Abstract
Background
The present study aimed to describe the genetic diversity of HIV-1, as well as the resistance profile of the viruses identified in HIV-1 infected pregnant women under antiretroviral therapy in the state of Pará, Northern Brazil.
Methods
Blood samples were collected from 45 HIV-1 infected pregnant to determine the virus subtypes according to the HIV-1 protease (PR) gene and part of the HIV-1 reverse transcriptase (RT) gene by sequencing the nucleotides of these regions. Drug resistance mutations and susceptibility to antiretroviral drugs were analyzed by the Stanford HIV Drug Resistance Database.
Results
Out of 45 samples, only 34 could be amplified for PR and 30 for RT. Regarding the PR gene, subtypes B (97.1%) and C (2.9%) were identified; for the RT gene, subtypes B (90.0%), F (6.7%), and C (3.3%) were detected. Resistance to protease inhibitors (PI) was identified in 5.8% of the pregnant, and mutations conferring resistance to nucleoside reverse transcriptase inhibitors were found in 3.3%, while mutations conferring resistance to non-nucleoside reverse transcriptase inhibitors were found in 3.3%.
Conclusions
These results showed a low frequency of strains resistant to antiretroviral drugs, the prevalence of subtypes B and F, and the persistent low transmission of subtype C in pregnant of the state of Pará, Brazil.
Similar content being viewed by others
Background
The extensive genetic diversity of HIV-1 is one of its main features and allows the classification of the strains isolated in various parts of the world into four groups (M, N, O, and P). The M group may be further divided into subtypes, subsubtypes, circulating recombinant forms (CRF), and unique recombinant forms (URF) [1]. The comprehensive description of the genetic diversity of HIV-1 in different geographic regions in Brazil has a great impact on the epidemiological surveillance and may influence the diagnosis as well as the development of vaccines.
In Brazil, 798,366 cases of AIDS were reported through June 2015 [2], which corresponds to one third of cases recorded in South and Central America. The North region of Brazil comprises 5.7% of the reported cases and has an AIDS incidence rate of 25,7/100,000 people, with the largest number of cases (43.47%) occurring in Pará [2].
In the period between 2000 and 2015, 92,210 cases of HIV-1 were reported among pregnant women in Brazil; of which, 6548 cases (7.1%) occurred in the North region and the highest number was observed in the state of Pará (39.92%), followed by the states of Amazonas (3.08%) and Rondônia (6.98%) [2].
The majority of the studies reporting the genetic diversity of HIV-1 in Brazil are associated with the South and Southeast regions of the country as these regions were the epicenter of the HIV/AIDS epidemic in the 1980s. The South region of Brazil presents a different pattern compared to other regions in the country, with a high prevalence of subtype C and several recombinant forms (BC, BF1, and CRF31_BC) [3]. In the South [4] and Northeast [5] regions, subtypes B and F1 are the most prevalent, although various recombinant forms have been found.
The North region of Brazil, the largest land area of the country, circulating HIV-1 subtypes have been described in the states of Amazonas (B and F) [6], Amapá (B and F), and Pará and have presented higher HIV-1 genetic diversity (B, F, C, D, and the recombinant form CRF02_AG). These data demonstrate the importance of Pará as a port of HIV entry in Northern Brazil [7]. The present study describes the circulating HIV-1 subtypes among HIV-1-infected pregnant women as well as the resistance profile of the strains isolated in the state of Pará, Northern Brazil.
Methods
Regions and population studied
The study participants were women from the Reference Unit for Special Infectious and Parasitic Diseases (URE-DIPE) and the Reference Unit for Maternal-Child and Adolescent care (UREMIA) in Belém, state of Pará, Brazil.
This study included the entire population of HIV-1-infected pregnant women (n = 45) who received care from April 2007 to October 2008. These women were referred to the URE-DIPE and UREMIA by the Health Care Units located in 14 cities: Belém, (18) São Miguel do Guamá, (1) Paragominas, (1) Moju, (1) Concórdia do Pará, (1) Abaetetuba, (2) Marabá, (1) Marituba, (3) Mãe do Rio, (1) Igarapé-Miri, (1) Icoaraci, (3) Castanhal, (2) Canaã dos Carajás, (1) and Ananindeua (9).
This study was approved by the Human Research Ethics Committee of the João de Barros Barreto University Hospital, protocol number 2090/05. All patients signed a free and informed consent form at the time of sample collection and answered a structured epidemiological questionnaire (through an interview) applied by either a physician of the Unit that performed the clinical monitoring or a member of the study. The questionnaire included demographic and epidemiological information such as the date of the first positive HIV-1 test, history of partners from other states or countries, use of the antiretroviral therapy (ART), date when the patients started ART, and the stage of pregnancy when HIV-1 was diagnosed (for those women who did not know they were HIV-1 positive). Moreover, other information was obtained from the participants’ medical records such as the T CD4+ lymphocyte (CD4TL) count and plasma HIV-1 viral load; information was obtained from the reference units protocols, using data from the most recent tests at the time of interview.
Blood samples (5 mL) were collected in two tubes containing EDTA using a vacuum system. The samples (plasma and cellular components) were transported to the Virology Laboratory at the Institute of Biological Sciences, Federal University of Pará, and stored at −70 °C until use.
Polymerase chain reaction (PCR)
DNA was extracted from the cellular components of the blood using a Puregene kit (Puregene, Gentra Systems, Inc., USA) according to the manufacturer’s instructions. The nucleic acid was eluted and stored at −20 °C until use. Subsequently, DNA was submitted to a nested PCR to amplify part of the region of the HIV-1 RT and HIV-1 PR. The first PCR cycle was performed using 4 μg of extracted DNA, 125 mM of each dNTP (Perkin-Elmer, USA), 20 pmol/μL of each of the two external primers, 2.5 mM MgCl2 and 10× buffer (Perkin-Elmer, USA) in a final volume of 50 μL. The reactions were performed in a thermocycler (Perkin-Elmer, USA) for 5 min at 94 °C followed by 35 cycles at 94 °C (40 s), 50 °C (40 s), and 72 °C (1 min) and a 10 min extension at 72 °C. Five microliters of the amplified product was used in the nested PCR along with a set of internal primers in a final volume of 50 μL. The same incubation times and temperatures of the first reaction were used. For the PR, DP10/DP11 were used as external primers, and DP16/DP17 were used as internal primers [8]. For amplification of the fragment of the RT gene, RT09/RT12 were used as external primers, and RT01/RT04 were used as internal primers [9]. The amplified products were analyzed by electrophoresis on a 2% agarose gel. After amplification, the fragments were purified using the QIA Quick Purification Kit (Qiagen, USA) prior to nucleotide sequencing.
HIV-1 genetic subtypes, nucleotide sequencing and phylogenetic analysis
A 2 μL aliquot of PCR product was sequenced using the Big Dye Terminator Kit (Applied Biosystems, Foster City, CA, USA) on an automatic sequencer (DNA sequencer model 310, Biosystems, Foster City, CA, USA).
Prior to the phylogenetic analysis, an HIV-1 gene subtyping tool available on the website (https://hivdb.stanford.edu/hivdb/by-mutations/) was used to determine whether the obtained sequences were pure subtypes or CRFs.
The nucleotide and amino acid alignments were performed using the BioEdit version 5.0.9 software program [10]. The evolutionary distance was estimated using Kimura’s two parameter model, and the phylogenetic trees were constructed using the neighbor-joining and maximum likelihood methods; the reliability of the tree topology was tested by bootstrap analysis with 1000 replicates using the MEGA 5.0 software program [11]. The sequences determined in this study were aligned with the reference sequences of the subtypes obtained from the Los Alamos database (https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html), which are representative of the current HIV-1 strains circulating around the world.
Resistance analysis
Drug resistance mutations (DRM) and susceptibility to antiretroviral drugs were analyzed using the Calibrated Population Resistance (CPR) tool (version 5.0 beta, available at http://cpr.stanford.edu/cpr.cgi). The antiretroviral susceptibility mutation profiles were analyzed using the Stanford HIV Drug Resistance Database (hivdb.stanford.edu).
Results
Blood samples were collected from 45 HIV-1-infected pregnant women receiving ART in the city of Belém, Pará, Brazil, from April 2007 to October 2008. The mean age was 25.06 years (ranging from 14 to 37 years), and the majority of the women (64.4%) were three to six months pregnant. Table 1 shows the main demographic characteristics of the group. Most women reported being single (64.4%), having only basic education (48.9%), without an occupation other than housewives (60%).
Regarding the behavioral aspects analyzed, 51.1% mentioned having a heterosexual partner, 20% assumed having relationships with HIV-1 positive partners, 24.4% had a partner who was a non-injecting drug user, and 60% denied having had anal intercourse. The vast majority of women (75.6%) reported having only one partner per month, 62.2% had a relationship with people from other states of Brazil, and 20% had already been diagnosed with a sexually transmitted infection (STI). Regarding ART, 29 women (64.4%) were using therapy at the time of sample collection, with time ranging between three months to four years, 12 women (26.7%) had not yet started therapy, and 3 women (6.7%) reported having stopped the treatment against HIV at least once.
Among the pregnant women, 31 (68.9%) had other children (mean 2.54 children), while 14 (31.1%) were experiencing their first pregnancy. It is noteworthy that 22 women who already had children (48.9%) reported that they did not attend a prenatal care for any of the pregnancies, and therefore, they breastfed their first children and stopped after the diagnosis of HIV infection. Only two children (4.4%) were found to be infected by HIV-1.
The values of CD4TL count were obtained from 28 participants, and plasma HIV-1 viral load was obtained from 23 participants (Table 2). There was no statistically significant association between the stage of pregnancy, use of ART, mean CD4TL count (365.04 ± 236.25 cells/μL, ranging from 122 to 1210 cells/μL), and mean plasma HIV-1 viral load (log10 3.699, ranging from 2.699 to 4.699).
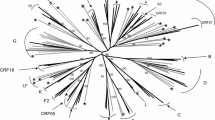
The PR gene region was successfully amplified in 34 samples, and the phylogenetic tree showed that 97.1% of the sam ples were subtype B and 2.9% were subtype C (Fig. 1). However, the RT region (Fig 2) was amplified in 30 samples (which were not necessarily the same samples amplified for the PR gene region), and showed that 90.0% were classified as subtype B, 6.7% as subtype F, and 3.3% as subtype C. Only 19 samples were amplified for both regions and no recombinant forms was observed. The information of CD4TL, viral load, PR mutation, RT mutations and ART treatment status as shown in Table 2.
Phylogenetic analysis based on the alignment of 249 nucleotides of the pro gene of samples from Pará (♦) with reference strains from the Los Alamos database. The sequence SIVCPZgab was used as an outgroup. The tree was constructed using the Kimura 2-parameter neighbor-joining (NJ) method. The numbers on the nodes of the tree indicate the bootstrap value obtained using 2000 replicates. The scale bar represents a difference of 2%
Phylogenetic tree based on the alignment of the 450 nucleotides of the reverse transcriptase gene of 24 samples from Pará (•) with reference strains from the Los Alamos database. The tree was constructed using the Kimura 2-parameter neighbor-joining (NJ) method. The numbers on the nodes of the tree indicate the bootstrap value obtained using 2000 replicates. The scale bar represents a difference of 1%
Four women presented HIV-1 strains with some sign of resistance to ARV, being two (5.8%; #14045 and #15278) to protease inhibitors (PIs), one (3.3%; #15275) to nucleoside reverse transcriptase inhibitors (NRTIs) (#15275), and one (3.3%; #16078) to non-nucleoside reverse transcriptase inhibitors (NNRTIs).
Woman #14045 (infected with HIV-1; subtype BPR/BRT; IP mutations M46I, G48E, L10I, A71V) was 26 years old, lived in Belém, had been using ART since 2005, had three children who were not infected with HIV-1, attended prenatal care during all pregnancies, but she reported stopping her treatment at some point. Another woman (#15278) (HIV-1; subtype BPR/BRT; IP mutation I84V) lived in Abaetetuba, which is a town close to Belém, and had been using ART since 2006, without interruption. She was 27 years old and had two children who were not infected with HIV-1; she and her partner were non-injecting drug users.
Woman #15275 (HIV-1, subtype BPR/BRT; NRTI mutation K70E) was 29 years old, lived in Belém, had two children, did not breastfeed, had only one partner, who was also HIV-positive, had been using ART since 2006, and reported previous interruption of treatment. Pregnant woman #16078 (HIV-1; subtype BPR/BRT; NNRTI mutation K103 N) was 19 years old, was living in Ananindeua, had only one child, did not breastfeed, was a housewife and single, had only one partner who was HIV positive, had been using ART since 2006, and reported previous interruption once.
Discussion
The lack of information associated with low educational level are risk factors for HIV-1 exposure in Brazil [3], and the present study was consistent with the findings that are usually observed: infected persons are young women with low educational level, which currently reflects the population more affected by HIV-1 infection in Brazil [2].
Other social and behavioral information obtained from the examined women also show variables that are often associated with higher risk of transmission of HIV-1 and other STIs. The consistent use of condoms during intercourse was low, and 20% of the surveyed women had contact with HIV-1-positive individuals. Although drugs were not used intravenously, this variable promotes other variables of risk, including unprotected sexual relations with anal intercourse, as previously shown in other studies [12, 13]. Even with the existence of a free treatment program widely distributed across the country, a large percentage of this population was not using ART, and even worse, nearly one third of this population had never used the treatment facilities. Difficulties with ART adherence by HIV-1-positive individuals are often reported, and this could be one explanation for the large number of women who do not receive treatment [14].
Although the immunological response is physiologically compromised during pregnancy and a decrease in the CD4TL count is usually observed [15], no significant difference was found between the mean value of these cells and the pregnancy stage in the present study, which has also been observed in other studies [16, 17].
Studies on the molecular epidemiology of HIV-1 in Brazil show that the B subtype is the most prevalent strain in all described geographical regions of the country, in addition to the decreasing prevalence of the C, D, and F subtypes and the recombinant forms [5, 18–21]. In the present study, the B subtype was also the most prevalent, followed by the F and C subtypes, which had been previously described in the cities of Belém and Macapá in the adult population [8]. It is important to mention that the F subtype could only be found when the RT gene was amplified, which demonstrates the need to amplify at least two genes in studies addressing the occurrence of subtypes and recombinant forms of the virus [22].
The prevalence of subtypes B and F demonstrated in the present study has also been observed in other areas of South America [23, 24]. Most of the women had only one partner, although 20% of them reported relationships with HIV-1-positive men. It is possible that this low activity for HIV-1 transmission did not facilitate the occurrence of recombinant forms of the virus. Moreover, the majority of the patients were already using ART.
Two pregnant women, one living in Belém (#14045) and another living in Abaetetuba (#15278), were carriers of strains with mutations in the protease region (M46I, G48E, L10I, A71V, and I84V). The low prevalence of HIV-1 strains that are resistant to this class of drugs reflects the low circulation of viruses with resistance to PIs in the North region of the country [25], differently from what was observed in Rio de Janeiro [26] and in Goias [27] with a prevalence level around 16%.
The most frequently occurring mutation that conferred resistance to PIs was the M46I mutation, which can be a good indicator of regional differences in HIV-1 strains circulating in the country. However, eight or more mutations are required for a strain to be considered resistant, while partial resistance can be observed by the accumulation of five to seven of these mutations in addition to the combination of several primary and secondary mutations that do not always confer resistance [28]. This resistance development process is complex and requires the accumulation of mutations, which was not observed in the present study. Three mutations that confer resistance to atazanavir were identified in sample #14045 (G16E, M46I, and I62V) and two in sample #15278 (I62V and I84V).
The NRTIs initially used as ART included regimens consisting of tenofovir/emtricitabine, zidovudine/lamivudine, and abacavir/lamivudine with a combination of doses [29]. The M41 L and D67N mutations confer resistance to zidovudine and stavudine [28, 30]. The M184 V mutation confers resistance to lamivudine as well as to abacavir and emtricitabine [28, 31]. The K70E mutation (#15275), which is associated with resistance to NRTIs, and the K103 N mutation (#16078), which is associated with resistance to NNRTIs, were observed in two strains. None of these mutations confers a degree of resistance that is noteworthy for the two classes of drugs used in ART.
Conclusions
The present study indicated that in Pará, Northern Brazil, the frequency of circulating ART-resistant HIV-1 strains is still low among pregnant women, which may suggest the success of the therapy currently used in the country, shown by the absence of mutations indicative of primary resistance in the investigated group. Although the study was performed with samples collected in 2007 and 2008, this is the first approach to study HIV-1 among pregnant women in the state of Para and it will certainly serve as a basis for future studies. The prevalence of subtype B demonstrated that these groups of pregnant women present the same epidemiological pattern as the rest of the population and, in fact, present a lower frequency of other subtypes observed in the region.
Abbreviations
- ART:
-
Antiretroviral therapy
- CD4TL:
-
T CD4+ lymphocyte
- CRF:
-
Circulating recombinant forms
- IDU:
-
Injectable drug user
- NIDU:
-
Non-injectable drug user
- NNRTI:
-
Non-nucleoside reverse transcriptase inhibitor
- NRTI:
-
Nucleoside reverse transcriptase inhibitor
- PI:
-
Protease inhibitor
- RT:
-
Reverse transcriptase
- STI:
-
Sexually transmitted infection
- URE-DIPE:
-
Reference Unit for Special Infectious and Parasitic Diseases
- UREMIA:
-
Reference Unit for Maternal-Child and Adolescent care
- URF:
-
Unique recombinant forms
References
Sharp PM, Hahn BH. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841.
Brasil. Ministério da Saúde. Secretaria de Vigilância em Saúde. Departamento de DST, Aids e Hepatites Virais. Boletim Epidemiológico: Aids e DST. 2014; ano III, n. 1.
Gräf T, Passaes CP, Ferreira LG, Grisard EC, Morgado MG, Bello G, Pinto AR. HIV-1 genetic diversity and drug resistance among treatment naïve patients from southern Brazil: an association of HIV-1 subtypes with exposure categories. J Clin Virol. 2011;51:186–91.
Alcalde R, Guimarães ML, Duarte AJ, Casseb J. Clinical, epidemiological and molecular features of the HIV-1 subtype C and recombinant forms that are circulating in the city of São Paulo, Brazil. Virol J. 2012;9:156.
Santos LA, Monteiro-Cunha JP, Araujo AF, Brites C, Galvao-Castro B, Alcantara LC. Detection of distinct human immunodeficiency virus type 1 circulating recombinant forms in northeast Brazil. J Med Virol. 2011;83:2066–72.
Vicente AC, Otsuki K, Silva NB, Castilho MC, Barros FS, Pieniazek D, Hu D, Rayfield MA, Bretas G, Tanuri A. The HIV epidemic in the Amazon Basin is driven by prototypic and recombinant HIV-1 subtypes B and F. J Acquir Immune Defic Syndr. 2000;23:327–31.
Machado LF, Ishak MO, Vallinoto AC, Lemos JA, Azevedo VN, Moreira MR, Souza MI, Fernandes LM, Souza LL, Ishak R. Molecular epidemiology of HIV type 1 in northern Brazil: identification of subtypes C and D and the introduction of CRF02_AG in the Amazon region of Brazil. AIDS Res Hum Retrovir. 2009;25:961–6.
Janini LM, Pieniazek D, Peralta JM, Schechter M, Tanuri A, Vicente AC, dela Torre N, Pieniazek NJ, Luo CC, Kalish ML, Schochetman G, Rayfield MA. Identification of single and dual infections with distinct subtypes of human immunodeficiency virus type 1 by using restriction fragment length polymorphism analysis. Virus Genes. 1996;13:69–81.
Cerqueira DM, Ramalho ED, Oliveira CP, Silva RR, Franchini M, Felipe MSS, Martins CRF. HIV-1 subtypes and mutations associated to antiretroviral drug resistance in human isolates from Central Brazil. Braz J Microbiol. 2004;35:187–92.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. 1999;S41:95–8.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9.
Nóbrega I, Dantas P, Rocha P, Rios I, Abraão M, Netto EM, Brites C. Syphilis and HIV-1 among parturient women in Salvador, Brazil: low prevalence of syphilis and high rate of loss to follow-up in HIV-infected women. Braz J Infect Dis. 2013;17:184–93.
Miranda AE, Pinto VM, McFarland W, Page K. HIV infection among young pregnant women in Brazil: prevalence and associated risk factors. AIDS Behav. 2014;18:S50–2.
Seidl EM, Melchíades A, Farias V, Brito A. Persons living with HIV/AIDS: factors associated with adherence to antiretroviral treatment. Cad Saude Publica. 2007;23:2305–16.
Hocke C, Morlat P, Chene G, Dequae L, Dabis F. Prospective cohort study of the effect of pregnancy on the progression of human immunodeficiency virus infection. The Groupe d’Epidémiologie Clinique du SIDA en Aquitaine. Obstet Gynecol. 1995;86:886–91.
Palacios R, Senise J, Vaz M, Diaz R, Castelo A. Short-term antiretroviral therapy to prevent mother-to-child transmission is safe and results in a sustained increase in CD4 T-cell counts in HIV-1-infected mothers. HIV Med. 2009;10:157–62.
Heffron R, Donnell D, Kiarie J, Rees H, Ngure K, Mugo N, Were E, Celum C, Baeten JM. Partners in Prevention HSVHIV transmission study. A prospective study of the effect of pregnancy on CD4 counts and plasma HIV-1 RNA concentrations of antiretroviral-naive HIV-1-infected women. J Acquir Immune Defic Syndr. 2014;65:231–6.
Cabral VP, Cunha CB, Magalhaes EF, Pinto-Neto LF, Couto-Fernandez JC, Dietze R, Morgado MG, Ribeiro-Rodrigues R. Human immunodeficiency virus type-1 subtypes of infected patients in Espírito Santo, Brazil. Mem Inst Oswaldo Cruz. 2006;101:881–5.
de Medeiros LB, Lacerda HR, Cavalcanti AM, de Albuquerque MF. Primary resistance of human immunodeficiency virus type 1 in a reference center in Recife. Pernambuco, Brazil. Mem Inst Oswaldo Cruz. 2006;101:845–9.
Monteiro JP, Alcantara LC, de Oliveira T, Oliveira AM, Melo MA, Brites C, Galvão-Castro B. Genetic variability of human immunodeficiency virus-1 in Bahia state, northeast, Brazil: high diversity of HIV genotypes. J Med Virol. 2009;81:391–9.
da Silveira AA, Cardoso LP, Francisco RB, de Araújo Stefani MM. HIV type 1 molecular epidemiology in pol and gp41 genes among naive patients from Mato Grosso do Sul state, central western Brazil. AIDS Res Hum Retrovir. 2012;28:304–7.
Cunha LK, Kashima S, Amarante MF, Haddad R, Rodrigues ES, Silva KL, Lima TA, Castro DB, Brito FC, Almeida EG, Covas DT, Malheiro A. Distribution of human immunodeficiency virus type 1 subtypes in the state of Amazonas, Brazil, and subtype C identification. Braz J Med Biol Res. 2012;45:104–12.
Carrión AG, Laguna-Torres VA, Soto-Castellares G, Castillo M, Salazar E, Negron E, Kolevic L, Montano SM, Sánchez JL, Bautista CT, Oberhelman RA, Kochel TJ. Molecular characterization of the human immunodeficiency virus type 1 among children in Lima, Peru. AIDS Res Hum Retrovir. 2009;25:833–5.
Aulicino PC, Bello G, Guimaraes ML, Ruchansky D, Rocco C, Mangano A, Morgado MG, Sen L. Longitudinal analysis of HIV-1 BF1 recombinant strains in vertically infected children from Argentina reveals a decrease in CRF12_BF pol gene mosaic patterns and high diversity of BF unique recombinant forms. Infect Genet Evol. 2011;11:349–57.
Soeiro CM, Miranda AE, Saraceni V, Lucena NO, Talhari S, Ferreira LC. Mother-to-child transmission of HIV infection in Manaus, state of Amazonas, Brazil. Rev Soc Bras Med Trop. 2011;44:537–41.
de Lourdes TM, Nafea S, Yeganeh N, Santos E, Gouvea MI, Joao E, Ceci L, Bressan C, Cruz ML, Sidi LC, Nielsen-Saines K. High rates of baseline antiretroviral resistance among HIV-infected pregnant women in an HIV referral centre in Rio de Janeiro, Brazil. Int J STD AIDS. 2015;26:922–8.
da Costa ZB, de Lima YA, Martelli CM, Stefani MM. Transmitted HIV resistance among pregnant young women infected with HIV-1 in Brazil. AIDS Patient Care STD. 2013;27:439–41.
Cardoso SW, Grinsztejn B, Velasque L, Veloso VG, Luz PM, Friedman RK, Morgado M, Ribeiro SR, Moreira RI, Keruly J, Moore RD. Incidence of modifying or discontinuing first HAART regimen and its determinants in a cohort of HIV-infected patients from Rio de Janeiro, Brazil. AIDS Res Hum Retrovir. 2010;26:865–74.
Shafer RW. Low-abundance drug-resistant HIV-1 variants: finding significance in an era of abundant diagnostic and therapeutic options. J Infect Dis. 2009;199:610–2.
Johnson JA, Li JF, Wei X, Lipscomb J, Irlbeck D, Craig C, Smith A, Bennett DE, Monsour M, Sandstrom P, Lanier ER, Heneine W. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008. doi:10.1371/journal.pmed.0050158.
Hammer SM, Saag MS, Schechter M, Montaner JS, Schooley RT, Jacobsen DM, Thompson MA, Carpenter CC, Fischl MA, Gazzard BG, Gatell JM, Hirsch MS, Katzenstein DA, Richman DD, Vella S, Yeni PG, Volberding PA. International AIDS society-USA panel. Treatment for adult HIV infection: 2006 recommendations of the international AIDS society-USA panel. JAMA. 2006;16:827–43.
Acknowledgment
We acknowledge all subjects enrolled in the present study.
Funding
This work was supported by grants from the Programa Nacional de DST e AIDS (CSV321/2006), Ministry of Health, Brazil, and Universidade Federal do Pará.
Availability of data and materials
All available data are described in the manuscript.
Authors’ contribution
LFAM, ACRV, RI and MOGI designed the study, analyzed, interpreted data, and wrote the paper. MNF participated in the clinical analysis. IBC and ALBL collected samples and participated in experiments of genotyping. ACRV and RI reviewed the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All work performed in this study was approved by the Human Research Ethics Committee of the Joao de Barros Barreto University Hospital, protocol number 2090/05 HUJBB/UFPA and all patients signed a free and informed consent form before enrolling in this study.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Machado, L.F.A., Costa, I.B., Folha, M.N. et al. Lower genetic variability of HIV-1 and antiretroviral drug resistance in pregnant women from the state of Pará, Brazil. BMC Infect Dis 17, 270 (2017). https://doi.org/10.1186/s12879-017-2392-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-017-2392-y