Abstract
Accurate sepsis diagnosis is paramount for treatment decisions, especially at the emergency department (ED). To improve diagnosis, clinical decision support (CDS) tools are being developed with machine learning (ML) algorithms, using a wide range of variable groups. ML models can find patterns in Electronic Health Record (EHR) data that are unseen by the human eye. A prerequisite for a good model is the use of high-quality labels. Sepsis gold-standard labels are hard to define due to a lack of reliable diagnostic tools for sepsis at the ED. Therefore, standard clinical tools, such as clinical prediction scores (e.g. modified early warning score and quick sequential organ failure assessment), and claims-based methods (e.g. ICD-10) are used to generate suboptimal labels. As a consequence, models trained with these “silver” labels result in ill-trained models. In this study, we trained ML models for sepsis diagnosis at the ED with labels of 375 ED visits assigned by an endpoint adjudication committee (EAC) that consisted of 18 independent experts. Our objective was to evaluate which routinely measured variables show diagnostic value for sepsis. We performed univariate testing and trained multiple ML models with 95 routinely measured variables of three variable groups; demographic and vital, laboratory and advanced haematological variables. Apart from known diagnostic variables, we identified added diagnostic value for less conventional variables such as eosinophil count and platelet distribution width. In this explorative study, we show that the use of an EAC together with ML can identify new targets for future sepsis diagnosis research.
Similar content being viewed by others
Introduction
Sepsis is a severe clinical illness, defined as a life-threatening organ dysfunction caused by a dysregulated host response to infection [1]. The diagnosis of sepsis still remains challenging due to the heterogeneity of clinical symptoms, and the lack of robust diagnostic tests [2,3,4]. Early recognition of sepsis in the emergency department (ED) can be particularly difficult as only limited time and measurements are available for accurate diagnosis. Therefore, numerous tools have been proposed in literature, such as the quick sequential organ failure assessment (qSOFA) score, early warnings scores (e.g. modified early warning score (MEWS)), and biomarkers (e.g. CD64, red blood cell distribution width and pancreatic stone protein) [1, 5,6,7,8]. However, as most of these tools were developed for prognostic purposes, using them for diagnostic endpoints often results in disappointing results [9].
Machine learning (ML) approaches may help doctors to integrate routine care data from EHR databases in diagnostic models. As some ICU databases are publicly available, many studies already used ML to develop diagnostic models for sepsis in the ICU [10, 11]. Though some of these models show high discriminative power, none of them have been broadly implemented in the clinic yet, because of important limitations, including lack of validation and differences in the used definition for sepsis, thereby limiting generalizability [12, 13].
The lack of reliable labels is a major challenge in sepsis research at the ED. By lack of a gold standard, diagnostic tools (e.g. MEWS, qSOFA) and claims-based methods (e.g. ICD-10) are currently used to generate labels for sepsis modelling. Nonetheless, these labelling methods are known to be suboptimal [14]. As the quality of the input defines the eventual quality of the model, i.e. the diagnostic performance, using high quality outcome labels is an essential factor for developing accurate models with ML algorithms. A good alternative is an endpoint adjudication committee (EAC) which is a proven method to gain consensus on outcome, and define labels for machine learning practices [12, 14].
For this study, we used ML and EAC generated labels to explore 95 variables from multiple types of variable groups; demographic and vital functions (1), laboratory tests (2) and advanced haematological variables from the Abbott CELL-DYN sapphire analyser (3), to find additional value in routinely stored EHR data in the ED. We estimated the importance of each variable to identify what variables are most discriminative for sepsis, generating hypotheses for future research.
Methods
Study design and setting
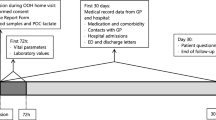
We included patients from the SPACE-cohort (SePsis in the ACutely ill patients in the Emergency department) in the University Medical Centre Utrecht (UMCU) [7]. In brief, ED patients with a suspicion of infection presenting who presented at either internal medicine or its subspecialties (endocrinology, geriatrics, haematology, immunology, infectious disease, nephrology, oncology, rheumatology and vascular medicine) were included in SPACE, no specific exclusion criteria were used. Suspicion of infection was automatically documented by the treating physician, via the electronic health system. An independent physician validated whether the suspicion of infection of each patient by evaluating anti-microbial treatment, microbiological culturing or other signs of infection. Demographic data and physiological variable measurements collected during ED visit were documented and validated by a team of physicians. In this study we used a subset of the SPACE cohort, namely all 375 ED visits that had been labelled for sepsis by the EAC with a blood sample within the first hours of ED visit.
Endpoint adjudication committee
An EAC with 18 independent experts was formed. The EAC consisted of ED specialists, internists (subspecialty infectiology or acute medicine) and ICU specialists. All EHR data including the ED, follow-up in the hospital and after discharge, was made available to the EAC. Members of the EAC were instructed to identify sepsis based on international guidelines (Sepsis-3). All available medical data was assessed by two independent experts. In case of disagreement, an additional expert was consulted followed by a majority vote to generate the final label.
Data collection and pre-processing
ED patients suspected of infection are routinely evaluated by a specific panel of laboratory variables related to infection measured in blood ("internal medicine laboratory panel” or “sepsis panel”). Standard haematology variables included in the panel are routinely collected by the Abbott CELL-DYN Sapphire haematology analyser (Abbott Diagnostics, Santa Clara, CA, USA) [13, 15]. In addition, the Abbott CELL-DYN measures advanced haematology variables that are automatically measured and stored in the laboratory information system, yet not shown in the electronic health record and available for research purposes. The Sapphire is a 5-diff haematological automated cell counter system equipped with an integrated 488-nm blue diode laser and uses multiple techniques, such as electrical impedance, spectrophotometry and laser light scattering, to measure morphological characteristics of leukocytes, RBCs, and platelets for classification and enumeration [16].
Laboratory and advanced haematology data were extracted for SPACE patients from the Utrecht Patient Oriented Database (UPOD) [17]. In brief, UPOD is an infrastructure of relational databases comprising data on patient characteristics, hospital discharge diagnoses, medical procedures, medication orders and laboratory tests for all patients treated at the UMCU since 2004. Only ED visits with measurements for both the laboratory panel and the advanced haematology variables were selected. We removed variables that showed high correlation (Pearson correlation > 0.8) to reduce multicollinearity. We did this for two reasons: (1) removal of highly correlated variables would remove noise from our results (2), advanced ML algorithms can arbitrarily select one of two highly correlated variables, thereby affecting the analysis what variables are important in the models. Moreover, variables with a low number of unique values (N < 5) were removed. Missing measurements were imputed with the variable’s median.
Model development
We were interested in what variables, and in particular what variable groups, had high discriminative accuracy. We used two approaches to identify predictors from the following three variable groups: demographic and vital, laboratory, and advanced haematology variables (Supplemental Table 1). First, we performed univariate analysis by fitting a logistic regression for each individual variable. To evaluate the discriminative power of each variable’s model, we computed the receiver operator characteristic curve (ROC) and precision recall curve (PRC), and subsequently the area under the ROC curve (AUROC). The ROC shows the trade-off between sensitivity and specificity, whereas the PRC shows the trade-off between the sensitivity and precision. In our study, the latter is more of interest, as we are more interested in identifying sepsis cases (sensitivity) and how certain we are of our prediction (precision). Secondly, we trained multiple ML models with the three variable groups. As there is no machine learning algorithm that ‘rules over all’ (no free lunch theorem) [18], we evaluated the diagnostic performance by training three machine learning models; logistic regression (LR), L1 regularization (L1) and Random Forest (RF) [19,20,21]. Both LR and L1 are able to find linear associations between the predictors and outcome, though L1 is able to reduce the coefficients of unimportant predictors to zero, thereby reducing the number of predictors. The RF algorithm trains a number of decision trees that, in comparison to LR and L1, are able to identify non-linear associations between predictors and outcome.
Double loop cross validation to estimate model performance
Training classifiers with too many variables on a small cohort may result in the identification of cohort-specific patterns that may reduce the generalizability of the results, also known as overfitting [23, 24]. Model performance was therefore assessed with k-fold cross validation (CV). With CV, all data is split into k partitions (folds) of which in sequence k-1 folds are used for training and 1 for testing the performance, resulting in k out-of-sample test estimates. To increase the model’s performance, we optimized several hyperparameters, such as the number of trees of the RF algorithm. Optimizing hyperparameters and creating a model with the same training data does not allow for a valid independent estimate of the model’s accuracy. Therefore, we applied a double loop cross validation (DLCV) scheme, also known as nested CV, to optimize several hyperparameters for each model (Supplemental Fig. 1, Supplemental Table 2). In brief, the k-1 folds used for training the algorithm are first used in the inner CV scheme to optimize the hyperparameters, after which the best hyperparameter configuration is used in the outer CV to train the model on the same k-1 training folds. Finally, the model is tested on the k fold (hold-out test set) to obtain an independent estimate of the model’s accuracy. The DLCV scheme was run once.
Receiver operator characteristic (ROC) and precision recall (PRC) curves of the three models with the best performance of each model configuration in the double loop cross validation (DLCV) scheme. Three model configurations are shown; base (logistic regression on demographic and vital data), extended L1 (lasso on demographic, vital, laboratory and sapphire data) and extended RF (random forest on demographic, vital, laboratory and sapphire data). For each of the three model configurations, ten models were trained on different data splits in the tenfold DLCV scheme. During each iteration, predictions were computed with on both training and test folds. Both ROC and PRC curves were drawn with the aggregated train and test data over 10 folds for each model configuration
Model performance and variable importance
Discriminative performance between classifiers was assessed by comparing both the ROC and PRC curves. Also, performance of the 10 models trained in the DLCV for each algorithm was estimated by computing the AUROC and computing the 95% confidence interval (95% CI) with the package cvAUC [25]. Variable importance of LR was assessed with the estimated coefficient. For the L1, the number of times a variable was selected in the ten iterations of the DLCV and the associated coefficients were reported. Shapley values were calculated for the RF to investigate variable importance among the several models and datasets. The Shapley value method takes the sepsis labelled patients along with the non-sepsis patients, and assesses to what extent differences in variables between the two groups contribute to the model’s output. Mean and standard deviation are shown for normally distributed variables whereas median and inter-quartile range (IQR) are shown for non-normally distributed variables. All analyses were done in RStudio (2021.09.0) using R version 4.1.2 [26].
Results
Sepsis patients were older and more often male
Between 2018–01-17 and 2018–04-07 we included 375 ED visits from 335 patients with available demographic, vital and laboratory data (Table 1, Supplemental Table 1). Of all visits, 75 (20.0%) visits were labelled as having sepsis by the EAC. There were slightly more males in both sepsis (53.0%) and non-sepsis (50.7%) groups. Patients with sepsis were slightly older (median of 65.0 versus 60.0 years old) and had worse outcome; longer median length of stay (7.0 versus 0.0 days), higher ICU admission (15.2% versus 2.7%) and mortality (14.7% versus 1.4%). 4% of sepsis patients did not have an infection according to the treating physician, but the EAC did label these patients as having sepsis.
Advanced haematology variables are predictive for sepsis
Supplemental Table 2 shows the discriminative power (AUROC) of all 95 variables used in this study. Of the vital variables, both respiratory rate (rr) and heart rate (hr) showed the highest discriminative performance, AUROC of 0.75. CRP showed the highest performance of the lab variables with an AUROC of 0.75 as well. Five advanced haematology variables; percentage of banded granulocytes (pbnd), banded granulocytes count (bnd), percentage neutrophilic granulocytes (pneu), percentage of immature granulocytes (pig) and immature granulocytes (ig), all had similar performances of AUROC values between 0.68 – 0.70.
Combining variable groups improves sepsis diagnostic performance
After removing the collinear variables (N = 25) and two variables with low number of unique values (nrbc, pnrbc), 70 variables from the three variable groups were used to train models in the DLCV pipeline (Supplemental Fig. 2). Regardless of algorithm, models trained with demographic and vital variables showed a higher average test AUC as compared to models trained with solely the laboratory or advanced haematology variables (Table 2). LR scored the highest test AUC when trained on the demographic and vital variables alone (0.84, 0.79–0.89 95% CI). In comparison, the L1 and RF algorithms had similar test AUC scores when trained on all 70 variables, 0.85 (0.80–0.90 95% CI) and 0.84 (0.78–0.89 95% CI), respectively.
Variable importance of both the base (logistic regression with demographic and vital variables) and the extended L1 (L1 lasso with all data) models. Dots represent the learned coefficient in the trained models in the outer loop of the DLCV. The numbers indicate the number of times a variable was selected in the trained model in the outer loop of the DLCV. Only variables selected more than 7 times are shown
For each algorithm, the model with the highest test performance was used to investigate variable importance. Figure 1 shows ROC and PRC curves based on the aggregated 10 test folds predictions of the three selected models: base (logistic regression with demographic and vital data), extended L1 (lasso with all data) and extended RF (random forest trained with all data).
Variable importance
Figure 2 shows the variable importance of both the base and the extended L1 models and Fig. 3 shows the SHAP values computed by the ten RF models. Of the nine demographic and vital variables, seven were also deemed as important by the extended L1 model, and 6 were also included in the 20 top variables of the extended RF model. Temperature, sex and GCS had on average the highest coefficient estimates for the base and extended L1 models, whereas the respiratory rate (rr), heart rate (hr) and systolic blood pressure (sbp) showed highest importance by the RF model. Of the laboratory variables, the L1 model selected glucose, ASAT, CRP, Gamma-GT and sodium in the DLCV, though with low coefficients. The extended RF identified Gamma-GT, ASAT and CRP. Concerning the advanced haematological variables, the L1 model identified percentage of immature granulocytes (pig), segmented granulocyte absolute count (seg), percentage of banded granulocytes (pbnd), mean lymphocyte size (lamn), platelet distribution width (pdw) and eosinophil granulocyte count (eos), as predictors for sepsis diagnosis, of which the last two had a negative coefficient and of which the latter showed the highest absolute coefficient of all variables. With SHAP, we found that the RF model also identified the same three variables as the L1 extended model; pig, pbnd and seg, with the same direction of importance: higher value is associated with sepsis.
Discussion
As far as we know, this is the first study to investigate the use of an EAC as sepsis labels for the identification of new predictors with ML analyses using routine care data at the ED. For sepsis at the ED, there is no diagnostic gold standard and without such gold standard, developing a clinically valuable ML model is extremely challenging. In studies on ML and sepsis, the definition of sepsis is often based on suboptimal labels such as claims-based methods (e.g. ICD-10 coding) [14]. Alternatively, some ML models are based on clinical prediction scores such as qSOFA or SIRS, that were not designed for diagnostic purposes [27, 28]. Furthermore, these ill-suited methods may imply major limitations for the development of ML models at the ED, and consequently the identification of new predictors by ML. An EAC is likely the best option to define sepsis at the ED, since every member of the EAC is fully aware of the Sepsis-3 definition and will be able to apply this definition in clinical practice. Especially for relatively small datasets, such as our own, an EAC is executable and reliable.
Accurate sepsis diagnosis relies on accurate diagnostic tools to improve treatment strategies and outcome of septic patients. With machine learning, clinical diagnostic models can be developed by integrating big sets of routine care. Instead of ‘silver’ sepsis labels, we showed that in a relatively small dataset with high quality sepsis labels, ML can be used as method to identify new sepsis predictors on a wide variety of routine care data in the ED. Apart from the standard variables that are measured during an ED visit, we found multiple advanced haematology variables that show univariate diagnostic importance for sepsis diagnosis that is similar as compared to vital, demographic and standard laboratory variables. Moreover, known variables, such as granulocytes, and in particular the immature, banded granulocytes, were identified as important variables by both L1- and RF-extended models, serving as a positive control. Even though not many advanced haematology variables were selected by the machine learning algorithms, combining them with the routinely available variables resulted in a better performance, hinting at their diagnostic value.
The vital variables had a better diagnostic performance as compared to the laboratory and advanced haematological variables. In particular, hr and rr showed high univariate performance and were found to have high variable importance in both extended models. These results are in line with previous research and they are also used in multiple clinical prediction scores (e.g. (q)SOFA, MEWS) [5, 22, 29]. One reason that vital variables had an overall high performance may have been because the EAC was instructed to use the sepsis-3 criteria which includes sbp and rr. Therefore, the EAC may have quickly labelled patients as sepsis as these standard variables are available at the start of the clinical assessment.
Of the laboratory variables, only CRP showed high diagnostic performance in the univariate analysis. In the extended models, CRP was the second most important in the RF model, though CRP had a very low coefficient in the L1 model. CRP is a known maker for inflammation in blood and known to be associated with infection [30,31,32]. As the model evaluated many haematological variables associated with infection, the effect of CRP may have been reduced in the L1 model. Of the remaining laboratory variables, only ASAT or Gamma-GT showed marginally importance in the extended models, which are biomarkers for liver dysfunction. Although regarding the sepsis criteria, only bilirubin is used in the context of liver dysfunction [33].
The majority of the found advanced haematological variables by the extended models were related to granulocytes. Immature granulocytes are prematurely released from the bone marrow when the body’s immune response is active [34]. Research has shown that elevated values for ig and band neutrophils are associated with infection and can be an early sign of sepsis [35]. As result, these markers have been used for sepsis diagnosis as either a rule-in or rule-out test for early sepsis diagnosis [36]. Most interesting, the eosinophiles were negatively associated with sepsis in the L1 extended model. Wilar (2019) found the same association in neonatal sepsis as well as Abidi (2008) [37, 38]. As we removed correlated variables, the following variables were likely as predictive: immature granulocytes count (ig), banded granulocytes count (bnd) and percentage eosinophil granulocyte (peos).
As blood is drawn for every ED visit who visits our internal medicine department, complete laboratory data, including the advanced haematology variables, were available for almost all ED visits. Only a few visits missed 1 or 2 laboratory variables (data not shown). Although we imputed missing vital data points, all used vital variables had missing data < 1%, except for rr (28.6%), fio2 (15.6%), and spo2 (15.6%). Measuring variables related to the respiratory tract is most often neglected as it is time-consuming, especially in the ED where time is limited. A median rr value of 17 was used, which is representative for our internal medicine population. As rr was positively related to sepsis outcome, it is important that rr should accurately be measured during ED visit.
During model development, all data was used for training and testing in the DLCV scheme instead of a single train-test split. Also, no data was reused for hyperparameter optimization or testing in the DLCV scheme due to the inner fold and outer fold. Moreover, the variable importance was evaluated of each of the 10 trained DLCV models, instead of retraining a final model thereby reusing all data, hence introducing bias. Lastly, for each of the three selected models, we found a high AUC performance that strengthen the validity of the identified predictors. Even though a big proportion of the variables was found by both the L1 and RF models, both models also identified different variables. For example, the RF model could have identified non-linear relationships between variables, though interestingly these relationships were not supported by the SHAP values. However, evaluating variable importance does not imply causation, and is not a 1-to-1 comparison. Even though the same set of variables were identified as important by both L1 and RF algorithms, these predictors should first be evaluated as univariate predictors before being used in prediction models.
This study has several limitations. First, as this is a single centre study the results are not validated and may not be generalizable to other centres. Moreover, ED population are very diverse in terms of illness, age and secondary problems which may hamper validation of our results. Secondly, although we are convinced that an EAC is the optimal choice for labelling possible sepsis patients, an EAC is not perfect: patients will still be labelled based on the concept of sepsis of the expert. As a consequence, predictors known to be associated with sepsis, were also found by the algorithms. However, the advanced haematological variables were not available for the experts during the labelling process. Another limitation of an EAC is that it is labour intensive and may limit the reproducibility of our results. Nonetheless, there are semi-supervised ML models that would be able to label large datasets based on a couple of hundred EAC labelled patients [39]. We found that the final diagnosis sometimes remains uncertain for treating physicians at the ED. Therefore, it is possible that the final diagnosis as determined by the treating physician may differ from the EAC’s opinion. Future studies should be performed to explore these possibilities. Finally, we used ML to identify associations between variables and sepsis, but did not look into any causal relationships. Future studies should explore causal inference to evaluate the causal relationship to determine the dependence of the found variables with sepsis [40].
Conclusions
Using ML and EAC generated labels we identified potential groups of predictors for sepsis diagnosis in routine EHR data, including new advanced haematological variables. Routinely collected variables from multiple sources already show predictive power for sepsis diagnosis, indicating that there is already value in data currently collected at the ED. Altogether, this method is an illustrative example of how machine learning can be utilized in data research. In our opinion, ML should not only be used to develop diagnostic tools, but also for other aspects of data research, such as the identification of diagnostic predictors and the related importance. This approach is not only applicable in the context of sepsis, but also for other syndromes that lack a reliable gold standard.
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due to patient privacy but are available from the corresponding author (s.haitjema@umcutrecht.nl) upon reasonable request.
References
Singer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA - Journal of the American Medical Association. 2016;315(8):801–10. https://doi.org/10.1001/jama.2016.0287.
HJ de Grooth, J Postema, SA Loer, JJ Parienti, HM Oudemans-van Straaten, AR Girbes. Unexplained mortality differences between septic shock trials: a systematic analysis of population characteristics and control-group mortality rates. Intensive Care Med. 2018;44(3):311–322. https://doi.org/10.1007/s00134-018-5134-8.
Vincent JL. The clinical challenge of sepsis identification and monitoring. PLoS Med. 2016;13(5): e1002022. https://doi.org/10.1371/journal.pmed.1002022.
Talisa VB, Yende S, Seymour CW, Angus DC. Arguing for adaptive clinical trials in sepsis. Front Immunol. 2018;9:1502. https://doi.org/10.3389/fimmu.2018.01502.
Subbe CP, Davies RG, Williams E, Rutherford P, Gemmell L. Effect of introducing the modified early warning score on clinical outcomes, cardio-pulmonary arrests and intensive care utilisation in acute medical admissions. Anaesthesia. 2003. https://doi.org/10.1046/j.1365-2044.2003.03258.x.
Crouser ED, et al. Monocyte distribution width: a novel indicator of sepsis-2 and sepsis-3 in high-risk emergency department patients. Crit Care Med. 2019;47(8):1018–25. https://doi.org/10.1097/CCM.0000000000003799.
Uffen JW, Oomen P, de Regt M, Oosterheert JJ, Kaasjager K. The prognostic value of red blood cell distribution width in patients with suspected infection in the emergency department. BMC Emerg Med. 2019;19(1):76. https://doi.org/10.1186/s12873-019-0293-7.
Davis BH. Improved diagnostic approaches to infection/sepsis detection. Expert Rev Mol Diagn. 2005. https://doi.org/10.1586/14737159.5.2.193.
Brunetti E, et al. Comparison of Diagnostic Accuracies of qSOFA, NEWS, and MEWS to Identify Sepsis in Older Inpatients With Suspected Infection. J Am Med Dir Assoc. 2022;23(5):865-871.e2. https://doi.org/10.1016/j.jamda.2021.09.005.
Johnson AEW, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:1–9. https://doi.org/10.1038/sdata.2016.35.
Fleuren LM, et al. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46(3):383–400. https://doi.org/10.1007/s00134-019-05872-y.
Rutjes AW, Reitsma JB, Coomarasamy A, Khan KS, Bossuyt PM. Evaluation of diagnostic tests when there is no gold standard. A review of methods. Heal Technol Assess. 2007;11(50):10–51. https://doi.org/10.3310/hta11500.
Kang SH, Kim HK, Ham CK, Lee DS, Cho HI. Comparison of four hematology analyzers, CELL-DYN Sapphire, ADVIA 120, Coulter LH 750, and Sysmex XE-2100, in terms of clinical usefulness. Int J Lab Hematol. 2008. https://doi.org/10.1111/j.1751-553X.2007.00981.x.
TAP de Hond, MSA Niemantsverdriet, WW van Solinge, JJ Oosterheert, S Haitjema, KAH Kaasjager. Sepsis labels defined by claims-based methods are ill-suited for training machine learning algorithms. Clin Microbiol Infect Off Publ Eur So. Clin Microbiol Infect Dis. 2022. https://doi.org/10.1016/j.cmi.2022.03.029.
Müller R, et al. European multi-center evaluation of the Abbott cell-dyn sapphire hematology analyzer. Lab Hematol. 2006. https://doi.org/10.1532/LH96.05041.
Gijsberts M et al., “Hematological Parameters Improve Prediction of Mortality and Secondary Adverse Events in Coronary Angiography Patients,” Medicine (United States). 2015. https://doi.org/10.1097/MD.0000000000001992.
ten Berg MJ, Huisman A, van den Bemt PM, Schobben AF, Egberts AC, van Solinge WW. Linking laboratory and medication data: new opportunities for pharmacoepidemiological research. Clin Chem Lab Med. 2007;45(1):13–9. https://doi.org/10.1515/CCLM.2007.009.
Wolpert DH, Macready WG. No free lunch theorems for optimization. IEEE Trans Evol Comput. 1997. https://doi.org/10.1109/4235.585893.
Cox DR. The regression analysis of binary sequences. J R Stat Soc Ser B. 1959. https://doi.org/10.1111/j.2517-6161.1959.tb00334.x.
R Tishbirani. Regression shrinkage and selection via the Lasso. Journal of the Royal Statistical Society. Series B (Methodological). 1996. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x.
TK Ho. Random decision forests. 1995. https://doi.org/10.1109/ICDAR.1995.598994
CW Seymour, VX Liu, TJ Iwashyna. Assessment of Clinical Criteria for Sepsis: For the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). 2016; (vol 315, pg 762, 2016). Jama-J Am Med Assoc. 2016;315(20):2237. https://doi.org/10.1001/jama.2016.5850.
Hawkins DM. The problem of overfitting. J Chem Inform Comp Sci. 2004. https://doi.org/10.1021/ci0342472.
Babyak MA. What you see may not be what you get: a brief, nontechnical introduction to overfitting in regression-type models. Psychosom Med. 2004. https://doi.org/10.1097/01.psy.0000127692.23278.a9.
Ledell E, Petersen M, Van Der Laan M. Computationally efficient confidence intervals for cross-validated area under the ROC curve estimates. Electron J Stat. 2015;9(1):1583–607. https://doi.org/10.1214/15-EJS1035.
RStudio Team, “RStudio: Integrated Development for R.” RStudio, PBC, Boston, MA, 2021. Available: http://www.rstudio.com/.
Ratzinger F, et al. “Machine learning for fast identification of bacteraemia in SIRS patients treated on standard care wards: a cohort study. Sci Rep. 2018. https://doi.org/10.1038/s41598-018-30236-9.
Kwon YS, Baek MS. Development and validation of a quick sepsis-related organ failure assessment-based machine-learning model for mortality prediction in patients with suspected infection in the emergency department”. J Clin Med. 2020. https://doi.org/10.3390/jcm9030875.
Vincent JL, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. Intensive Care Med. 1996. https://doi.org/10.1007/BF01709751.
Sabel KG, Hanson L. The clinical usefulness of c-reactive protein (CRP) determinations in bacterial meningitis and septicemia in infancy”. Acta Pædiatrica. 1974. https://doi.org/10.1111/j.1651-2227.1974.tb04814.x.
Matson A, Soni N, Sheldon J. C-reactive protein as a diagnostic test of sepsis in the critically ill. Anaesth Intensive Care. 1991. https://doi.org/10.1177/0310057x9101900204.
Póvoa P, et al. C-reactive protein as an indicator of sepsi. Intensive Care Med. 1998. https://doi.org/10.1007/s001340050715.
Singer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA - J Am Med Assoc. 2016;315(8):801–10. https://doi.org/10.1001/jama.2016.0287.
Van der Geest PJ, Mohseni M, Brouwer R, van der Hoven B, Steyerberg EW, Groeneveld ABJ. Immature granulocytes predict microbial infection and its adverse sequelae in the intensive care unit. J Crit Care. 2014. https://doi.org/10.1016/j.jcrc.2014.03.033.
Drifte G, Dunn-Siegrist I, Tissières P, Pugin J. Innate immune functions of immature neutrophils in patients with sepsis and severe systemic inflammatory response syndrome. Crit Care Med. 2013. https://doi.org/10.1097/CCM.0b013e318274647d.
Ayres LS, Sgnaolin V, Munhoz TP. Immature granulocytes index as early marker of sepsis. Int J Lab Hematol. 2019;41(3):392–6. https://doi.org/10.1111/ijlh.12990.
Wilar R. Diagnostic value of eosinopenia and neutrophil to lymphocyte ratio on early onset neonatal sepsis. Korean J Pediatr. 2019;62(6):217–23. https://doi.org/10.3345/kjp.2018.06723.
Abidi K, et al. Eosinopenia is a reliable marker of sepsis on admission to medical intensive care units. Crit Care. 2008;12(2):1–10. https://doi.org/10.1186/cc6883.
de Vries S, Thierens D. A reliable ensemble based approach to semi-supervised learning. Knowledge-Based Syst. 2021. https://doi.org/10.1016/j.knosys.2021.106738.
Pearl J. Causal inference in statistics: an overview. Stat Surv. 2009;3:96–146. https://doi.org/10.1214/09-SS057.
Acknowledgements
Not applicable.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Conception and design: Michael Niemantsverdriet, Titus de Hond, Jan Jelrik Oosterheert, Karin Kaasjager, Saskia Haitjema. Collection and assembly of data: Michael Niemantsverdriet, Titus de Hond, Saskia Haitjema. Data analysis and interpretation: Michael Niemantsverdriet, Titus de Hond, Domenico Bellomo, Saskia Haitjema. Manuscript writing: all authors. Final approval of manuscript: all authors. Accountable for all aspects of the work: all authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed according to the declaration of Helsinki and the ethical guidelines of our institution. The SPACE cohort was reviewed and approved by the Medical Ethical Committee of the UMCU and waived the need for informed consent as only pseudonymized data were used for this study under number 16/594 and registered in the Dutch Trial Register (NTR) under number 6916. Data collection and handling was conducted in accordance with European privacy legislation (GDPR).
Consent for publication
Not applicable.
Competing interests
M.S.A. Niemantsverdriet is supported by a PhD fellowship from SkylineDx, Rotterdam.
D. Bellomo is employed by SkylineDx, Rotterdam.
All others have no potential conflict of interest to report.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Supplemental Table 1. Definitions of all variables for the three variable types used in this study. Supplemental Table 2. Algorithms used in this study and the associated hyperparameters that were optimized in the double loop cross validation (DLCV). Supplemental Table 3. Immunocompromised definitions. Supplemental Figure 1. scheme of the double loop cross validation. HP: hyperparameter, CV: cross validation. Supplemental Figure 2. dendrogram of all 95 variables computed with Euclidean distance.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Niemantsverdriet, M.S.A., de Hond, T.A.P., Hoefer, I.E. et al. A machine learning approach using endpoint adjudication committee labels for the identification of sepsis predictors at the emergency department. BMC Emerg Med 22, 208 (2022). https://doi.org/10.1186/s12873-022-00764-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12873-022-00764-9