meropenem MIC distribution in the non-infection group (n = 161). C Imipenem MIC distribution in the infection group. D Imipenem MIC distribution in the non-infection group. (E) Ertapenem MIC distribution in the infection group. (F) Ertapenem MIC distribution in the non-infection group
Relationship between different colonized bacterial factors
The distributions of species and carbapenem MICs in different carbapenemase types are shown in Fig. 2. Approximately 97.2% of KPC-positive strains were K. pneumoniae, among which ST11-K. pneumoniae (95.7%) was the most prevalent (P < 0.001) (Fig. 2A). The most frequently observed MICs of meropenem and ertapenem in KPC-positive strains was ≥ 64 mg/L (93.0 and 91.6%). Over 90% of KPC-CRE had high-level imipenem MICs (Fig. 2B). For strains producing other carbapenemases, the distributions of MICs focused on 16 to 128 mg/L for meropenem and ertapenem (91.7 and 87.0%), and 15.5% strains had a high level of imipenem MICs (Fig. 2C). Few NCP-CRE had high-level carbapenem MICs (Fig. 2D). In brief, rates of high-level carbapenem MICs were much higher in KPC-CRE (P < 0.001).
The distribution of species and carbapenem MICs among CRE-colonizing strains producing different carbapenemases. A Species distribution among strains producing different carbapenemases. B Carbapenem MICs distribution among KPC-producing strains (n = 71). C Carbapenem MIC distribution among strains producing other carbapenemases (n = 84). D Carbapenem MIC distribution among NCP-CRE strains (n = 64). Abbreviations: MEM, meropenem; IPM, imipenem; ETP, ertapenem; MIC, minimum inhibitory concentration; CRE, carbapenem-resistant Enterobacteriaceae; KPC, K. pneumoniae carbapenemase; NCP-CRE, non-carbapenemase-producing CRE
Relationship between colonized bacterial factors and risk of CRE infection
Univariate analyses revealed that K. pneumoniae, meropenem MIC ≥ 64 mg/L, imipenem MIC ≥ 32 mg/L, ertapenem MIC ≥ 64 mg/L, CP-CRE and KPC-CRE were risk factors for subsequent CRE infection in CRE intestinal carriers. When combined different factors, there were no obvious improvements in the predictive ability (Table 3).
When dichotomizing carbapenemase by if it was KPC production or not, KPC-CRE (adjusted odds ratio [aOR], 4.507; 95% CI, 1.339–15.171; P = 0.015) was independently associated with a subsequent infection in a multivariable analysis. On the other hand, when dichotomizing carbapenemase by if it was CP-CRE or NCP-CRE, imipenem MIC ≥ 32 mg/L (aOR, 9.515; 95% CI, 1.617–55.977; P = 0.013) was the only independent factor (Supplementary Table 3).
Discussion
CRE has been classified as an urgent threat, and CRE colonization was significantly associated with the increased risk of subsequent CRE infection [2,3,4,5,6]. The need to identify patients as having a high risk for CRE infection has been recognized. Many reports have evaluated risk factors for CRE infection, and some have proposed risk factor scoring models; however, these reports were focused on demographic data and clinical information, such as comorbid medical conditions, colonization history and prior antibiotic exposures [3, 15,16,17,18, 29]. In the current study, we innovatively analysed microbiological parameters of colonizing strains to search for risk factors for subsequent infection after CRE colonization.
According to report of China Antimicrobial Surveillance Network (CHINET), CR-KP increased from 2.4 to 13.4% between 2005 and 2014 [30]. K. pneumoniae accounted for the largest percentage of CRE strains (66.7%) and 64% of K. pneumoniae isolates were ST11-KPC [31]. 85.7% of CRE strains were found to produce carbapenemases, among which KPC was predominant in K. pneumoniae isolates (77%) and NDM was predominant in E. coli isolates (75%) [31]. In accordance with domestic changing trend, CR-KP in our hospital increased significantly in recent two decades, for example, the detection rate of CR-KP in bloodstream infections was below 5% in 1998–2012 and increased to 34.9% in 2013–2017 [32]. ST11-KPC K. pneumoniae has caused a series of nosocomial outbreaks in China, including our hospital [33, 34]. There was an outbreak of CR-KP in the neonatal ward in 2015 in our hospital, among which ST11-KPC-2, ST20-NDM-1 and ST888-NDM-1 K. pneumoniae was 81.48% (22/27), 14.81% (4/27) and 3.70%(1/27), respectively [33]. Likewise, ST11-KPC K. pneumoniae was predominant in community-onset CRE (CO-CRE) infection. According to a tertiary hospital in China, K. pneumoniae accounted for 53.6% among 28 CO-CRE isolates, and 86.7% of K. pneumoniae strains belonged to ST11 containing blaKPC-2 [35]. Consistent with the prevalence of CRE strains from clinical specimens, K. pneumoniae was the most common rectal strain in our study, 51.9% of which were KPC-positive. Somewhat differently, approximately 30% of rectal strains were NCP-CRE; moreover, the proportions of KPC and NDM enzymes were approximately the same (31.5 and 35.2%, respectively). MLST analysis revealed a rich genetic diversity among intestinal K. pneumoniae strains, of which 36 distinct STs were identified, and ST11 was the most prevalent. The results of our study indicated that approximately all KPC-producing strains were ST11-K. pneumoniae, and half of NDM-producing strains were E. coli. Moreover, KPC-CRE had a high carbapenem MIC, and NCP-CRE had low imipenem and meropenem MICs.
Univariate analyses showed that there were differences in species, STs of K. pneumoniae strains, carbapenemase production status and carbapenemase-encoding genes between the two groups. Distributions of the above-mentioned microbiological parameters were concentrated in the infection group, in which KPC-2 K. pneumoniae was major strain type; by contrast, distributions were diverse and dispersed in the non-infection group, in which NCP-K. pneumoniae was the most prevalent, followed by NDM-5 E. coli and KPC-2 K. pneumoniae. After dichotomizing carbapenem MICs, the proportion of high-level carbapenem MICs was remarkably different between the two groups, in which the infection group was much higher. Among these significant variables, the OR of KPC-CRE was the highest, followed by high-level MICs of imipenem and ertapenem. Tamma P.D. et al. reported that ICU patients colonized with CP-CRE were more likely than NCP-CRE-colonized patients to develop CRE infections (36% vs. 5%) [19]. Our data showed that OR of CP-CRE was far less than that of KPC-CRE (3.993 vs. 13.028), suggesting that the correlation between CP-CRE colonization and subsequent infection was not as strong as KPC-CRE. In our previous study, carbapenem resistance score which based on the inhibition zone diameters of meropenem and imipenem was an independent risk factor for CRE bloodstream infection in intestinal carriers [36]. In line with previous report, high-level carbapenem MICs were remarkably relevant to subsequent infection in CRE-colonized patients in this study. It was noteworthy that carbapenem inhibition zone diameters of these CRE strains were usually 6 mm, hence the MICs value were more accurate. In addition, we analysed ertapenem resistance, which was shown to be a risk factor for subsequent infection.
We tried to improve the predictive ability by combining different indicators and found that they were not as good as KPC-CRE alone. Finally, we conducted multivariable analyses and found that only KPC-CRE or high-level imipenem MIC was an independent risk factor for infection when we included different significant variables. Our findings suggest that patients colonized with KPC-CRE or strains with imipenem MIC ≥ 32 mg/L may be at particularly high risk of subsequent CRE infections during their hospital stay. It is noteworthy that the precise epidemiology of carbapenemase is diverse across countries and regions. CR-KP strains harbouring KPC are prevalent in the United States, some parts of Europe and the Mediterranean region [37, 38], and most regions of China [31]; some countries are more affected by other carbapenemases, including Spain (VIM), India (NDM), most regions of the Middle East (except Israel) and north Africa (OXA-48) [38]; moreover, the NDM type is reported to be the key carbapenemase responsible for the carbapenem resistance phenotypes in children in some parts of China, including Shanghai [39, 40]. The predictive ability of KPC-CRE may not be generalizable to other hospitals and people.
Decolonization demonstrated a decline in CRE carriage rates and may be potentially useful for the prevention of a subsequent infection [10,11,12]. Common strategies for decolonization are selective digestive decontamination and faecal microbiota transplantation, which are promising but costly and invasive [10,11,12,13]. Various regimens for digestive decontamination have been investigated, including oral aminoglycosides (e.g. gentamicin), colistin and a combination of both [13]; however, it was reported that gut decontamination has been associated with the development of colistin and gentamicin resistance [41]. Our antibiotic susceptibility testing of rectal CRE strains showed a high susceptibility to colistin, and the susceptibility to gentamicin varied among species and carbapenemase types. Only 7.2% of strains producing KPC were susceptible to gentamicin, suggesting that phenotypic or genotypic testing of CRE colonizing strains is needed. Early identification of KPC-CRE-colonized patients is important because it may facilitate the targeted use of interventions and limit antimicrobial use.
This study also had several limitations. First, our study was conducted in the department of haematology and ICU, and the prevalence of CRE colonizing strains and risk factors may not be generalizable to other institutions or departments. Second, we did not confirm that the colonizing and clinical infection CRE isolates were the same. It is of great clinical significance to match the CRE colonization isolate and subsequent infected isolate on all available microbial parameters, including bacterial species, phenotypic antimicrobial resistance profile, antimicrobial resistance genes, virulence genes, etc., which is the theoretical basis for secondary infection caused by CRE colonization. In the follow-up study, we will analyse the homology of CRE colonization strains and infection strains by multiple technique, such as pulsed field gel electrophoresis (PFGE) and whole-genome sequencing (WGS). Moreover, we performed MLST of K. pneumoniae isolates only, MLST analyses of other species were not evaluated. In the future, we will accumulate more cases and analyse the association between MLST stratified by bacterial species and subsequent CRE infection. Finally, patients’ clinical information, which was associated with a subsequent infection to a certain extent, was not included. Combined analysis of bacterial and clinical factors may improve the predictive ability.
Conclusions
In summary, this was an innovatively study to investigate colonizing isolates on all available microbiological parameters, expanding our understanding of the crucial factors of colonizing strains in the incidence of a CRE infection. Our findings suggest that phenotypic or genotypic testing of colonizing CRE strains is needed, and patients colonized with KPC-CRE or strains with imipenem MIC ≥ 32 mg/L are more likely to develop subsequent CRE infections during their hospital stay.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- CRE:
-
Carbapenem-resistant Enterobacteriaceae
- ICU:
-
Intensive care unit
- MLST:
-
Multilocus sequence typing
- KPC:
-
K. pneumoniae carbapenemase
- NCP-CRE:
-
Non-carbapenemase-producing CRE
- NDM:
-
New Delhi metallo-β-lactamase
- MIC:
-
Minimum inhibitory concentration
- aOR:
-
Adjusted odds ratio
- CI:
-
Confidence interval
- CDC:
-
Centers for Disease Control and Prevention
- CP-CRE:
-
Carbapenemase-producing CRE
- MALDI-TOF MS:
-
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry
- CLSI:
-
Clinical and Laboratory Standards Institute
- EUCAST:
-
European Committee on Antimicrobial Susceptibility testing
- mCIM:
-
Modified carbapenem inactivation method
- PCR:
-
Polymerase chain reaction
- ST:
-
sequence types
- OR:
-
Odds ratio
- CR-KP:
-
Carbapenem-resistant K. pneumoniae
References
Tzouvelekis LS, Markogiannakis A, Piperaki E, Souli M, Daikos GL. Treating infections caused by carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2014;20(9):862–72. https://doi.org/10.1111/1469-0691.12697.
Tischendorf J, de Avila RA, Safdar N. Risk of infection following colonization with carbapenem-resistant Enterobactericeae: a systematic review. Am J Infect Control. 2016;44(5):539–43. https://doi.org/10.1016/j.ajic.2015.12.005.
Dickstein Y, Edelman R, Dror T, Hussein K, Bar-Lavie Y, Paul M. Carbapenem-resistant Enterobacteriaceae colonization and infection in critically ill patients: a retrospective matched cohort comparison with non-carriers. J Hosp Infect. 2016;94(1):54–9. https://doi.org/10.1016/j.jhin.2016.05.018.
Ben-David D, Masarwa S, Fallach N, Temkin E, Solter E, Carmeli Y, et al. Success of a National Intervention in controlling Carbapenem-resistant Enterobacteriaceae in Israel's long-term care facilities. Clin Infect Dis. 2019;68(6):964–71. https://doi.org/10.1093/cid/ciy572.
Schwaber MJ, Lev B, Israeli A, Solter E, Smollan G, Rubinovitch B, et al. Israel Carbapenem-resistant Enterobacteriaceae working G: containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin Infect Dis. 2011;52(7):848–55. https://doi.org/10.1093/cid/cir025.
French CE, Coope C, Conway L, Higgins JP, McCulloch J, Okoli G, et al. Control of carbapenemase-producing Enterobacteriaceae outbreaks in acute settings: an evidence review. J Hosp Infect. 2017;95(1):3–45. https://doi.org/10.1016/j.jhin.2016.10.006.
Centers for Disease Control and Prevention (CDC). Facility Guidance for Control of Carbapenem-resistant Enterobacteriaceae (CRE). CDC Guidance, 2015.
Magiorakos AP, Burns K, Rodriguez Bano J, Borg M, Daikos G, Dumpis U, et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European Centre for Disease Prevention and Control. Antimicrob Resist Infect Control. 2017;6(1):113. https://doi.org/10.1186/s13756-017-0259-z.
World Health Organization. Guidelines for the prevention and control of carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii and Pseudomonas aeruginosa in health care facilities. Geneva: World Health Organization, 2017.
Tascini C, Sbrana F, Flammini S, Tagliaferri E, Arena F, Leonildi A, et al. Oral gentamicin gut decontamination for prevention of KPC-producing Klebsiella pneumoniae infections: relevance of concomitant systemic antibiotic therapy. Antimicrob Agents Chemother. 2014;58(4):1972–6. https://doi.org/10.1128/AAC.02283-13.
Davido B, Batista R, Michelon H, Lepainteur M, Bouchand F, Lepeule R, et al. Is faecal microbiota transplantation an option to eradicate highly drug-resistant enteric bacteria carriage? J Hosp Infect. 2017;95(4):433–7. https://doi.org/10.1016/j.jhin.2017.02.001.
Kesecioglu J, Eggimann P. What is new in selective decontamination of the digestive tract? Intensive Care Med. 2016;42(8):1270–5. https://doi.org/10.1007/s00134-015-4009-5.
Bar-Yoseph H, Hussein K, Braun E, Paul M. Natural history and decolonization strategies for ESBL/carbapenem-resistant Enterobacteriaceae carriage: systematic review and meta-analysis. J Antimicrob Chemother. 2016;71(10):2729–39. https://doi.org/10.1093/jac/dkw221.
Gysin DV, Cookson B, Saenz H, Dettenkofer M, Widmer AF. Infections ESGfN: variability in contact precautions to control the nosocomial spread of multi-drug resistant organisms in the endemic setting: a multinational cross-sectional survey. Antimicrob Resist Infect Control. 2018;7(1):81. https://doi.org/10.1186/s13756-018-0366-5.
Daikos GL, Vryonis E, Psichogiou M, Tzouvelekis LS, Liatis S, Petrikkos P, et al. Risk factors for bloodstream infection with Klebsiella pneumoniae producing VIM-1 metallo-beta-lactamase. J Antimicrob Chemother. 2010;65(4):784–8. https://doi.org/10.1093/jac/dkq005.
Swaminathan M, Sharma S, Poliansky Blash S, Patel G, Banach DB, Phillips M, et al. Prevalence and risk factors for acquisition of carbapenem-resistant Enterobacteriaceae in the setting of endemicity. Infect Control Hosp Epidemiol. 2013;34(8):809–17. https://doi.org/10.1086/671270.
Chiotos K, Tamma PD, Flett KB, Naumann M, Karandikar MV, Bilker WB, et al. Multicenter Study of the Risk Factors for Colonization or Infection with Carbapenem-Resistant Enterobacteriaceae in Children. Antimicrob Agents Chemother. 2017;61(12). https://doi.org/10.1128/AAC.01440-17.
van Loon K, Voor In’ t Holt AF, Vos MC. A systematic review and meta-analyses of the clinical epidemiology of carbapenem-resistant enterobacteriaceae. Antimicrob Agents Chemother. 2018;62(1). https://doi.org/10.1128/AAC.01730-17.
Tamma PD, Kazmi A, Bergman Y, Goodman KE, Ekunseitan E, Amoah J, et al. The Likelihood of Developing a Carbapenem-Resistant Enterobacteriaceae Infection during the Hospital Stay. Antimicrob Agents Chemother. 2019;63(8). https://doi.org/10.1128/AAC.00757-19.
Wang X, Wang Q, Cao B, Sun S, Zhang Y, Gu B, et al. Retrospective observational study from a Chinese network of the impact of combination therapy versus monotherapy on mortality from carbapenem-resistant enterobacteriaceae bacteremia. Antimicrob Agents Chemother. 2018;63(1). https://doi.org/10.1128/AAC.01511-18.
Solter E, Adler A, Rubinovitch B, Temkin E, Schwartz D, Ben-David D, et al. Israeli National Policy for Carbapenem-resistant Enterobacteriaceae screening, carrier isolation and discontinuation of isolation. Infect Control Hosp Epidemiol. 2018;39(1):85–9. https://doi.org/10.1017/ice.2017.211.
Institute CaLS. Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI supplement M100. 28th ed. PA: Clinical and Laboratory Standards InstituteWayne; 2018.
Tamma PD, Simner PJ. Phenotypic detection of carbapenemase-producing organisms from clinical isolates. J Clin Microbiol. 2018;56(11). https://doi.org/10.1128/JCM.01140-18.
Kiaei S, Moradi M, Hosseini Nave H, Hashemizadeh Z, Taati-Moghadam M, Kalantar-Neyestanaki D. Emergence of co-existence of blaNDM with rmtC and qnrB genes in clinical carbapenem-resistant Klebsiella pneumoniae isolates in burning center from southeast of Iran. Folia Microbiol (Praha). 2018;64(1):55–62.
Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–5. https://doi.org/10.1093/jac/dkp498.
Aubron C, Poirel L, Ash RJ, Nordmann P. Carbapenemase-producing Enterobacteriaceae, U.S. rivers. Emerg Infect Dis. 2005;11(2):260–4. https://doi.org/10.3201/eid1102.030684.
Radice M, Power P, Gutkind G, Fernandez K, Vay C, Famiglietti A, et al. First class a carbapenemase isolated from enterobacteriaceae in Argentina. Antimicrob Agents Chemother. 2004;48(3):1068–9. https://doi.org/10.1128/AAC.48.3.1068-1069.2004.
Queenan AM, Torres-Viera C, Gold HS, Carmeli Y, Eliopoulos GM, Moellering RC Jr, et al. SME-type carbapenem-hydrolyzing class a beta-lactamases from geographically diverse Serratia marcescens strains. Antimicrob Agents Chemother. 2000;44(11):3035–9. https://doi.org/10.1128/AAC.44.11.3035-3039.2000.
Miller BM, Johnson SW. Demographic and infection characteristics of patients with carbapenem-resistant Enterobacteriaceae in a community hospital: development of a bedside clinical score for risk assessment. Am J Infect Control. 2016;44(2):134–7. https://doi.org/10.1016/j.ajic.2015.09.006.
Hu FP, Guo Y, Zhu DM, Wang F, Jiang XF, Xu YC, et al. Resistance trends among clinical isolates in China reported from CHINET surveillance of bacterial resistance, 2005-2014. Clin Microbiol Infect. 2016;22(Suppl 1):S9–14. https://doi.org/10.1016/j.cmi.2016.01.001.
Wang Q, Wang XJ, Wang J, Ouyang PW, Jin CM, Wang RB, et al. Phenotypic and genotypic characterization of Carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin Infect Dis. 2018;67(suppl_2):S196–205. https://doi.org/10.1093/cid/ciy660.
Tian L, Zhang Z, Sun Z. Antimicrobial resistance trends in bloodstream infections at a large teaching hospital in China: a 20-year surveillance study (1998-2017). Antimicrob Resist Infect Control. 2019;(8):86.
Yu J, Tan K, Rong Z, Wang Y, Chen Z, Zhu X, et al. Nosocomial outbreak of KPC-2- and NDM-1-producing Klebsiella pneumoniae in a neonatal ward: a retrospective study. BMC Infect Dis. 2016;16(1):563. https://doi.org/10.1186/s12879-016-1870-y.
Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. https://doi.org/10.1016/S1473-3099(17)30489-9.
Hu H, Mao J, Chen Y, Wang J, Zhang P, Jiang Y, et al. Clinical and microbiological characteristics of community-onset Carbapenem-resistant Enterobacteriaceae isolates. Infect Drug Resist. 2020;13:3131–43. https://doi.org/10.2147/IDR.S260804.
Wang Y, Lin Q, Chen Z, Hou H, Shen N, Wang Z, et al. Construction of a risk prediction model for subsequent bloodstream infection in intestinal carriers of Carbapenem-resistant Enterobacteriaceae: a retrospective study in hematology department and intensive care unit. Infect Drug Resist. 2021;14:815–24. https://doi.org/10.2147/IDR.S286401.
Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–75. https://doi.org/10.1093/femsre/fux013.
Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–96. https://doi.org/10.1016/S1473-3099(13)70190-7.
Pan F, Tian D, Wang B, Zhao W, Qin H, Zhang T, et al. Fecal carriage and molecular epidemiology of carbapenem-resistant Enterobacteriaceae from outpatient children in Shanghai. BMC Infect Dis. 2019;19(1):678. https://doi.org/10.1186/s12879-019-4298-3.
Zhu J, Sun L, Ding B, Yang Y, Xu X, Liu W, et al. Outbreak of NDM-1-producing Klebsiella pneumoniae ST76 and ST37 isolates in neonates. Eur J Clin Microbiol Infect Dis. 2016;35(4):611–8. https://doi.org/10.1007/s10096-016-2578-z.
Lubbert C, Faucheux S, Becker-Rux D, Laudi S, Durrbeck A, Busch T, et al. Rapid emergence of secondary resistance to gentamicin and colistin following selective digestive decontamination in patients with KPC-2-producing Klebsiella pneumoniae: a single-Centre experience. Int J Antimicrob Agents. 2013;42(6):565–70. https://doi.org/10.1016/j.ijantimicag.2013.08.008.
Acknowledgments
The authors would like to thank the clinical and laboratory staff of the Tongji hospital for facilitating the collection of the rectal samples, and Doctor Na Shen for assistance with statistical analysis.
Funding
This work was supported by the National Mega Project on Major Infectious Disease Prevention (2017ZX10103005–007) and Natural Science Foundation of Hubei (2019CFB666). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
QL performed experiments. YW designed the study. JY helped in literature survey and conducting the experiments. SL, YZ, HW, XL and DL were responsible for the acquisition of data and reviewed the final draft of the manuscript. YL and GT analysed data. LM and ZC drafted and revised the manuscript. ZS conceived the idea and interpreted the results. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Consent to publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Tongji Hospital ethics committee for research in health. Written informed consent was waived by the Tongji Hospital ethics committee for research in health due to the anonymized retrospective nature of the analysis.
Competing interests
The authors have no competing interests to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lin, Q., Wang, Y., Yu, J. et al. Bacterial characteristics of carbapenem-resistant Enterobacteriaceae (CRE) colonized strains and their correlation with subsequent infection. BMC Infect Dis 21, 638 (2021). https://doi.org/10.1186/s12879-021-06315-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-021-06315-0