Abstract
Background
DAL-1 gene was reported to inhibit proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT) of gastric cancer (GC) cells in our previous study. The association between the genomic variants in DAL-1 gene with risk of GC is still unclear.
Methods
In this study, 505 GC cases and 544 healthy controls (HCs) were collected to evaluate the association between six single nucleotide polymorphisms (SNPs) (rs7240736, rs73937194, rs3817466, rs8082898, rs73381527, rs9953490) of DAL-1 gene and GC risk in the Han population in Northeast China.
Results
The TA + AA genotypes of rs9953490 were significantly associated with an increased risk in N3 compared with N0 subgroup (adjusted OR = 4.56, 95% CI = 1.49–13.98, P = 0.008), and also showed evident association with an increased risk in TNM stage III compared with stage I-II (adjusted OR = 2.33, 95% CI = 1.16–4.67, P = 0.017).
Conclusion
The rs9953490 of DAL-1 gene may play an important role in the occurrence and development of GC in the Han population in Northeast China.
Similar content being viewed by others
Background
Gastric cancer (GC) is the fifth most common neoplasia and the third leading cause of cancer-related death worldwide [1]. The histological heterogeneity classified GC into different subtypes as cardia carcinoma and noncardia carcinoma, which show distinct clinical, epidemiological and molecular features among [2], and seriously hinder the diagnosis and treatment [3]. Moreover, the incidence of GC is highly determined by multiple environmental factors as: microbial infections, the host genetic background (age, gender, lifestyle, dietary regime) [4, 5], alcohol, processed meat, and obesity [6]. Notably, effective diagnostic markers and targeted therapy against GC are still lacking. Currently, GC remains to be a serious fatal disease with poor prognosis throughout Asia, especially in China [7].
DAL-1 (differentially expressed in adenocarcinoma of the lung-1) gene is located on human chromosome 18p 11.3 and belongs to the 4.1 protein superfamily, which is isolated and detected in lung adenocarcinoma for the first time by Tran et al. [8]. The expression of DAL-1 is significantly reduced or lost in various tumors such as breast [9], hepatocellular [10], colon [11], and ovarian [12]. Several studies have identified that DAL-1 is significantly associated with cancer cell differentiation, lymph node metastasis, disease progression, and TNM stage [13, 14]. Our previous study first identified the loss of heterozygosity (LOH) at 18p11.3 region (DAL-1 loci) in 45 sporadic GCs, suggesting DAL-1 might be a candidate tumor suppressor gene [15]. Afterwards, we found promoter methylation-mediated down-regulation of DAL-1 in four GC cell lines and 94.6% (35 of 37) of surgically resected primary GCs. We also demonstrated that DAL-1 effectively inhibited the malignant transformation of GC cells [16], and was significantly associated with cancer progression and poor survival of GC patients [17].
Previous studies indicated that genomic variations may have great impact on the liability for GC [18]. Patients who carry the GG genotype of rs1049174 of NKG2D gene have a higher incidence of GC in Fujian Province of China [19]. Xu et al. [20] found that SNP rs4880 of SOD2 gene and SNP rs1695 of GSTP1 gene may increase cancer progression and tumor aggressiveness. Recently, we found that two SNPs (rs34700818 and rs61482741) of TOB1 gene were significant associated with increased GC progression [21]. Therefore, we investigated the association between six SNPs of DAL-1 gene and GC risk in a cohort of 505 GC patients and 544 healthy controls (HCs). Our data suggested that rs9953490 of DAL-1 might be an important marker of GC risk in the Han population of Northeast China.
Material and methods
Study population
505 unrelated Han Chinese primary GC patients who were recruited from Harbin Medical University Cancer Hospital, and 544 age- and sex-matched HCs who were recruited from the Second Affiliated Hospital of Harbin Medical University between January 2015 and June 2016, were included in the study. The ethics committee of the Second Affiliated Hospital of Harbin Medical University approved the trial protocol. Each participant was interviewed face-to-face by trained interviewers using a standardized questionnaire and signed a written informed consent. Peripheral blood was collected and stored at −80 °C for DNA extraction.
SNP selection
The candidate SNPs were selected using the dbSNP database (http://www.ncbi.nlm.nih.gov/snp) and the 1000 Genomes database (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/). These SNPs were selected based on the criterion of the minor allele frequency (MAF) greater than 0.05 in the Chinese Han population [21]. Ultimately, six SNPs (rs7240736, rs73937194, rs3817466, rs8082898, rs73381527 and rs9953490) of DAL-1 gene met the requirement and were included in our study in Table 1.
Genotyping
Genomic DNA was extracted from the blood samples using the Qiagen Blood DNA Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. The improved Multiple Ligase Detection Reaction (iMLDR) method was used to acquire the genotypes of all the SNPs. The primer sequences of the six SNPs are shown in Table 2. Polymerase chain reactions (PCRs) were performed in a 20 μl reaction solution containing 1 µl of template DNA, 0.5 µl of each primer, 0.2 μl Taq enzyme (Qiagen, Germany) and 10 μl PCR reaction buffer (Takara, Japan). The original data were collected by ABI3730XL sequencer (Applied Biosystems, USA) and analyzed by GeneMapper 4.1 software (Applied Biosystems, USA). Genotyping data were confirmed with 10% randomly-selected samples which showed 100% concordance in repeated tests.
Statistical analysis
Continuous variables with a normal distribution were described as mean ± SD and compared with the Student’s t-test [21]. Discrete variables we re described as frequency (percentage) and compared using the Chi-square (χ2) test [21]. Genotype frequencies for each polymorphism in the control group were tested by Hardy–Weinberg equilibrium using the χ2 test. Associations between the genotypic or allelic frequency and the risk of GC were estimated by odds ratios (ORs) and 95% confidence intervals (CIs). The age of the two groups at the stratified analysis was dichotomized according to the median age (58 years) of the control group [22]. Linkage disequilibrium (LD) and haplotype analyses were performed with Haploview 4.2 software (http://sourceforge.net/projects/haploview/). P values and ORs with 95% CIs were calculated using multiple regression analysis adjusted for age, gender, smoking status, pack-years, and drinking status. All statistical analyses were conducted using the SAS 9.3 software. All P values were two-sided, and P < 0.05 was considered statistically significant.
Results
Population characteristics
The data in Table 3 (at the end of the manuscript) showed the general information of all subjects of 505 GCs and 544 HCs. The mean age was 59.08 (59.08 ± 10.55 years) in GC group and 58.33 (58.33 ± 11.55 years) in the HCs. According to the 7th Edition of the American Joint Committee on Cancer (AJCC) [23], 87 cases (17.2%), 156 cases (30.9%), 128 cases (25.3%), 65 cases (12.9%) and 69 cases (13.7%) were classified as TNM I, II, III, IV and other stages, respectively. The number of subjects with a history of GC, no family history of cancer, and other cancers were 398 (78.8%), 42 (8.3%) and 65 (12.9%), respectively. There was no significant difference in age or gender between the two groups (P = 0.105 and P = 0.404), indicating no sample matching bias between groups. However, there was a significant difference in smoking status, pack-years and drinking status between the two groups (P < 0.0001).
Distribution of the genotypic and allelic frequencies of DAL-1 polymorphisms and their association with GC susceptibility
The genotypic frequencies of six SNPs except rs7240736 of the controls were all in accordance with Hardy–Weinberg equilibrium (P > 0.05). Hence the rs7240736 was eliminated in the next analysis. The genotypic distributions of the other five SNPs among the cases and controls and their associations with GC risk are summarized in Additional file 1: Table S1. All the allelic frequencies were not significantly different between the GCs and HCs. There was no evident association between the five SNPs with GC risk in the homozygotes, heterozygotes or two genetic models (dominant genetic model and recessive genetic model) after adjusting for age, gender, smoking status, pack-years, and drinking status (P > 0.05). The above comparisons were all not statistically significant by using multiple test correction.
Haplotype analysis and GC risk
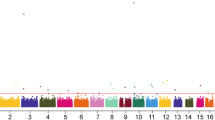
No strong LD for the selected SNPs of the DAL-1 were identified by the Haploview software (Fig. 1). One LD block was found in the DAL-1 gene, which contained five haplotypes in the block. The associations between haplotype frequencies and GC risk were shown in Additional file 1: Table S2. The most common haplotypes within this block were determined as GTTC (0.777), ACTC (0.074), GTCC (0.068), followed by GTTG (0.044), and ATTC (0.020). No apparent association between each haplotype and GC risk within this block was observed.
Stratified analysis between DAL-1 polymorphisms and GC risk
We conducted stratified analyses for all candidate SNPs according to age, gender, smoking status, pack-years, and drinking status, which were reported to have potential influences on GC occurence. As was shown in Table 4 (at the end of the manuscript), the TA + AA genotypes of rs9953490 were associated with a significantly higher GC risk in smoker than nonsmoker before adjustment by age, gender, smoking status, pack-years, and drinking status (TA + AA vs TT, OR = 2.33, 95% CI = 1.11–4.87, P = 0.025). However, this significant association was compromised after adjustment by age, gender, smoking status, pack-years, and drinking status (TA + AA vs TT, adjusted OR = 1.62, 95% CI = 0.61–4.27, P = 0.333). Meanwhile, for other SNPs, no significant association was found between genotypes and GC risk in the stratified analyses (Additional file 1: Tables S3–S7).
We further conducted stratified analyses in 274 patients with available clinicopathological information such as family history of cancer, tumor size, neoplasia location, depth of invasion, lymph metastasis, TNM stage, and Lauren’s classification. As shown in Table 5 (at the end of the manuscript), the TA + AA genotypes of rs9953490 were significantly associated with increased GC risk in N3 compared with N0 (TA + AA vs TT adjusted OR = 4.56, 95% CI = 1.49–13.98, P = 0.008), and with a obviously increased GC risk in TNM stage III compared with stage I-II (TA + AA vs TT adjusted OR = 2.33, 95% CI = 1.16–4.67, P = 0.017). No associations were found between other SNPs and the clinicopathological characteristics of GC (Additional file 1: Tables S8–S12). The above comparisons showed no statistical significance using multiple test correction analysis.
Discussion
In the present study, we investigated the associations between six SNP polymorphisms of the DAL-1 gene and GC risk in the Han population in Northeast China. Stratification analyses based on smoking revealed that the TA + AA genotypes of rs9953490 were significantly associated with a significantly higher GC risk in smoker than nonsmoker. However, this association was abolished after adjustment by age, gender, smoking status, pack-years, and drinking status.
The reason might be as the following: First, although many studies have shown that smoking can increase the risk of GC [24, 25], the relationship between genomic polymorphisms and susceptibility to smoking-related GC has not yet been defined. A Meta-analysis about CYP1A1 polymorphisms with GC susceptibility pointed out that the m1 genotypes (CC + CT) decreased the susceptibility of GC among ever-smokers, but there wasn’t any association between the m2 genotypes (GG + AG) and GC risk among the smokers [26]. Second, the mechanism by which tobacco smoke facilitates cancer development is not completely elucidated. Moreover, most smoke-derived procarcinogens and their bioactivation are not organ-specific. When all tobacco-related cancers are considered together, smoking tends to increase the risk for developing cancers which are cancer-prone [24, 26, 27]. Third, the reason may also be due to the smaller sample size caused by subgrouping in stratified analysis, and the great discrepancy of sample number between subgroups. Last but not the least, GC is a heterogeneous disease involved with multiple etiological factors including age, gender, geography, lifestyle, dietary regime [4, 5, 28]. To confirm the results of this study, larger sample size will facilitate the statistic power so as to rule out the possibility of population stratification and “observation bias” [26] in future study.
Interesting, we found that the TA + AA genotypes of the rs9953490 were associated with the significant increased GC risk in N3 and TNM stage III, representing its possible correlation with lymph node metastases and poor prognosis of GC. We speculated that the TA + AA genotype, located in the 3’UTR region of the DAL-1 gene, might be able to regulate DAL-1 expression or its promoter methylation status, which may inhibit its original tumor suppressive function. In this case, the association of DAL-1 gene polymorphism with GC would further consolidate our previous study, which has demonstrated that DAL-1 gene is involved in anti-proliferation, inhibition of metastasis, associated with poor survival in GC [15,16,17].
Altogether, we have identified the association between the DAL-1 gene polymorphisms and GC susceptibility. The study has provided new evidence for DAL-1 gene as a potential target in GC clinical practice.
Conclusion
This study demonstrated that the rs9953490 TA + AA genotypes of DAL-1 gene is significantly associated with the occurrence and development GC in the Han population of Northeast China.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional file 1.
Abbreviations
- GC:
-
Gastric Cancer
- DAL-1:
-
Differentially expressed in adenocarcinoma of the lung-1
- LOH:
-
Loss of heterozygosity
- EMT:
-
Epithelial to mesenchymal transition
- SNP:
-
Single nucleotide polymorphism
- MAF:
-
Minor allele frequency
- iMLDR:
-
Improved Multiple Ligase Detection Reaction
- PCRs:
-
Polymerase chain reactions
- ORs:
-
Odds ratios
- CIs:
-
Confidence intervals
- LD:
-
Linkage disequilibrium
- AJCC:
-
American Joint Committee on Cancer
References
World Health Organization. (2018). A report about cancer.Retrieved from http://www.iarc.fr/en/mediacentre/pr/2018/pdfs/pr263_E.pdf,2018-09-12.
Kim MA, Lee HS, Yang H-K, et al. Clinicopathologic and protein expression differences between cardia carcinoma and noncardia carcinoma of the stomach. Cancer. 2005; 103: 1439–46.
Russo AE, Strong VE. Gastric Cancer Etiology and Management in Asia and the West. Annu Rev Med. 2019; 70 (1).
Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–8.
Fang WL, Huang KH, Chen JH, et al. Comparison of the survival difference between AJCC 6th and 7th editions for gastric cancer patients. World J Surg. 2011;35:2723–9.
Joanna K, Urszula C. Physical activity and its relation to cancer risk: updating the evidence. Asian Pac J Cancer Prev. 2013;14:3993–4003.
Strong VE, Wu AW, Selby LV, et al. Differences in gastric cancer survival between the U.S. and China. J Surg Oncol. 2015; 112: 31–7.
Tran YK, Bögler O, Gorse KM, et al. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 1999; 59: 35–43.
Heller G, Ziegler B, Brandstetter A, et al. CDK10 is not a target for aberrant DNA methylation in breast cancer. Anticancer Res. 2009;29:3939–44.
Chen X, Cheung ST, So S, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–39.
Ohno N, Terada N, Murata SI, et al. Immunolocalization of protein 4.1B/DAL-1 during neoplastic transformation of mouse and human intestinal epithelium. Histochem Cell Biol. 2004; 122: 579–86.
Dafou D, Grun B, Sinclair J, et al. Microcell-mediated chromosome transfer identifies EPB41L3 as a functional suppressor of Epithelial Ovarian Cancers. Neoplasia. 2010;12:579–89.
Chen X, Guan X, Zhang H, et al. DAL-1 attenuates epithelial-to mesenchymal transition in lung cancer. J Exp Clin Cancer Res. 2015;34:3.
Liu Z, Zhao Q, Li D, et al. Expression and clinical significance of TSLC1 and DAL-1 / 4.1B in pancreatic cancer. WCJD. 2008; 16: 3585–89.
Yu JC, Sun KL, Liu B, et al. Allelotyping for loss of heterozygosity on chromosome 18 in gastric cancer. World J Gastroenterol. 2004;10:1964.
Wang H, Xu M, Cui X, et al. Aberrant expression of the candidate tumor suppressor gene DAL-1 due to hypermethylation in gastric cancer. Sci Rep. 2016;6:21755.
Haonan Guo, Rui Zhang, Justice Afrifa, et al. Decreased expression levels of DAL-1 and TOB1 are associated with clinicopathological features and poor prognosis in gastric cancer. Pathol Res Pract. 2019; 215: 152403.
Siddavaram N. Carcinoma of the stomach: a review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointestinal Oncol. 2012;4:156–69.
Zheng W, Li H, Liu B, et al. Association between the SNPs in trace element-related metabolic genes and the risk of gastric cancer: a case–control study in Xianyou of China. J Genet. 2019; 98(3).
Xu Z, Zhu H, Luk J, et al. Clinical significance of SOD2 and GSTP1 gene polymorphisms in Chinese patients with gastric cancer. Cancer. 2012;118(22):5489–96.
Wang H, Hao H, Guo H, et al. Association between the SNPs of the TOB1 gene and gastric cancer risk in the Chinese Han population of northeast China. J Cancer. 2018;9:1371–8.
Guan X, Hui Z, Niu J, et al. The VEGF -634 G>C promoter polymorphism is associated with risk of gastric cancer. BMC Gastroenterol. 2009;9(1):77.
Mcghan LJ, Pockaj BA, Gray RJ, et al. Validation of the updated 7th edition AJCC TNM staging criteria for gastric adenocarcinoma. J Gastrointestinal Surg. 2012; 16: 53–61.
Ladeiras-Lopes R, Pereira AK, Nogueira A, et al. Smoking and gastric cancer: systematic review and meta-analysis of cohort studies. Cancer Causes Control. 2008;19:689–701.
Mahapatra NR, Kesh K, Swarnakar S, et al. Association of MMP7–181 A G promoter polymorphism with gastric cancer risk: Influence of nicotine in differential allele-specific transcription via increased phosphorylation of cAMP-response element-binding protein (CREB). 2015; 290: 14391–406.
Han F, Wang X, Wang X, et al. Meta-analysis of the association of CYP1A1 polymorphisms with gastric cancer susceptibility and interaction with tobacco smoking. Mol Biol Rep. 2012;39:8335–44.
Ozlü T, Bülbül Y. Smoking and lung cancer. Tuberk Toraks. 2005;53(2):200–9.
Daniyal M, Ahmad S, Ahmad M, et al. Risk factors and epidemiology of Gastric Cancer in Pakistan. Asian Pac J Cancer Prev. 2015;16:4821–4.
Acknowledgements
Not applicable.
Funding
This work was supported by the Research fund of key laboratory of preservation of human genetic resources and disease control to Jingcui Yu.
Author information
Authors and Affiliations
Contributions
Study design and concept: Jingcui Yu, Hui Wang; Data acquisition and sample collection: Hui Wang, Yuling Jiang, Lina Yu; SNP selection: Yuling Jiang, Kexian Dong; DNA extraction: Hui Wang, Yuling Jiang, Rongwei Guan; Statistical analyses: Lina Yu, Lidan Xu, Mengdi Cai, Xiao Liang; Drafting manuscript: Hui Wang, Kexian Dong; Critical Revision of the manuscript and supervision: Jing Bai, Jingcui Yu. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All subjects gave their informed consent for inclusion before they participated in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the Second Affiliated Hospital of Harbin Medical University (KY2018-370).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplementary Table 1: Distribution of genotype and allele frequencies and their association with GC susceptibility. Supplementary Table 2: The frequencies of haplotypes of five SNPs in DAL-1 in cases and controls. Supplementary Table 3: Stratified analyses for the rs73937194 genotypes of DAL-1 gene in cases and controls. Supplementary Table 4: Stratified analyses for the rs3817466 genotypes of DAL-1 gene in cases and controls. Supplementary Table 5: Stratified analyses for the rs8082898 genotypes of DAL-1 gene in cases and controls. Supplementary Table 6: Stratified analyses for the rs73381527 genotypes of DAL-1 gene in cases and controls. Supplementary Table 7: Stratified analyses for the rs9953490 genotypes of DAL-1 gene in cases and controls. Supplementary Table 8: Association between DAL-1 (rs73937194) genotypes and clinicopathologic features of GC. Supplementary Table 9: Association between DAL-1 (rs3817466) genotypes and clinicopathologic features of GC. Supplementary Table 10: Association between DAL-1 (rs8082898) genotypes and clinicopathologic features of GC. Supplementary Table 11: Association between DAL-1 (rs73381527) genotypes and clinicopathologic characteristics features of GC. Supplementary Table 12: Association between DAL-1 (rs9953490) genotypes and clinicopathologic features of GC.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, H., Jiang, Y., Yu, L. et al. The rs9953490 polymorphism of DAL-1 gene is associated with gastric cancer risk in the Han population in Northeast China. BMC Gastroenterol 21, 354 (2021). https://doi.org/10.1186/s12876-021-01929-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-021-01929-9