Abstract
Background
The most common cause of Gram-negative bacterial neonatal meningitis is E. coli K1. It has a mortality rate of 10–15 %, and neurological sequelae in 30–50 % of cases. Infections can be attributable to nosocomial sources, however the pre-colonisation of enteral feeding tubes has not been considered as a specific risk factor.
Methods
Thirty E. coli strains, which had been isolated in an earlier study, from the residual lumen liquid and biofilms of neonatal nasogastric feeding tubes were genotyped using pulsed-field gel electrophoresis, and 7-loci multilocus sequence typing. Potential pathogenicity and biofilm associated traits were determined using specific PCR probes, genome analysis, and in vitro tissue culture assays.
Results
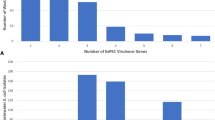
The E. coli strains clustered into five pulsotypes, which were genotyped as sequence types (ST) 95, 73, 127, 394 and 2076 (Achman scheme). The extra-intestinal pathogenic E. coli (ExPEC) phylogenetic group B2 ST95 serotype O1:K1:NM strains had been isolated over a 2 week period from 11 neonates who were on different feeding regimes. The E. coli K1 ST95 strains encoded for various virulence traits associated with neonatal meningitis and extracellular matrix formation. These strains attached and invaded intestinal, and both human and rat brain cell lines, and persisted for 48 h in U937 macrophages. E. coli STs 73, 394 and 2076 also persisted in macrophages and invaded Caco-2 and human brain cells, but only ST394 invaded rat brain cells. E. coli ST127 was notable as it did not invade any cell lines.
Conclusions
Routes by which E. coli K1 can be disseminated within a neonatal intensive care unit are uncertain, however the colonisation of neonatal enteral feeding tubes may be one reservoir source which could constitute a serious health risk to neonates following ingestion.
Similar content being viewed by others
Background
Liu et al. [1] reported that worldwide the estimated mortality in children younger than 5 years in 2010 was 7,600,000. Neonates, in particular those with very low birth-weight, are of particular concern due to their weak immune system [2]. The risk of infection in neonates increases with low birth weight and additionally prolonged hospitalization [3, 4]. Mortality among neonates is attributed to infectious causes, preterm birth complications, intrapartum-related complications, sepsis and meningitis.
The most common cause of Gram-negative bacterial neonatal meningitis is E. coli K1. It has a mortality rate of 10–15 %, and neurological sequelae in 30–50 % of cases [5–8]. Other bacteria responsible for neonatal and infant morbidity and mortality include Group B streptococci (GBS), Enterobacter spp., Citrobacter koseri, Neisseria meningitidis, Serratia spp., and Cronobacter spp. [9, 10].
E. coli serotypes associated with neonatal meningitis are primarily O18:K1:H7, O1:K1, O7:K1, O83:K1 and the more recently reported O45:K1:H7 [11–13]. These are in the E. coli extraintestinal pathogenic (ExPEC) subgroup B2, and are sequence type (ST) 95 (Achtman scheme) [14]. Bacterial invasion across the blood–brain barrier is multifactorial, requiring several genes for binding, invasion and intracellular survival. Proposed virulence genetic determinants include ibeA, sfaS, cnf1, gimA, and ompA [15, 16]. However these are not always supported in experimental assays and are not demonstrable in all E. coli strains isolated from cerebral spinal fluid [12, 13, 17].
Although there have been some concerns regarding bacterial biofilm formation inside neonatal nasogastric feeding tubes by opportunistic pathogens, few systematic studies have been undertaken [18–20]. Mehall et al. [18] reported both feeding intolerance and a link to necrotizing enterocolitis in neonates following the bacterial colonised of their feeding tubes. Previous studies by Hurrell et al. [19] have revealed that in situ the inside of such tubes can be colonised by a variety of fungi and various opportunistic bacterial pathogens producing a biofilm of mixed microbial composition; Candida spp., E. coli, Enterobacter hormaechei, Klebsiella pneumoniae, Cronobacter sakazakii, Yersinia enterocolitica, and Pseudomonas fluorescens. The aim of this study was to investigate the diversity of the E. coli strains previously isolated by Hurrell et al. [19] from the residual liquid in the lumen and biofilm from 30 neonatal nasogastric feeding tubes, which had been collected from among 129 neonates on two intensive care units.
Methods
Bacterial strains used
Thirty isolates of E. coli were included in this study; Table 1. All isolates had previously been isolated by Hurrell et al. [19] from the residual lumen liquid and biofilms of nasogastric enteral feeding tubes on neonatal intensive care units at Hospital 1 (n = 3) and 2 (n = 27).
Pulsed-field gel electrophoresis
Pulsotypes were determined using pulsed-field gel electrophoresis (PFGE) with XbaI and SpeI restriction enzymes as described by PulseNet [21]. Salmonella enterica serovar Typhimurium H9812 was used as the reference strain. Dendrogram construction and band assignment was achieved using BioNumerics software version 3.5. Dice coefficient, unweighted pair group method with arithmetic mean (UPGMA) were used for cluster analysis. Less than 95 % of band similarity value was used to consider the isolates to be non-clonal [22]. The tolerance and optimization of the bands was 1.5 %.
Multilocus sequence typing (MLST)
Sequence type (ST) of the E. coli isolates used the 7-loci MLST Achtman scheme (http://mlst.warwick.ac.uk/mlst/mlst/dbs/Ecoli). Seven housekeeping genes were amplified by PCR using the primers for adk (adenylate kinase), fumC (fumarate hydratase), gyrB (DNA gyrase), icd (isocitrate/isopropylmalate dehydrogenase), mdh (malate dehydrogenase), purA (adenylosuccinate dehydrogenase), and recA (ATP/GTP binding motif). The sequences were aligned using CLC Sequence Viewer 6.6 (http://www.clcbio.com). The trimmed allele sequences were compared against the E. coli MLST database (http://mlst.warwick.ac.uk/mlst/mlst/dbs/Ecoli) and the sequence types were subsequently determined.
Serotyping
The O-antigen serotype was determined using comparative genomic analysis, and confirmed by laboratory analysis (Statens Serum Institut) [23].
Motility determination
Motility was determined by measuring the zones of growth in semi-solid agar [24]. A single colony from each strain was used to inoculate 3 ml of TSB which was then incubated at 37 °C with shaking incubator at 200 rpm. The culture was diluted to 104 CFU/ml and 3 μl of the suspension used to stab inoculate TSB supplemented with 0.4 % agar. The inoculated plates were incubated overnight at 37 °C. Strains were analysed twice, each time in triplicate.
Haemolysis reaction
Haemolysis was examined by streaking on TSA-blood agar plates containing 5 % sheep blood (Oxoid Thermo Fischer Scientific, UK), and then incubating for 24 h for at 37 °C. The resultant colony morphology was recorded after 24 h to determine the formation of either α- or β-haemolysis.
Antibiotic resistance determination
Antibiograms were determined using the disc diffusion method. Antibiotic discs were obtained from MAST Group (UK). For each antibiotic the diameter of the zone was measured and then compared with standard measurements to determine if the strains were resistant or sensitive to the antibiotic [25]. The control strains were E. coli NCTC 13351, E. coli NCTC 13352, E. coli NCTC 13353 and E. coli NCTC 10418. The presence of the β-lactamase resistance genes SHV, TEM, CTX-M, and OXA were screened for by a multiplex PCR assay [26]. Strains with known β-lactamase types were included as reference strains. These were E. coli NCTC 13351 (TEM-3), E. coli NCTC 13353 (CTX-M-15, TEM, OXA), K. pneumoniae NCTC 13368 SHV-18. Genomes were analysed using the Comprehensive Antibiotic Database (CARD; http://arpcard.mcmaster.ca) for genes encoding antibiotic resistance [27].
PCR detection of virulence factor genes
The presence of 30 virulence factor genes was determined using 5 multiplex PCR–based assays [28]. The gene classes included adhesins (papAH, papC, papEF, papG, sfa/focDE, sfaS, focG, afa/draBC, bmaE, gafD, nfaE and fimH), toxins (hlyA, cnf1, cdtB), siderophores (fyuA, iutA), polysaccharide coatings (kpsMT II kpsMT III, kpsMT K1, kpsMT K5), invasins (ibeA) and others (rfc, cvaC, traT, malX).
Attachment and invasion assay
Attachment and invasion assays to examine the capability of selected bacterial strains to attach and invade mammalian cells (Caco-2, rBCEC4 and HBMEC) were as previously described [29]. Bacterial strains were investigated for their uptake and persistence in macrophages (U937) obtained from the American Type Culture Collection [30].
Adherence pattern determination
The Giemsa stain was used to determine the adherence pattern of the E. coli strains. Caco-2 and Hep-2 cell monolayers were grown on tissue culture coverslips in six-well tissue culture plates [31]. The slides were seeded with 2 × 104 cells and then incubated at 37 °C with 5 % CO2 for 48 h. After the incubation period, the monolayers were infected with 108 per well of overnight bacterial culture and further incubated at 37 °C under 5 % CO2 for 2 h. The coverslips were washed three times with sterile PBS, fixed with absolute methanol for 5 min and allowed to air dry. The cells were stained with 5 % of Giemsa stain (Life Technologies™, UK) for 15 min, washed with sterile PBS and allowed to air dry. The slide was examined using light microscopy.
Genomic analysis
Bacterial DNA was extracted from 1-day old cultures of selected strains using GenElute™ bacterial genome kit (Sigma Aldrich®, USA). The genome sequences were generated on an Illumina MiSeq using v3 chemistry and 300 bp paired end reads using dual indexed Nextera XT libraries. The de novo assembly was performed using SPAdes assembly program and Quast [32]. Genome annotation used the SEED-based automated annotation system provided by the RAST server (http://rast.nmpdr.org) and prokaryotic genome annotation system PROKKA [33].
The genome sequences obtained were compared to published chromosomal, plasmid and O-antigen sequences for E. coli APEC O1 (Accession number CP000468), E. coli CE10 O7:K1 (Accession number GCA_000227625), E. coli S88 O45:K1:H7 (Accession numbers: chromosome CU928161, plasmid CU928146), O-antigens O1 and O2 (Accession numbers GU299791 and GU299792, respectively), and plasmids E. coli O1 pAPEC-O1-CelBM (Accession number DQ381420) and E. coli O18:K1 pRS218 (Accession number CP007150) [1, 13, 17, 34–37]. The genomes were also searched for various biofilm formation associated traits. Whole genome alignment used Parsnp from the Harvest Tools software v1.1.2 with a reference genome selected at random. Tree visualisation used FigTree v1.4.2, to construct a midpoint rooted tree.
Nucleotide sequence accession numbers
The Whole Genome Shotgun projects have been deposited at DDBJ/EMBL/GenBank under accession JQFB00000000, JQFC00000000, JQFD00000000, JQFQ00000000, JQFR00000000, JQFE00000000, JQFF00000000, JQFS00000000, JQFG00000000, JQFH00000000, JQFI00000000 and JQFT00000000 for isolates 904, 910, 913, 923, 926, 929, 934, 937, 939, 943, 947 and 949 respectively.
Results
Pulsed-field analysis of E. coli strains
As shown in Fig. 1, the PFGE analysis of thirty of E. coli isolates from neonatal enteral feeding tubes from hospital 1 (n = 3) and hospital 2 (n = 27) showed the strains clustered into four pulsotypes (PT1-4) and one unique (U) strain. These strains were isolated from the residual liquid in the tube and from biofilms on the inner wall of 30/129 feeding tubes (Table 1). Three strains (1047, 1050, 1051) belonging to PT1 had been isolated from the same neonate on the same day (22 October 2007). These were from both the lumen contents and biofilm within the tubes. This neonate had been fed ‘ready to feed’ formula. Four strains (1009, 1010, 1015, 1016), previously isolated from both the residual liquid and biofilm of feeding tubes, formed PT2. These had been isolated on the same day (12 June 2007) from 3 neonates fed either breast milk or fortified breast milk. Three strains (786, 780, 796) from feeding tube biofilms belonged to PT4 and were isolated over a one month period (16 January to 16 February 2007) from 2 different neonates fed breast milk, fortified breast milk, and reconstituted infant formula. There was one unique (U) strain (1008) from a neonate who had been fed both fortified breast milk and reconstituted infant formula. Of particular interest were the nineteen strains belonging to PT3. These had isolated over a two week period (24 April to 8 May 2007) from 11 different neonates; Table 1. These neonates had been fed during the sampling period breast milk, fortified breast milk, reconstituted infant formula, and ready to feed formula. Again, indistinguishable strains were isolated from both tube lumen contents and biofilms.
Multilocus sequence typing
The 7-loci MLST sequence types (ST) obtained for eight selected strains are given in Table 2. Five STs were identified across the pulsotype groups, and were internally consistent within the clonal group. Pulsotypes 1 to 4 and the unique isolate corresponded with sequence types ST2076, ST73, ST95, ST127 and ST394, respectively. Sequence types 394 and 2076 differ by one nucleotide in the parA allele. These strains had been isolated from two hospitals at different times; 1 October 2007, PT4 January and February 2007. All the sequence types belong to the extra-intestinal (ExPEC) pathogenic E. coli group B2; [38].
Antibiograms
Antimicrobial susceptibility of the E. coli strains is given in Table 3. The two E. coli ST2076 strains 1047 and 1050, which belong to PT1, showed resistance to the penicillin antibiotics and were susceptible to all other antibiotics. The PT2 strain 1009, belonging to ST73, was susceptible to all antibiotics. The ST95 (PT3) E. coli strains 904, 923 and 939 were susceptible to all antibiotics, except ampicillin. E. coli strain 780 (ST394, PT4) was resistance to ampicillin, and augmentin. E. coli strain 1008 (ST127, U) was susceptible to all antibiotics. The two closely related STs 394 and 2076 differed in their susceptibility to piperacillin and meropenem; Table 3. The bla tem gene, conferring resistance to ampicillin, was found in E. coli ST95, ST394 and ST2076 isolates and none harboured bla SHV , bla CTX-M or bla OXA. E. coli strains 1008 (ST127, U) and 1009 (ST73, PT2) did not encode any of these genes.
Physiological traits
Eight strains were selected as representatives of the initial thirty E. coli strains for further detailed study; 1047 (ST2076), 1050 (ST2076), 1009 (ST73), 904 (ST95), 923 (ST95), 939 (ST95), 780 (ST127), and 1008 (ST394). The E. coli strains were non-motile and showed α-haemolysis on sheep blood agar, except for strain 1008 ST394 which was motile and β-haemolytic; Table 2. The serotype of pulsotype representatives were determined by laboratory analysis (Statens Serum Institute). This revealed a range of O-types including O1:K1 (ST95) and O25:K5 (ST73); Table 2.
Genome analysis of E. coli ST95 isolates
The E. coli K1 phylogenetic group B2 ST95 strains with indistinguishable pulsotypes (PT3) had been isolated from the feeding tubes of 11 neonates in an intensive care unit over a two week period (Fig. 1). As given above, the representative PT3 strain (904) was laboratory determined to be O1:K1; Table 2. Given the significant association of E. coli K1 with neonatal meningitis, all the PT3 strains were genome sequenced. Their genome size was in the order of 4997507 bp, average G + C content was 50.7 % (ranging from 49.2 to 51.3 %). The genome annotation indicated that all the E. coli ST95 strains were serotype O1 and capsular type K1. Whole genome alignment was performed for conformational purposes with a variety of publically available genomes of E. coli strains expressing different K antigens; Fig. 2. The genomes also revealed the presence of genes encoding for curli fimbriae and colanic acid which are associated with extracellular matrix production and biofilm formation.
The range of antibiotic resistance encoding genes or ORFs was predicted using the Comprehensive Antibiotic Database (CARD; http://arpcard.mcmaster.ca) and has been summarised in Table 4. This analysis revealed the E. coli ST95 strains had two streptomycin resistance associated genes (strA and strB) in the aminoglycoside resistance class. The number of predicted antibiotic efflux genes in the 12 E. coli ST95 strains varied slightly. The majority (10/12) of strains had 52 genes, whereas one had 51 and the remainder had 53. This was the only trait which distinguished between the ST95 strains. Further studies will provide a more in-depth analysis of this small variation. Also the E. coli ST95, ST2076, and ST127 had an additional β–lactamase gene compared with ST73 and ST394. Further detailed analysis is given in the Additional file 1: Figure S1.
Virulence traits
The presence of 30 virulence related traits were screened using PCR. The 30 virulence genes investigated, included a range of traits including genes encoding for adhesins, invasins, capsule, toxins, siderophores and others. The presence of these genes differed depending on the strain sequence type; Table 5. For example, E. coli K1 ST95 strains encoded adhesin genes fimH, papACEFG1, siderophores fyuA, traT and UPEC PAI. In contrast, the ST127 E. coli K5 strain 1008 encoded sfaS, haemolysin hlyA and cnf. The UPEC PAI marker, malX from archetypal ExPEC strain CFT073 (serotype O6:K2:H1), was only present in STs 73, 95, and 127. The aerobactin receptor gene (iutA) was only in ST73 (PT2). The two closely related STs 394 and 2076 differed in the possession of fimH and fvtA; Table 5.
Adhesion, invasion and persistence in mammalian cell types
In vitro tissue culture assays showed that E. coli sequence types varied in their ability to attach and invade mammalian cell lines; Fig. 3. E. coli K1 ST95 attached and invaded intestinal cells (Caco-2), and both human and rat brain cell lines; HBMCE and rBCEC4. They also persisted for 48 h in U937 macrophages; Fig. 3. E. coli STs 73, 394 and 2076 also persisted in macrophages and invaded Caco-2 and human brain cells, but only ST394 invaded rat brain cells. E. coli ST127 was notable as it did not invade any cell lines. Nearly all strains of E. coli showed an aggregative attachment pattern on Caco-2 and Hep-2 cell lines. The exception was E. coli ST127 strain 1008 which showed a diffuse attachment pattern on both cell lines. This strain is also O-rough antigen type; Table 2. One-way ANOVA statistical analysis demonstrated that all strains attached significantly more than E. coli K12 (p ≤ 0.001).
Discussion and conclusions
Microbial colonisation of the neonate starts at birth, or even sooner through the meconium [39]. This initial flora is largely commensal but may include E. coli pathovars [40]. Hurrell et al. [19] has already given a general overview of the Enterobacteriacae isolates from bacterial biofilms inside neonatal nasogastric feeding tubes collected from these two hospitals. Those studies included electron micrographs of multi-organism (bacteria and fungi) biofilms inside the used feeding tubes. E. coli was isolated from 29 % (n = 129) of these tubes. However, in that study, the E. coli strains had not been genotyped to determine if there was a common source.
Our follow-up study shows, as expected, that the same pulsotype strains were isolated from both the residual liquid of the lumen and inner surface biofilm from 66 % of tubes; Table 1. For example, PT1 were three strains from one neonate, which had been isolated on the same day. This neonate had been fed ready-to-feed formula. Given that this is a sterile product, the source of the E. coli strains inside the feeding tube is uncertain. This issue has already been considered by Hurrell et al. [19] who proposed that a possible secondary source of the enteral tube flora was the throat due to gastroesophageal reflux. In preterm neonates this occurs 3–5 times per hour when the lower oesophageal sphincter relaxes. This would increase the exposure of the feeding tube to the throat flora.
However, Fig. 1 also shows that the 30 E. coli isolates only formed five pulsotypes, and therefore multiple indistinguishable strains had been isolated from different neonates over a one year period. PT4 strains had been isolated from three neonates over a 4 week period. These neonates had all received breast milk, and two had also received reconstituted infant formula. This demonstrates the possible dissemination of strains in the neonatal intensive care unit. Of particular significance was pulsotype 3 which was composed of 19 indistinguishable E. coli strains from 11 neonates on different feeding regimes, over a two week period; Table 1. This reinforces the probability that strains were acquired due to dispersion within the intensive care unit by carers and the environment and not a specific feed source such as contaminated infant formula.
MLST revealed that each pulsotype corresponded with a unique sequence type. It is noted that although ST394 and ST2076 only differ in one nucleotide in the parA allele, that the strains differed in their antibiotic susceptibilities and virulence; Table 2. The E. coli ST127 strain 1008 was the only strain which was motile, and also showed β-haemolysis on sheep blood agar; Table 2.
Since the clinical representation of the neonates in the study was not available, the potential pathogenicity of the strains was assessed using both genetic analysis for virulence traits (PCR-probes, and genome sequence analysis) as well as in vitro tissue culture. E. coli K1 translocates from the neonatal intestines to the bloodstream, where they multiply and cross the blood–brain barrier by invading the brain microvascular endothelial cells. These steps were investigated using attachment and invasion studies of human colonic carcinoma epithelial cells (Caco-2), rat blood brain barrier cells (rBECE4) and human brain microvascular endothelial cells (HBMEC) tissue culture cells; Fig. 3. Macrophage survival was studied using the (U937) cell line of human monocyte cells. These assays revealed there was considerable variation in the presence of virulence traits and in vitro pathogenicity according to the E. coli sequence type.
The three E. coli K1 ST95 strains were notable for their ability to attach and invade intestinal and both human and rat brain cells at levels comparable to Salmonella enterica and C. koseri, respectively; Fig. 3a, c, d. Macrophage uptake and persistence was comparable to C. koseri; Fig. 3b. The three other sequence types (ST394, ST73, ST2076) also attached and invaded human intestinal and brain cells, and ST394 was also able to invade rat brain cells.
These five sequence types are in the ExPEC biogroup B2, and combining the results of the motility assay with serotyping showed that the ST95 strains were E. coli O1:K1:NM. This group is of high significance due to their strong association with neonatal meningitis, and on this occasion 19 indistinguishable strains had been isolated from the tubes of 11 neonates. E. coli phylogroup B2 ExPEC strains of serotypes O1, O2, O18, and O45 are most frequently in ST95, and have been a focus of considerable research in recent years [12–14, 41,42]. In order to assess the virulence potential of the strains, a total of 30 virulence genes were screened for. These included adhesins, invasins, capsule, toxins, siderophores and others commonly associated with neonatal meningitic E. coli (NMEC), avian pathogenic E. coli (APEC) and uropathogenic E. coli (UPEC) [28].
The presence of the virulence related genes differed depending on the sequence type; Table 5. For example, E. coli O1:K1:NM ST95 strains encoded adhesin genes fimH, papACEFGI, papG allele II, siderophores fyuA (yersiniabactin receptor), and UPEC pathogenicity associated island (PAI) marker (malX) as well as the serum resistance associated gene traT. However despite the attachment and invasion of human and rat brain cells (Fig. 3), the ST95 strain however did not encode for ibeA or sfaS. Similarly Johnson et al. [17] reported their occurrence in only 33 % and 59 % of NMEC strains (n = 70), respectively. In contrast, the β–haemolytic ST127 E. coli K5 strain 1008 which did not attach or invade any cell line encoded for the sfaS adhesin as well as the haemolysin encoded by hlyA and the cytotoxic necrotizing factor (cnf). MalX, a marker for a UPEC PAI from the archetypal ExPEC strain CFT073 (serotype O6:K2:H1) [43] was present in sequence types 73, 95, and 127. A fuller description of the E. coli genomes derived from this study will be given in a separate publication.
Mora et al. [38] reviewed the source and virulence profiles of 59 ExPEC O1:K1:H7/NM ST95 strains of animal and human origin, recovered from different dates and geographic sources. They reported that some APEC isolates may act as potential pathogens for humans from poultry, suggesting no host specificity for this type of isolate. In contrast, the strains in this study had been isolated from preterm neonates in isolation units who had been fed breast milk and infant formula. Microbiological analysis of the feeds and microbial carriage by staff was not assessed at the time by Hurrell et al. [19]. Therefore the source of these E. coli K1 strains is currently uncertain.
E. coli K1 are the second most common cause of severe neonatal infections after Group B streptococcal (GBS) meningitis [44]. Although 85 % of infected neonates recover, this is often not as full as would occur with older infants and children. Sources and dissemination of such pathogenic organisms needs further investigation, especially since E. coli K1 causes 80 % of neonatal meningitis cases. Neonates acquire their initial flora at birth from the mother, environment and other carers. Maternal to child transmission of E. coli has been reported, and has been linked to late-onset neonatal infection [45–47]. There is also the possible transmission of E. coli K1 by nurses’ hands [40]. In addition, recent microbiome studies have indicated the possible dispersion of bacteria in the neonatal intensive care units [48]. It should also be noted that E. coli ST73, ST394 and ST2076 strains demonstrated the ability to invade human cells lines and therefore their occurrence in nasogastric feeding tubes may additionally pose a pathogenic risk towards neonates.
Due to confidentiality reasons, the clinical condition of the specific neonates from whom the E. coli strains in this study were isolated is not available. However during this collection period there were four cases of E. coli infection. It should be noted that the genomic analysis showed the E. coli K1 strains (ST95) had two streptomycin resistance genes belonging to the aminoglycoside class antibiotics (Table 4). This could be of clinical significance since aminoglycoside antibiotics such as gentamicin are regularly used as 1st and 2nd line combinations on NICUs. Since the patients’ details and isolates were not available for analysis, no direct causal infection route from the nasogastric tube can be made. Attribution of the source of the E. coli K1 ST95 is not feasible as there no environmental sampling or screening of carriage by staff or mothers. Nevertheless given the indistinguishable strains were obtained from neonates on different feeding regimes it seems probable that strains were disseminated in the NICU by carers and the environment, and not directly from a single feeding source.
Ethics statement
Isolates from this study were obtained by culturing stock isolates. All clinical data are taken from a previous publication [19].
Abbreviations
- Caco-2:
-
Human colonic carcinoma epithelial cells
- CFU/ml:
-
Colony forming units per millilitre
- ExPEC:
-
Extra-intestinal pathogenic E. coli
- GBS:
-
Group B streptococci
- HBMEC:
-
Human brain microvascular endothelial cells
- MLST:
-
Multilocus sequence typing
- NICU:
-
Neonatal intensive care unit
- NM:
-
Non-motile
- NMEC:
-
Neonatal meningitic E. coli
- PAI:
-
Pathogenicity associated island
- PCR:
-
Polymerase chain reactions
- PFGE:
-
Pulsed-field gel electrophoresis
- PT:
-
Pulsotype
- rBECE4:
-
Rat blood brain barrier cells
- ST:
-
Sequence type
- UPEC:
-
Urinary pathogenic E. coli
- UPGMA:
-
Unweighted pair group method with arithmetic mean
References
Li D, Liu B, Chen M, Guo D, Guo X, Liu F, et al. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Meth. 2010;82:71–7.
Levy O, Zarember KA, Roy RM, Cywes C, Godowski PJ, Wessels MR. Selective impairment of TLR-mediated innate immunity in human newborns: neonatal blood plasma reduces monocyte TNF-{alpha} induction by bacterial lipopeptides, lipopolysaccharide, and imiquimod, but preserves the response to R-848. J Immunol. 2004;173:4627–34.
Stoll BJ, Hansen NI, Higgins RD, Fanaroff AA, Duara S, Goldberg R, et al. Very low birth weight preterm infants with early onset neonatal sepsis: the predominance of gram-negative infections continues in the National Institute of Child Health and Human Development Neonatal Research Network, 2002–2003. Ped Infect Dis J. 2005;24:635–9.
Klinger G, Levy I, Sirota L, Boyko V, Lerner-Geva L, Reichman B. Outcome of early-onset sepsis in a national cohort of very low birth weight infants. Pediatrics. 2010;125:e736–40.
de Louvois J, Halket S, Harvey D. Neonatal meningitis in England and Wales: sequelae at 5 years of age. Euro J Pediatr. 2005;164:730–4.
Kim KJ, Chung JW, Kim KS. 67-kDa laminin receptor promotes internalization of cytotoxic necrotizing factor 1-expressing Escherichia coli K1 into human brain microvascular endothelial cells. J Biol Chem. 2005;280:1360–8.
Zhu L, Pearce D, Kim KS. Prevention of Escherichia coli K1 penetration of the blood–brain barrier by counteracting the host cell receptor and signaling molecule involved in E. coli invasion of human brain microvascular endothelial cells. Infect Immun. 2010;78:3554–9.
Logue CM, Doetkott C, Mangiamele P, Wannemuehler YM, Johnson TJ, Tivendale KA, et al. Genotypic and phenotypic traits that distinguish neonatal meningitis-associated Escherichia coli from fecal E. coli isolates of healthy human hosts. Appl Environ Microbiol. 2012;78:5824–30.
Kaufman D, Fairchild KD. Clinical microbiology of bacterial and fungal sepsis in very-low-birth-weight infants. Clin Microbiol Rev. 2004;17:638–80.
Forsythe SJ, Dickins B, Jolley KA. Cronobacter, the emergent bacterial pathogen Enterobacter sakazakii comes of age; MLST and whole genome sequence analysis. BMC Genomics. 2014;15:1121.
Bonacorsi S, Bingen E. Molecular epidemiology of Escherichia coli causing neonatal meningitis. Intl J Med Microbiol. 2005;295:373–81.
Bonacorsi S, Clermont O, Houdouin V, Cordevant C, Brahimi N, Marecat A, et al. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates; identification of new virulent clone. J Infect Dis. 2003;187:1895–906.
Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Médigue C. The plasmid of Escherichia coli strain S88 (O45: K1: H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun. 2009;77:2272–84.
Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51.
Huang S-H, Jong AY. Cellular mechanisms of microbial proteins contributing to invasion of the blood–brain barrier. Cell Microbiol. 2001;3:277–87.
Wang Y, Kim KS. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Ped Res. 2002;51:559–63.
Johnson JR, Oswald E, O’Bryan TT, Kuskowski MA, Spanjaard L. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J Infect Dis. 2002;185:774–84.
Mehall JR, Kite CA, Saltzman DA, Wallett T, Jackson RJ, Smith SD. Prospective study of the incidence and complications of bacterial contamination of enteral feeding in neonates. J Ped Surg. 2002;37:1177–82.
Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Hilton A, Armstrong R, et al. Neonatal enteral feeding tubes as loci for colonisation by members of the Enterobacteriaceae. BMC Infect Dis. 2009;9:46.
Hurrell E, Kucerova E, Loughlin M, Caubilla-Barron J, Forsythe SJ. Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Intl J Food Microbiol. 2009;136:227–31.
Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67.
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.
Ørskov F, Ørskov I. Serotyping of Escherichia coli. Methods Microbiol. 1984;14:43–112.
Reller LB, Mirrett S. Motility-indole-lysine medium for presumptive identification of enteric pathogens of Enterobacteriaceae. J Clin Microbiol. 1975;2:247–52.
British Society for Antimicrobial Chemotherapy. BSAC methods for antimicrobial susceptibility testing, version 14. 2015. http://bsac.org.uk/susceptibility/methodologylatestversion/.
Fang H, Ataker F, Hedin G, Dornbusch K. Molecular epidemiology of extended-spectrum β-lactamases among Escherichia coli isolates collected in a Swedish hospital and its associated health care facilities from 2001 to 2006. J Clin Microbiol. 2008;46:707–12.
McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57:3348–57.
Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181:261–72.
Townsend SM, Hurrell E, Gonzalez-Gomez I, Lowe L, Frye JG, Forsythe S, et al. Enterobacter sakazakii invades brain capillary endothelial cells, persists in human macrophages influencing cytokine secretion and induces severe brain pathology in the neonatal rat. Microbiology. 2007;153:3538–47.
Townsend SM, Pollack HA, Gonzalez-Gomez I, Shimada H, Badger JL. Citrobacter koseri brain abscess in the neonatal rat: survival and replication within human and rat macrophages. Infect Immun. 2003;71:5871–80.
Ruttler ME, Yanzon CS, Cuitino MJ, Renna NF, Pizarro MA, Ortiz AM. Evaluation of a multiplex PCR method to detect enteroaggregative Escherichia coli. Biocell. 2006;30:301–8.
Prjibelski AD, Vasilinetc I, Bankevich A, Gurevich A, Krivosheeva T, Nurk S, et al. ExSPAnder: a universal repeat resolver for DNA fragment assembly. Bioinformatics. 2014;30:i293–301.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Johnson TJ, Johnson SJ, Nolan LK. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J Bacteriol. 2006;188:5975–83.
Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, et al. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol. 2007;189:3228–36.
Lu S, Zhang X, Zhu Y, Kim KS, Yang J, Jin Q. Complete genome sequence of the neonatal-meningitis-associated Escherichia coli strain CE10. J Bacteriol. 2011;193:7005.
Wijetunge DS, Karunathilake KH, Chaudhari A, Katani R, Dudley EG, Kapur V, et al. Complete nucleotide sequence of pRS218, a large virulence plasmid, that augments pathogenic potential of meningitis-associated Escherichia coli strain RS218. BMC Microbiol. 2014;14:203.
Mora A, López C, Dabhi G, Blanco M, Blanco JE, Alonso MP, et al. Extraintestinal pathogenic Escherichia coli O1:K1:H7/NM from human and avian origin: detection of clonal groups B2 ST95 and D ST59 with different host distribution. BMC Microbiol. 2009;9:132.
Moles L, Gómez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One. 2013;8, e66986.
Alos JI, Lambert T, Courvalin P. Comparison of two molecular methods for tracing nosocomial transmission of Escherichia coli K1 in a neonatal unit. J Clin Microbiol. 1993;31:1704–9.
Bidet P, Metais A, Mahjoub-Messai F, Durand L, Dehem M, Aujard Y, et al. Detection and identification by PCR of a highly virulent phylogenetic subgroup among extraintestinal pathogenic Escherichia coli B2 strains. Appl Environ Microbiol. 2007;73:2373–7.
Wang S, Meng Q, Dai J, Han X, Han Y, Ding C, et al. Development of an allele-specific PCR assay for simultaneous sero-typing of avian pathogenic Escherichia coli predominant O1, O2, O18 and O78 strains. PLoS One. 2014;9, e96904.
Guyer DM, Kao JS, Mobley HLT. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect Immun. 1998;66:4411–7.
Simonse KA, Anderson-Berry AL, Delair SF, Davies HD. Early-onset neonatal sepsis. Clin Microbiol Rev. 2014;27:21–47.
Bingen E, Denamur E, Brahimi N, Elion J. Genotyping may provide rapid identification of Escherichia coli K1 organisms that cause neonatal meningitis. Clin Infect Dis. 1996;22:152–6.
Raymond J, Lopez E, Bonacorsi S, Poyart C, Moriette G, Jarreau PH, et al. Evidence for transmission of Escherichia coli from mother to child in late-onset neonatal infection. Pediatr Infect Dis J. 2008;27:186–8.
de Muinck EJ, Oien T, Storrø O, Johnsen R, Stenseth NC, Rønningen KS, et al. Diversity, transmission and persistence of Escherichia coli in a cohort of mothers and their infants. Environ Microbiol Rep. 2011;3:352–9.
Hewitt KM, Mannino FL, Gonzalez A, Chase JH, Caporaso JG, Knight R, et al. Bacterial diversity in two neonatal intensive care units (NICUs). PLoS One. 2013;8;e54703.
Acknowledgements
The authors thank the Libyan Ministry of Higher Education and Nottingham Trent University for their financial support. We also acknowledge the following support: Wellcome Trust Institutional Strategic Support Fund (WT097835MF), Wellcome Trust Multi User Equipment Award (WT101650MA) and BBSRC LOLA award (BB/K003240/1).
Funding
This study was funded by the Libyan Ministry of Higher Education (AA, NR, MS), and Nottingham Trent University (PO, NM), Wellcome Trust Institutional Strategic Support Fund, Wellcome Trust Multi User Equipment Award and BBSRC (KM, AF, KP).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors have no conflict of interests to declare.
Authors’ contributions
AA (PCR & tissue culture), PO & NM (bioinformatics), MS (PFGE), NR (phenotyping), KM, AF, KP (genome sequencing). SF wrote the first draft of the manuscript and managed the project. PO contributed to the writing of the final version of the manuscript. All authors read and approved the final manuscript.
Additional file
Additional file 1: Figure S1.
Genomic analysis of E. coli sequence types using the Comprehensive Antibiotic Database (CARD: http://arpcard.mcmaster.ca). (DOC 1740 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Alkeskas, A., Ogrodzki, P., Saad, M. et al. The molecular characterisation of Escherichia coli K1 isolated from neonatal nasogastric feeding tubes. BMC Infect Dis 15, 449 (2015). https://doi.org/10.1186/s12879-015-1210-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-015-1210-7