Abstract
Background
Enterobacter cloacae complex (ECC) is one of the most common extended-spectrum β-lactamase and carbapenemase-producing pathogen that threatens millions of the elderly and vulnerable sick persons. The objective of this study was to perform the molecular characteristics of the carbapenem-resistant E. cloacae complex (CREC) emerged in Heilongjiang Province of China.
Methods
Six CREC strains were isolated from the patients with infectious diseases. The identities of ECC isolates were confirmed by sequencing the polymerase chain reaction (PCR) products of 16S rRNA gene. The characterization of the CREC isolates were analyzed by sequencing PCR products of the carbapenemase, ampC and fluoroquinolone resistance genes and performing multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE) and whole genome sequencing.
Results
All 6 isolates harbored multiple resistance genes. Of them, 5 carried metallo-β-lactamases and one was blaKPC-2-positive. The levofloxacin and ciprofloxacin-resistant strains had substitutions of gyrA83, gyrA87, and parC80 in the quinolone-resistance determining regions. The MLST analyses revealed that 6 isolates belonged to five sequence types (ST520, ST528, ST1119, ST1120, and ST93) while the PFGE patterns of the isolates fallen into four clusters. The strain ST1120 was found to carry two separated plasmids that encode blaNDM-1 and blaIMP-4.
Conclusions
Our study, for the first time, identified a CREC strain that co-produces blaNDM-1 and blaIMP-4 in the Northeast China. Our finding emphasizes an urgent need for more intensive surveillance and precaution measures to prevent the CERC spread.
Similar content being viewed by others
Background
The carbapenem-resistant Enterobacteriaceae (CRE) may cause serious health problems for the elderly and sick persons because the germ has become resistant to most of antibiotics. Among all Enterobacteriaceae, Enterobacter cloacae complex (ECC) ranks the third in its ability to cause infections in wounds, urinary tract, intra-abdominal foci, blood, lungs, skin, and other soft tissues [1]. In clinics, Carbapenems are used as a last resort to treat severe Gram-negative bacterial infections when other antibiotics have failed. However, some strains of ECC could develop Carbapenem-resistance through the mechanisms by which the bacteria either constitutively overexpress AmpC β-lactamases and make mutations that reduce its membrane permeability or enhance efflux of antibiotics, or, more commonly, they gain a plasmid that encodes carbapenemase genes to degrade the antibiotics. While the membrane defect associated resistance may be disseminated via clonal expansion, bacteria carrying cabapenemase genes can be spread among different bacterial strains and species by horizontal transfer of large conjugative plasmids. The significance of the global spread of carbapenemase-producing ECC isolates through patient transfer or colonized travelers in Europe, North America, the Far East and Australia has been well described in two reviews [2, 3]. Recent meta-analysis and multicenter clinical study demonstrated a more than two-fold increased risk of mortality in patients infected with carbapenem-resistant gram-negative, MDR-positive bacteria, and a significant increase in healthcare cost [4, 5]. It has been reported that mortality rate in the patients with CRE infections was as high as 44% [6]. The global spread of drug-resistant bacteria also has a significant impact on the environment and on wild animals and livestock since they can acquire MDR bacteria in hospitals and other waste sources [7]. Because multiple mechanisms are involved in the development of drug-resistance, accurate identification of species and subspecies within the CRECs is needed to monitor the outbreaks, control the infections, and develop better strategies to fight the bacteria.
Several lines of evidence have shown that high-risk bacterial clones were resulted from the global spread of antimicrobial resistance genes in ECC. It has been reported that the extended-spectrum beta-lactamase (ESBL)-producing ECC belonged to ST66, ST78, ST108, and ST114; ST78, and ST114 clusters, and they carried a resistant gene encoding the CTX-M-15 β-lactamase [8]. Peirano et al. reported that ST93 cluster spread in Australia, Belgium, China, Romania, Spain, Thailand, United States, and Vietnam could express a various of carbapenemases encoded by blaIMP-8, blaIMP-14, blaVIM-1, blaNDM-1, blaKPC-2, and blaOXA-48 genes [9]. Another study also highlighted the important role of ST182 in the dissemination of blaNDM-4 [10]. These studies demonstrated that ECC dissemination exists with geographical and seasonal variation in specific regions. However, which carbapenemase genes in ECC clones are passed on in the Heilongjiang Province is unknown. In this study, we isolated 6 CREC strains from patients with infectious diseases in our local hospital and carried out a molecular characterization.
Methods
Collection and identification of bacterial isolates
Six CRECs were collected from a total 169 of clinical ECC isolates found from the patients with infection diseases in the First Affiliated Hospital of Jiamusi University during the study period from September 2016 to March 2018. The minimum inhibitory concentration (MIC) of antibiotic assay was carried out with the Vitek 2 Compact system (BioMerieux, France). MICs of Meropenem (MEM), colistin (COL) and tigecycline (TGC) were determined by the broth microdilution method, and results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) M100-S25 interpretive criteria (CLSI, 2015). The identities of the six isolates were further confirmed by sequencing 16S rRNA as described previously [11]. The study protocol was approved by the Ethics Committee of Jiamusi University Clinical Medical College for research (20180326).
Detection of β-lactamase genes
The modified Hodge test (MHT) and modified carbapenem inactivation test (mCIM) were performed according to CLSI (2015) guidelines to identify the phenotypes of the bacterial strains. Bacterial DNA was extracted using the boiling method. The ß-lactamases type A encoding genes (blaTEM, blaSHV, blaCTX-M-15, blaCTX-M-, blakpc9), Type B encoding genes (blaNDM − 1, blaIMP-4, and blaVIM), Type D encoding genes (blaOXA-23, blaOXA-24, blaOXA-51, blaOXA − 48 and blaOXA-58), and AmpC enzyme encoding genes (blaACC, blaDHA, blaCMY) were detected by polymerase chain reaction (PCR) assays, followed by sequencing PCR product as described previously [12].
Identification of the drug resistant determinants in the quinolone resistance determining region
The determinants in the plasmid-mediated quinolone resistance (PMQR) (qnrA, qnrB, qnrS, aac (6)-Ib, and qepA) and the quinolone resistance determining region (QRDR) (gyrA and parC) were examined by PCR as described previously [13, 14].
Multilocus sequence typing (MLST) and pulsed-field gel electrophoresis (PFGE)
Multilocus sequence types (MLST) of seven housekeeping genes (dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB) were conducted to investigate the genetic relationships of CREC isolates. Briefly, PCR products were sequenced as described [12], and the DNA sequences were aligned with those in the MLST database (http://pubmlst.org/ecloacae) to identify allelic numbers and sequence types (ST). Molecular phylogenetic analyses were performed by using the Maximum Likelihood method and evolutionary analyses in MEGA7 software. CREC isolates were further investigated by the PFGE. Briefly, extracted bacterial DNAs were digested with XbaI restriction enzyme, and the digested DNA fragments were then electrophoresed on agarose gel as previously described [15]. The results were interpreted according to international criteria and allocated into clusters using a cut-off value of 80% genetic similarity [15].
Plasmid analysis
Plasmid DNA was extracted from the CREC isolates with QIAamp DNA Mini prep Kit (Qiagen, Hilden, Germany) and sequenced [12]. A 10-kb fragment library was prepared with PacBio System (Pacific Biosciences, Menlo Park, CA, USA) and assembled using canu software (https://canu.readthedocs.io/en/latest/quickstart.html). The genomic sequences were annotated using Glimmer 3.02 software. The expression of rRNA and tRNA was predicted using Barrnap 0.4.2 and tRNAscan-SE (ver 1.3.1) software, respectively. The annotated information for the predicted genes was obtained using BLAST (NCBI) and alignment with NR, Gene string, and GO Databases. (Additional file 1).
Results
Clinical data assessments
The clinical characteristics of the six patients with CREC infections are summarized in Table 1. The bacterial strains were isolated from the patients in the intensive care unit (ICU), the emergency room, and the infectious disease ward. Two strains were recovered from diabetic patients and other strains were recovered from the patients with fractures, cirrhotic livers, urinary tract infections, and cerebrovascular attacks. The patient carrying strain ECLC6 was a teenager with diabetic ketoacidosis, who had been transferred to another hospital. Most of the patients had increased risk factors for infection or colonization with ECC. All patients were treated with antimicrobials including cefoperazone-tazobactam, aztreonam, and levofloxacin, and etimicin. Out of 6, 5 patients recovered were discharged after successful treatments, one died from bacterial infection.
Species and antibiotic susceptibility
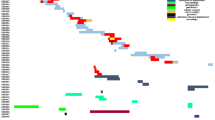
All six isolates were not susceptible to ampicillin-sulbactam, cefazolin, ceftazidime, ceftriaxone, cefepime, meropenem, or imipenem. However, some isolates were susceptible to amikacin, tigecycline, and levofloxacin, and only strain ECLC4 was susceptible to gentamicin and tobramycin (Fig. 1). Because three isolates were resistant to colistin (MIC = 4), we examined whether resistant clones carry the mcr-1 gene. We performed PCR with primers specific to the mcr-1 as described previously [16]. We failed to detect the presence of the mcr-1 gene in 6 isolates.
Genetic analyses of drug-resistance determinants in Enterobacter cloacae complex isolates. Drug-resistant gene presence was labeled as black shading and absence was presented as white shading for each isolate. The corresponding antimicrobial susceptibility was profiled with the VITEK2 automated system; PMICs for levofloxacin (LVX), meropenem (MEM), colistin (COL), and tigecycline (TGC) were determined using the broth microdilution method. ST: sequence type determined with the MLST. PFGE: pulsed-field gel electrophoresis showed four (A, B, C, D) clusters. R: resistance. S: sensitive
β-Lactam and fluoroquinolone resistance determinants
Because the resistance to fluoroquinolone (FQ) and β-lactam has been increasing, we examined if these strains bear FQ resistance genes by PCR assays. We found that all six isolates had multiple resistance genes including blaNDM − 1, blaIMP-4, blaTEM, blaSHV, blaCTX-M-15, blaCTX-M-9, blaKPC, blaACC, and blaDHA. Strains ECLC1-ECLC5 but not strain ECLC6 carried a metallo-β-lactamase. Interestingly, strain ECLC3 possessed both blaNDM-1 and blaIMP-4. All six strains neither harbor blaOXA − 48, blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-58 genes that encode class D carbapenemases, nor carry blaCMY gene. We next examined PMQR determinants (qnrA, qnrB, qnrS, aac (6)-Ib, and qepA) as well as QRDR genes (gyrA and parC) by PCR assays. We found that all clones harbored qnrB and qnrS but not qnrA. All 6 strains contained aac (6′)-Ib-cr but not qepA. Subsequent DNA sequence analyses revealed that the levofloxacin and ciprofloxacin-resistant strains carried gyrAS83F, gyrAS87V, and parCS80I mutations in gyrA83, gyrA87, and parC80 genes in the QRDR (Fig. 1).
Epidemiological links in CREC
To identify the possible phylogenetic relationships of the CREC strains, we performed molecular characterization and genotyping of the drug-resistant bacterial strains. By MLST analyses, we found that the six isolates could be clustered into five STs. The phylogenetic tree built from the sequence data of the seven MLST genes showed 4 phylogenies of the isolates including ST93, ST520, ST1119 and ST1120 (data not shown). Of them, ST1119 and ST1120 appeared new. It should be noticed that CREC3, CREC4 and CREC6 isolated different departments of the hospital were highly similar. By PFGE analyses, the isolates were grouped into clusters A (isolate numbers 2 and 5), B (isolate number 1), C (isolate numbers 3 and 6), and D (isolate number 4) with a cutoff of 80% genetic similarity. Consistent with MLST results, the bacterial strains isolated from different departments (isolate numbers 3, 4, and 6) were highly similar in their PFGE band patterns.
Genetic characterization of NDM-1 and IMP-4 in ST1120
To characterize the genetic environment of the resistance genes, we isolated the plasmids and sequenced the DNA. Our sequence analyses revealed that the blaNDM-1 gene was in a plasmid also containing blaNDM-1, blaSHV-12 and blaSHV-35 resistance genes (Fig. 2a). A partial insertion sequence element, IS5, was located upstream of the blaNDM-1, bleMBL, and tat genes. The blaSHV-12 was downstream of the DeoR gene (Fig. 2a). The characterized structure of blaNDM-1- carrying plasmid is like the blaNDM-13 containing plasmid, PZHDC33 as indicated in Figs. 2a & b. An insertion sequence element, IS4, was located upstream of the blaNDM-14 and bleMBL genes in PZHDC33 plasmid. The blaSHV-12 and DeoR genes were localized at the same positions as PC66-NDM-1 plasmid identified in ST1120. Interestingly, the BlaIMP-4 gene was localized in a separate plasmid. A detailed map of the plasmid harboring blaIMP-4 in ST1120 strain was not constructed yet because of short-read sequencing and insufficient DNA sequences. Thus, it is impossible to compare the sequence of blaIMP-carrying plasmid with previously characterized plasmids yet. It was likely that the Tn3 contributed to the transport of blaIMP-4 (Fig. 2c). The plasmid from isolate ST1120, designated as pC66-NDM-1, formed a 75 kb circular construct which is highly homologous to 54 kb plasmid, KX094555.1 as shown in Figs. 3a &b. The pC66-NDM-1 harbored three antimicrobial drug resistance genes (blaSHV-35, blaNDM-1, and blaSHV-12) in a region of approximately 40 kb from nucleotide position 271 to 42,784, and a heavy metal resistance locus between nucleotide positions 35,333–35,004 encoding the periplasmic divalent cation tolerance protein CutA.
The genetic characterization of blaNDM-1- and blaIMP-4- plasmids in ST1120. The blaNDM-1 gene was in a plasmid containing blaNDM-1, blaSHV-12, and blaSHV-35 resistance genes. The blaNDM-1- containing plasmid (a) was compared with previous published plasmid PZHDC33 carrying blaNDM-1 (b). A partial insertion sequence element, IS5, was located upstream of the blaNDM-1, bleMBL, and tat genes. The blaSHV-12 was downstream of the DeoR gene. The genetic characterization of blaIMP-carrying plasmid (c) was partially sequenced and presented
Schematic maps of blaNDM–carrying plasmids. A schematic mapping of plasmid pC66-NDM-1 (a). The first ring indicates the coordinates of the complete plasmid circle. The 2outer rings represent the forward and reverse open reading frames, respectively. A similar blaNDM–carrying plasmid of KX094555.1(b) was also presented
Discussion
In China, the first CREC was a KPC-producing strain isolated from Shanghai in 2010 [17]. Subsequently, blaNDM, blaIMP, and blaVIM of ECC were identified from various geographical regions in China as well as in other countries [18, 19]. It has been reported that strain BlaNDM-1 was the predominant drug-resistant pathogen of CPE in both animals and humans in China and that the bacteria was spread either through tourist traveling or horizontal transmission of plasmid DNA from one bacterial strain to others [19, 20]. Consistent with the previous reports, we demonstrated in this study that two out of six CREC isolates (33%) carried blaNDM-1. The ST1120 was highly homologous to high-risk ECC clone of ST93 isolated from the Ningxia Province of Western China, where the risk of bacterial transmissions was low because of low population and few tourist travels as compared to the primary global endemic regions of the eastern China [21]. Our studies found that patients carrying the blaNDM gene have not traveled to any epidemic areas before becoming sick, suggesting these isolates might acquire and share the carbapenem resistance genes via horizontal transfer between bacterial species.
We also revealed a new CREC strain co-produced both blaNDM-1 and blaIMP-4 from two separated plasmids isolated from ST1120 strain from the patient with a urinary tract infection in the Northeast China. Because the CREC isolate was resistant to most current antibiotics and was resistant to colistin too (MIC = 4), we assumed that the bacterial might carry the mobilized colistin resistance-1 (mcr-1) gene that can horizontally transfer between different strains of a bacterial species. Thus, we performed PCR with primers specific to mcr-1. In contrast to our prediction, we failed to detect the presence of the mcr-1 in the isolate and therefore excluded the possibility. Our studies indicate that the molecular mechanism of colistin-resistance of this strain is independent on mcr-1 gene and the genes that confer colistin-resistance remain to be identified.
In the present study, we found both the blaCTX-M-3 gene and the blaTEM-1 gene were co-present with qnrS genes in a single plasmid in the ST1120 strain (data not shown). This plasmid bearing blaCTX-M-3, blaTEM-1 and qnrS genes appeared independent on the hosts and was able to transform into a variety of organisms [22], resulting in high-level resistance to quinolones and other antibiotics such as tetracycline and florfenicol. Our studies indicate that the quinolone resistance was not resultant from overexpression of bacterial efflux pumps since the qepA gene was not present in the CREC strains.
Mutations of genes encoding DNA gyrase (gyrA, gyrB, gyC, parC. And parE) and topoisomerase IV (parC and parE) on bacterial chromosomes were also associated with FQ resistance [23, 24]. In our study, we found existence of mutations in three loci of ECLC5 strain and two loci of ECLC6 strain. Our broth microdilution assay revealed that the levofloxacin MIC for these two isolates was 16 μg/mL and 32 μg/mL, respectively, suggesting that the genetic mutations at these loci contributed to high levels of resistance to levofloxacin.
ST93 of CREC has been spread in other 11 cities but has not been reported in Heilongjiang Province of China before [19]. In this study, we were not only able to detect two ST93 strains present in Heilongjiang region but also uncovered the new strain of ST1120 that is distinct from ST93 in two housekeeping genes (gyrB and pyrG), emphasizing that more epidemiological tests are needed to prevent the spread of the high-risk clones, ST93 and novel clone, ST1120 in China.
Conclusions
In conclusion, we isolated 6 CREC strains from patients with infectious diseases and carried out a molecular characterization. We found all 6 isolates harbored multiple resistance genes. Five of them carried metallo-β-lactamases and one strain had blaKPC-2. The levofloxacin and ciprofloxacin-resistant strains had substitutions of gyrA83, gyrA87, and parC80 in the QRDR. Our molecular genetic analyses revealed that 6 isolates belonged to five sequence types (ST520, ST528, ST1119, ST1120, and ST93). We also discovered a new strain of ST1120 that carried two plasmids encoding blaNDM-1 and blaIMP-4. Our study has some limitations. First, the study involves a small number of isolates analyzed at a single medical center. Second, although we have characterized the plasmid containing blaNDM-1, a detailed map of the plasmid harboring blaIMP-4 in ST1120 strain was not constructed yet because of short-read sequencing and insufficient DNA sequences [25]. Thus, it is impossible to compare the sequence of blaIMP-4-containing plasmid with previously characterized plasmids yet. Third, we did not examine the colistin-resistant isolates for the presence of other mcr alleles (2 to 9) and chromosomal mutations mediating colistin-resistance in our future studies. The molecular mechanisms by which the mcr-1-negative isolates were resistant to colistin but remained susceptible to amikacin and tigecycline remain unanswered yet. Fourth, we did not sequence and analyze the all 6 isolates, and the other 5 strains remain analyzed in future.
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files.
Abbreviations
- CLSI:
-
Clinical and Laboratory Standards Institute
- COL:
-
Colistin
- CPE:
-
Carbapenemase- producing Enterobacteriaceae
- CRE:
-
Carbapenem-resistant Enterobacteriaceae
- CREC:
-
Carbapenem-resistant E. cloacae complex
- ECC:
-
Enterobacter cloacae complex
- ESBL:
-
Extended-spectrum beta-lactamase
- FQ:
-
Fluoroquinolone
- ICU:
-
Intensive care unit
- mCIM:
-
Modified carbapenem inactivation test
- MEM:
-
Meropenem
- MHT:
-
Modified Hodge test
- MLST:
-
Multilocus sequence typing
- MLST:
-
Multilocus sequence typing
- PCR:
-
Polymerase chain reaction
- PFGE:
-
Pulsed-field gel electrophoresis
- PMQR:
-
Plasmid-mediated quinolone resistance
- QRDR:
-
Quinolone resistance determining region
- ST:
-
Sequence types
- TGC:
-
Tigecycline
References
Temkin E, Adler A, Lerner A, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann N Y Acad Sci. 2014;1323:22–42.
Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15–21.
Brink AJ. Epidemiology of carbapenem-resistant gram-negative infections globally. Curr Opin Infect Dis. 2019;32(6):609-616. https://doi.org/10.1097/QCO.0000000000000608.
McCann E, Srinivasan A, DeRyke CA, Ye G, DePestel DD, Murray J, Gupta V, et al. Open Forum Infect Dis. 2018;5(10):ofy241.
Nathwani D, Raman G, Sulham K, Gavaghan M, Menon V. Clinical and economic consequences of hospital-acquired resistant and multidrug-resistant Pseudomonas aeruginosa infections: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2014;3(1):32.
Falagas ME, Tansarli GS, Karageorgopoulos DE, Vardakas KZ. Deaths attributable to carbapenem-resistant Enterobacteriaceae infections. Emerg Infect Dis. 2014;20(7):1170–5.
Dolejska M, Papagiannitsis CC. Plasmid-mediated resistance is going wild. Plasmid. 2018;99:99–111.
Annavajhala MK, Gomez-Simmonds A, Uhlemann AC. Multidrug-resistant Enterobacter cloacae Complex emerging as a global. Diversifying Threat Front Microbiol. 2019;10:44.
Peirano G, Matsumura Y, Adams MD, Bradford P, Motyl M, Chen L, Kreiswirth BN, Pitout JDD. Genomic epidemiology of global Carbapenemase-producing Enterobacter spp., 2008-2014. Emerg Infect Dis. 2018;24(6):1010–9.
Paskova V, Medvecky M, Skalova A, Chudejova K, Bitar I, Jakubu V, Bergerova T, Zemlickova H, Papagiannitsis CC, Hrabak J. Characterization of NDM-encoding plasmids from Enterobacteriaceae recovered from Czech hospitals. Front Microbiol. 2018;9:1549.
Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62(2):625–30.
Gong X, Zhang J, Su S, Fu Y, Bao M, Wang Y, Zhang X. Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the eastern region of Heilongjiang Province. China BMC Infect Dis. 2018;18(1):417.
Rodriguez-Martinez JM, Velasco C, Pascual A, Garcia I, Martinez-Martinez L. Correlation of quinolone resistance levels and differences in basal and quinolone-induced expression from three qnrA-containing plasmids. Clin Microbiol Infect. 2006;12(5):440–5.
Zhuang N, Chen X, Yue L, Liao X, Liu Y. Detection of PMQR gene in Escherichia coli isolated from animals. Sci Agric Sin. 2012;45(10):2052–7.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–9.
Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–8.
Wu Q, Liu Q, Han L, Sun J, Ni Y. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 and ArmA 16S rRNA methylase conferring high-level aminoglycoside resistance in carbapenem-resistant Enterobacter cloacae in China. Diagn Microbiol Infect Dis. 2010;66(3):326–8.
Dai W, Sun S, Yang P, Huang S, Zhang X, Zhang L. Characterization of carbapenemases, extended spectrum beta-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infect Genet Evol. 2013;14:1–7.
Jin C, Zhang J, Wang Q, Chen H, Wang X, Zhang Y, Wang H. Molecular characterization of Carbapenem-resistant Enterobacter cloacae in 11 Chinese cities. Front Microbiol. 2018;9:1597.
Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, Tyrrell JM, Zheng Y, Wang S, Shen Z, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2:16260.
Shi Z, Zhao H, Li G, Jia W. Molecular characteristics of Carbapenem-resistant Enterobacter cloacae in Ningxia Province. China Front Microbiol. 2017;8:94.
Galani I, Souli M, Mitchell N, Chryssouli Z, Giamarellou H. Presence of plasmid-mediated quinolone resistance in Klebsiella pneumoniae and Escherichia coli isolates possessing blaVIM-1 in Greece. Int J Antimicrob Agents. 2010;36(3):252–4.
Hooper DC. Mechanisms of fluoroquinolone resistance. Drug Resist Updat. 1999;2(1):38–55.
Hooper DC. Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin Infect Dis. 2001;32(Suppl 1):S9–s15.
Arredondo-Alonso S, Willems RJ, van Schaik W, Schurch AC. On the (im) possibility of reconstructing plasmids from whole-genome short-read sequencing data. Microb Genom. 2017;3(10):e000128.
Acknowledgements
We thank Dr. Ming-Gui Wang from Huashan Hospital Affiliated to Fudan University and Dr.Yun-Song YU from Sir Run Run Shaw Hospital affiliated to Zhejiang University for their assistances.
Funding
This study was supported by Excellent Team of Young Teachers Foundation of Heilongjiang Province (2018-KYYWF-0916), Heilongjiang Provincial Health and Family Planning Commission on Scientific Research Project (2018–290 and 2018–291) and Jiamusi University President Innovation and Entrepreneurship Fund Project (XZYF2018–43). They are financial support in this study.
Author information
Authors and Affiliations
Contributions
XZ conceived and designed the study. YZ and JZ wrote this paper. YZ, JZ, YF1, and CL performed the experiments. KH, SS, LY, YG, YF2 analyzed the data. All authors approved the final version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of Jiamusi University Clinical Medical College for research. The committee’s reference number is 20180326. Individual informed consent was waived by the ethics committee listed above because this study used currently existing sample collected during the course of routine medical care and did not cause any additional risks to the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1.
The plasmid sequence with NDM
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhao, Y., Zhang, J., Fu, Y. et al. Molecular characterization of metallo-β-lactamase- producing carbapenem-resistant Enterobacter cloacae complex isolated in Heilongjiang Province of China. BMC Infect Dis 20, 94 (2020). https://doi.org/10.1186/s12879-020-4768-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-020-4768-7