Abstract
Background
Primary carnitine deficiency (PCD) denotes low carnitine levels with an autosomal recessive pattern of inheritance. Cardiomyopathy is the most common cardiac symptom in patients with PCD, and early diagnosis can prevent complications. Next-generation sequencing can identify genetic variants attributable to PCD efficiently.
Objective
We aimed to detect the genetic cause of the early manifestations of hypertrophic cardiomyopathy and metabolic abnormalities in an Iranian family.
Methods
We herein describe an 8-year-old boy with symptoms of weakness and lethargy diagnosed with PCD through clinical evaluations, lab tests, echocardiography, and cardiac magnetic resonance imaging. The candidate variant was confirmed through whole-exome sequencing, polymerase chain reaction, and direct Sanger sequencing. The binding efficacy of normal and mutant protein-ligand complexes were evaluated via structural modeling and docking studies.
Results
Clinical evaluations, echocardiography, and cardiac magnetic resonance imaging findings revealed hypertrophic cardiomyopathy as a clinical presentation of PCD. Whole-exome sequencing identified a new homozygous variant, SLC22A5 (NM_003060.4), c.821G > A: p.Trp274Ter, associated with carnitine transport. Docking analysis highlighted the impact of the variant on carnitine transport, further indicating its potential role in PCD development.
Conclusions
The c.821G > A: p.Trp274Ter variant in SLC22A5 potentially acted as a pathogenic factor by reducing the binding affinity of organic carnitine transporter type 2 proteins for carnitine. So, the c.821G > A variant may be associated with carnitine deficiency, metabolic abnormalities, and cardiomyopathic characteristics.
Similar content being viewed by others
Introduction
Primary carnitine deficiency (PCD) is characterized by low levels of carnitine in the body and is inherited as an autosomal recessive trait predominantly [1]. The incidence of PCD varies by ethnicity, with a frequency ranging from 1 in 142,000 in the United States [2] to 1 in 300 in the Faroe Islands [3]. The clinical manifestations of PCD vary from asymptomatic to sudden cardiac death [4]. Cardiomyopathy with or without skeletal muscle weakness starting from 1 to 4 years of age is the most frequent cardiac involvement in patients with PCD [5]. Among types of cardiomyopathies, dilated cardiomyopathy is the most common [6].
The SLC22A5 gene on 5q31.1 is the sole gene responsible for systemic PCD and a member of the organic cation transporter family, which encodes organic carnitine transporter type 2 (OCTN2) [7]. SLC22A5 spans 25,910 base pairs and contains 10 exons [8]. The final product of this gene is a 63-kilodalton protein composed of 557 amino acids, enhancing the uptake of carnitine [9]. SLC22A5 encodes the OCTN2 transporter, responsible for carnitine uptake into cells. This process is crucial for transporting long-chain fatty acids into mitochondria for energy production. Various mutations ranging from missense and nonsense to splice-site mutations and deletions can be found in the SLC22A5 gene, leading to the impaired function or expression of the OCTN2 transporter. The consequence is diminished cellular carnitine transport, which manifests in metabolic crises or cardiomyopathy due to disrupted fatty acid oxidation. The prevalence of specific mutations can vary across populations [10].
While PCD can induce dangerous complications and even death, early diagnosis and treatment can prevent many complications. New diagnostic techniques, including next-generation sequencing, contribute to advances in diagnosing hereditary heart diseases and identifying new genes related to those conditions [11]. Next-generation sequencing helps investigate the whole genome or exome to identify the genetic cause of diseases [12]. In the present study, we detected a novel pathogenic variant, SLC22A5 (NM_003060.4), c.821G > A: p.Trp274Ter, in an Iranian family with early manifestations of hypertrophic cardiomyopathy and metabolic abnormalities, corresponding to the clinical and paraclinical symptoms of PCD. Although hypertrophic cardiomyopathy manifests in some patients with PCD, its precise etiology needs elucidation [13, 14]. Further in-depth information concerning the different variants of the genes involved in PCD can expedite its diagnosis and prevention.
Methods
Family recruitment and clinical evaluation
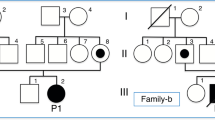
The proband in the current case study was an 8-year-old boy (Fig. 1A: IV-3) referred to Rajaie Cardiovascular Medical and Research Center, affiliated with Iran University of Medical Sciences, Tehran, Iran, with complaints of weakness and lethargy. The patient’s consanguineous parents (first cousins) had no cardiomyopathy or metabolic disorders. The father and mother were 45 and 34 years of age, respectively. The boy was the third offspring and had 2 brothers, aged 26 and 19, without cardiac or metabolic diseases. The mother had a miscarriage for unknown reasons in the second month of pregnancy. The proband’s symptoms started 2 weeks prior to his hospital admission. The initial evaluation raised suspicions of glucose storage disease due to hypotonia and hepatomegaly. During the patient’s hospital stay, his weakness and lethargy responded to the prescription of bicarbonate and carnitine, and his symptoms disappeared gradually. The response to carnitine therapy bolstered the evidence of carnitine deficiency. The ammonia level was 155 µmol/L, indicating hyperammonemia, a consistent finding in patients with PCD. The boy underwent echocardiography, cardiac magnetic resonance imaging, and genetic testing for the confirmation of the diagnosis.
A: The image illustrates the pedigree of the family with primary carnitine deficiency (PCD). The proband (IV-3), indicated by the arrow, is the case of the study. B: The image presents the chromatogram of the change position. The individual marked with a black dot is a homozygous carrier. + sign: the reference allele; − sign: the mutant allele
Sampling and whole-exome sequencing (WES)
Informed consent was obtained from the proband and all his available family members, who subsequently provided peripheral blood samples. Genomic DNA was extracted from the peripheral blood via standard salting-out methods, following the manufacturer’s protocol. WES was performed on the proband (Fig. 1A: IV-3). Exome capture was conducted with an Agilent SureSelect All Exon V6 kit, and library sequencing was performed with the Illumina HiSeq 6000. An in-house setup bioinformatics pipeline, including quality checks and primary filtering of reads, aligning the reads to the human reference genome (GRCh37/hg19), and calling and annotating the variants, was applied. All variants with minor allele frequencies below 1% in the 1000 Genomes Project, the Genome Aggregation Database (gnomAD), and Iranome were considered.
Mutations that were regarded as likely pathogenic or pathogenic in ClinVar and the Human Gene Mutation Database (HGMD) and had a related phenotype were considered priorities. The remaining variants were subjected to bioinformatics analysis via online tools, such as MutationTaster, Polymorphism Phenotyping v2 (PolyPhen-2), Protein Variation Effect Analyzer (PROVEAN), Sorting Intolerant From Tolerant (SIFT), and Combined Annotation-Dependent Depletion (CADD), to predict the effects of the variants on the structure and function of the protein. Further, ENTPRISE-X was employed to predict the consensus of nonsense variants. The variants predicted as pathogenic by most of the tools were selected for segregation analysis. The standards of the American College of Medical Genetics and Genomics (ACMG) were applied to interpret the variants. Finally, the candidate variant was subjected to confirmation and segregation analysis.
Polymerase chain reaction (PCR) and Sanger sequencing
Primer sequences surrounding the candidate variant were designed using the Primer3 (v. 0.4.0) server. PCR was performed with the SimpliAmp Thermal Cycler (Thermo Fisher Scientific) using 10 pmol/L of primers (forward primer: TATGGCGCCTTGGTCTTAGTA and reverse primer: TCAGCACACAGCCAGAACT), 100 ng of DNA, 200 mmol/L of dNTP, 1.5 mmol/L of MgCl2, and 1 U of Taq DNA polymerase (Amplicon). The PCR schedule was incubation at 95 °C for 5 min, followed by 35 cycles (40 s at 95 °C, 30 s at 59 °C, and 30 s at 72 °C). Subsequently, the PCR products were sequenced on the ABI Sequencer 3500XL PE (Applied Biosystems, USA) and the Codon Code Aligner (v. 7.1.2) (CodonCode Corp, USA).
Structural modeling and docking study
The 3D structure of the human OCTN2 protein (SLC22A5) was not available in the protein data bank (PDB: http://www.rcsb.org/pdb). Consequently, the protein sequence (FASTA format) was downloaded from the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/), and the 3D structure was constructed with AlphaFold2 using the MMseqs2 server [15]. For the generation of the 3D structures in the AlphaFold2 server, each protein sequence (the wild type and mutant [p.Trp274Ter]) was loaded in FASTA format into the AlphaFold2 server. Next, molecular modeling was initiated to generate the PDB format of each protein. The 3D structure of the L-carnitine molecule (vitamin BT, PubChem CID: 10,917) was accessed from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and processed to a PDB file format. The structures of the proteins were corrected with ViewerLite (v.5), and polar hydrogens were added. Further, the YASARA Energy Minimization Server was utilized to minimize energy during molecular docking (http://www.yasara.org/minimizationserver.htm) [16]. The 3D formation of the compounds was imported as an SCE file into the YASARA View (v.20.12.24) to deliver low-energy structures of the compounds and was then saved in a PDB file format. Molecular docking was performed with AutoDock Vina to evaluate the binding efficacy of normal and mutant protein-ligand complexes [17, 18]. Post-docking analyses were visualized using PyMOL (v.2.5.2) and LigPlus+ (v.2.2.4), [19] providing details of the size and location of the binding sites and the hydrogen-bond interaction of the docked ligand in various confirmations.
Ethical considerations
The study complies with the Declaration of Helsinki. Ethical approval was obtained from the Ethics Committees of Rajaie Cardiovascular Medical and Research Center, affiliated with Iran University of Medical Sciences, Tehran, Iran (IR.RHC.REC.1402.004). Written informed consent was obtained from the participants, who were ensured of their right to withdraw from the study at any point with no negative implications.
Results
Echocardiography and Cardiac magnetic resonance imaging findings
Echocardiography revealed mild left and right atrial enlargement, mild to severe left ventricular enlargement, mild left ventricular hypertrophy, left ventricular ejection fraction of 20%, severe right ventricular failure, tricuspid valve regurgitation, tricuspid annular plane systolic excursion of about 17 mm, and no atrial or ventricular septal defect and patent ductus arteriosus, favoring hypertrophic cardiomyopathy, an expected finding in carnitine deficiency. Cardiac magnetic resonance imaging showed cardiomegaly primarily due to left ventricular enlargement, increased left ventricular myocardial mass (mass index = 137 g/m2), left ventricular dyssynchrony, a severely reduced ejection fraction (≈ 17%), a cardiac index of 4 L/min/m2, moderate right ventricular enlargement without hypertrophy, a severely reduced systolic function (ejection fraction ≈ 24%), a right cardiac index of 2.9 L/min/m2, and no evidence of myocardial edema or enhancement (Fig. 2).
The image shows the proband’s cardiac magnetic resonance imaging. A, B, and C: They present 2- and 4-chamber views and short-axis short tau inversion recovery (STIR) images, respectively. Severe left ventricular enlargement is observed. D: The 4-chamber phase-sensitive inversion recovery (PSIR) with a 10-minute delay demonstrates no abnormal enhancement. Increased signal intensity adjacent to the subendocardial muscles in the STIR images is secondary to a significantly reduced ejection fraction and subsequent blood stasis
Genetic findings
WES detected a novel pathogenic variant, c.821G > A: p.Trp274Ter, in a homozygous state in exon 4 of the carnitine transporter gene, SLC22A5 (NM_003060). The identified variant was confirmed by Sanger sequencing. The variant was segregated in the pedigree with an autosomal recessive pattern. In other words, his healthy parents and 2 brothers had the heterozygous form of the variant (Fig. 1B). According to the ACMG/AMP guidelines (PVS1, PM2, and PM3), the identified variant was pathogenic. This variant has not been reported in any databases, and it appears to be the primary cause of PCD with hypertrophic cardiomyopathy manifestation in this pedigree.
Modeling and Docking
The best models of human OCTN2 (normal and p.Trp274Ter) were downloaded from AlphaFold2 with predicted local distance difference test (pLDDT) scores of 85.7 and 90.2. The pLDDT score (0-100) indicates the identity of the reproduced model with the reference protein structure. Docking analysis was performed on OCTN2-ligand compounds in mutated and wild-type modes with root-mean-square deviations of 0 and 2.413 and docking scores of − 3.7 and − 4.4. The OCTN2 wild-type ligand compounds had a binding energy of − 4.4 kcal/mol and 4 hydrogen bonds between L-carnitine and Ile43, Thr45, and Asn367. The OCTN2 mutant ligand compounds (− 3.7 kcal/mol) had no connection with each other (Fig. 3 [A and B]). The buried surface area for the normal and mutant OCTN2-ligand compounds with L-carnitine is depicted in Fig. 3 (C and D).
The images illustrate (A) the interaction between the wild-type protein and L-carnitine, (B) the interaction between the mutant protein and L-carnitine, (C) the 3D structure of normal organic carnitine transporter type 2 (OCTN2) (green) in interaction with L-carnitine, and (D) the 3D structure of the mutant OCTN2 (blue) in interaction with L-carnitine
Discussion
An 8-year-old boy was referred to us with a metabolic disorder and cardiomyopathic features. Paraclinical examinations and laboratory biochemical tests were compatible with carnitine deficiency. To confirm the diagnosis, we performed WES and detected a novel pathogenic variant in SLC22A5 (NM_003060.4), c.821G > A: p.Trp274Ter. This stop-gain variant led to an interruption in OCTN2 synthesis in the 274th amino acid of the peptide, and our bioinformatics analysis revealed no affinity between the transporter and carnitine as its ligand. Indeed, our docking results suggested that the truncated OCTN2 protein had no affinity with carnitine; therefore, the mutant SLC22A5 (p.Trp274Ter) had significantly reduced carnitine transport activity compared with the wild-type SLC22A5.
The clinical features of SLC22A5 variants and carnitine deficiency can vary widely. The reported variants in SLC22A5 associated with carnitine deficiency phenotypes are presented in Table 1. In some cases, affected individuals may be asymptomatic, whereas others may present with life-threatening metabolic crises. The diagnosis of carnitine deficiency is based on clinical presentation, biochemical testing, and genetic analysis, with early diagnosis playing a crucial role in preventing side effects [20]. Spiekerkoetter et al. [21] concluded that patients with a similar genotype had different ages of onset and various types of clinical manifestations, including weakness, cardiac presentations, and metabolic abnormalities. Even siblings who possess the same variant in the SLC22A5 gene have different onset ages and progressions [22]. SLC22A5 stop-gain pathogenic variants are associated with carnitine deficiency or cardiomyopathy as the only clinical phenotype without metabolic abnormalities [23]. The primary treatment for carnitine deficiency is carnitine supplementation, [5] and affected individuals may require treatment for cardiomyopathy, liver dysfunction, and other complications [24]. Early diagnosis with precise diagnostic tools and prompt treatment with carnitine supplementation can improve outcomes and prevent complications; however, in severe cases, the disorder can give rise to life-threatening metabolic crises and lifelong disorders [24]. Carnitine deficiency, particularly PCD, is associated with cardiomyopathy, specifically dilated cardiomyopathy, as the most common type linked to PCD. In patients with PCD, dilated cardiomyopathy is more frequent, [25] but it does not mean that other cardiomyopathies do not exist in these patients. Hypertrophic cardiomyopathy is less commonly associated with PCD than dilated cardiomyopathy. In hypertrophic cardiomyopathy, the heart muscle thickens, obstructing blood flow and causing cardiac dysfunction [26].
Research on the SLC22A5 gene is limited to patients with hypertrophic cardiomyopathy. Still, the literature contains studies on the coincidence of PCD and hypertrophic cardiomyopathy in SLC22A5 variants. Lahrouchi et al. [13] conducted WES on the members of a consanguineous family with a history of childhood hypertrophic cardiomyopathy and sudden cardiac death and reported a homozygous stop variant in the SLC22A5 gene, typically associated with PCD, as the probable genetic culprit. Ino et al. [27] retrospectively assessed 11 children with abnormal carnitine metabolism and cardiomyopathy to determine the prognosis of cardiomyopathy correlated with hypocarnitinemia. The children exhibited various carnitine-related disorders: 6 of them showed hypertrophic cardiomyopathy, and the rest suffered from dilated cardiomyopathy. Echocardiography revealed inconsistent left ventricular function and wall thickness. In histologic evaluations, Ino and colleagues observed significant lipid buildup within enlarged heart cells. The authors concluded that carnitine supplementation conferred echocardiographic improvements for 6 out of 8 treated patients over periods of 3 months to 2 years. García-Vielma et al. [28] sought to identify genetic variants related to hypertrophic cardiomyopathy in Mexican patients. The authors maintained that while most global research primarily pointed to genes like MHY7 and MYBPC3, there was a dearth of data on other genes. They evaluated 37 patients with heart disease and sudden death and focused on 168 genes. Among the identified pathogenic variants, the SLC22A5 gene was an affected gene, along with others like MYH7 and MYBPC3. Notably, 5 variants, including potentially the one in SLC22A5, had not been previously documented in public databases. In a case report, Deswal et al. [29] described a 9-month-old boy suffering from hypertrophic cardiomyopathy, significant hepatomegaly, and jaundice with extremely low free carnitine levels. Genetic analysis identified compound heterozygous mutations in the SLC22A5 gene. One of these mutations had been previously reported, while the other was a novel frame-shift mutation. After 3 months of oral carnitine supplementation, the patient showed significant improvement, including an ejection fraction of 75% and regular liver size and enzymes. It is noteworthy that the proband in our study had no other hepatic-related signs. Mutlu-Albayrak et al. [41] described a 9-year-old boy with a dysmorphic appearance and hypertrophic cardiomyopathy. Both the patient and his 4-year-old sister, who also had cardiomyopathy and fatigue, were diagnosed with a carnitine uptake defect through tandem mass spectrometry. In addition, the patient’s other sister died suddenly at 19 months. Genetic sequencing identified a new mutation in the SLC22A5 gene in both siblings, responsible for their carnitine uptake defect. The parents were found to be carriers of this mutation.
In conclusion, the paucity of information on PCD patients with hypertrophic cardiomyopathy involvement in SLC22A5 variants warrants future studies to reveal the role of this gene in the development of coincident PCD and hypertrophic cardiomyopathy.
Limitations
The findings of the present study should be interpreted in light of its limitations. Firstly, structural modeling and docking studies, albeit informative, cannot replace direct functional assays since they render the actual effects of the mutation on carnitine transport function speculative until these impacts are verified by in vivo studies. Nonetheless, in vivo or clinical interventions are not feasible due to ethical or cost-benefit considerations. Secondly, we did not consider other potential comorbidities and environmental factors that might have influenced the observed clinical manifestations. Thirdly, it is essential to recognize the potential phenotypic variability since not every individual with the same mutation may display the same clinical features. Fourthly, a study focusing on an Iranian family raises questions about the generalizability of the results to other ethnic or racial groups. Finally, while WES is powerful, it concentrates only on exonic areas, possibly missing causative variants in other crucial genomic regions and creating a potential detection bias. Further studies are, therefore, required to elucidate the pathogenicity of the variant identified in the current investigation.
Conclusions
The present study detected a novel genetic variant, SLC22A5 (NM_003060.4), c.821G > A: p.Trp274Ter by WES and in-silico study demonstrated a lower affinity to carnitine. The variant likely caused the abnormal manifestations and lab tests in the studied 8-year-old boy with symptoms of weakness and lethargy.
Data Availability
All data generated or analyzed during this study are included in this published article.
References
Longo N. Primary Carnitine Deficiency and Newborn Screening for disorders of the Carnitine cycle. Ann Nutr Metab. 2016;68(Suppl 3):5–9.
Therrell BL Jr., Lloyd-Puryear MA, Camp KM, Mann MY. Inborn errors of metabolism identified via newborn screening: ten-year incidence data and costs of nutritional interventions for research agenda planning. Mol Genet Metab. 2014;113(1–2):14–26.
Rasmussen J, Nielsen OW, Janzen N, Duno M, Gislason H, Køber L, et al. Carnitine levels in 26,462 individuals from the nationwide screening program for primary carnitine deficiency in the Faroe Islands. J Inherit Metab Dis. 2014;37(2):215–22.
Tomlinson S, Atherton J, Prasad S. Primary Carnitine Deficiency: a rare, reversible metabolic cardiomyopathy. Case Rep Cardiol. 2018;2018:3232105.
Shibbani K, Fahed AC, Al-Shaar L, Arabi M, Nemer G, Bitar F, et al. Primary carnitine deficiency: novel mutations and insights into the cardiac phenotype. Clin Genet. 2014;85(2):127–37.
Kilic M, Ozgül RK, Coşkun T, Yücel D, Karaca M, Sivri HS, et al. Identification of mutations and evaluation of cardiomyopathy in Turkish patients with primary carnitine deficiency. JIMD Rep. 2012;3:17–23.
Tamai I. Pharmacological and pathophysiological roles of carnitine/organic cation transporters (OCTNs: SLC22A4, SLC22A5 and Slc22a21). Biopharm Drug Dispos. 2013;34(1):29–44.
(OMIM®) OMIiM. SOLUTE CARRIER FAMILY 22 (ORGANIC CATION TRANSPORTER). MEMBER 5; SLC22A5.
Zong NC, Li H, Li H, Lam MP, Jimenez RC, Kim CS, et al. Integration of cardiac proteome biology and medicine by a specialized knowledgebase. Circ Res. 2013;113(9):1043–53.
Longo N, Frigeni M, Pasquali M. Carnitine transport and fatty acid oxidation. Biochim Biophys Acta. 2016;1863(10):2422–35.
Mazzaccara C, Lombardi R, Mirra B, Barretta F, Esposito MV, Uomo F et al. Next-generation sequencing gene panels in Inheritable cardiomyopathies and channelopathies: prevalence of pathogenic variants and variants of unknown significance in uncommon genes. Biomolecules. 2022;12(10).
Zhao Y, Fang LT, Shen TW, Choudhari S, Talsania K, Chen X, et al. Whole genome and exome sequencing reference datasets from a multi-center and cross-platform benchmark study. Sci data. 2021;8(1):296.
Lahrouchi N, Lodder EM, Mansouri M, Tadros R, Zniber L, Adadi N, et al. Exome sequencing identifies primary carnitine deficiency in a family with cardiomyopathy and sudden death. Eur J Hum Genetics: EJHG. 2017;25(6):783–7.
Matsuishi T, Hirata K, Terasawa K, Kato H, Yoshino M, Ohtaki E, et al. Successful carnitine treatment in two siblings having lipid storage myopathy with hypertrophic cardiomyopathy. Neuropediatrics. 1985;16(1):6–12.
Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19(6):679–82.
Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins. 2009;77(Suppl 9):114–22.
Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python Bindings. J Chem Inf Model. 2021;61(8):3891–8.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61.
Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–34.
Stanley CA. Carnitine deficiency disorders in children. Ann N Y Acad Sci. 2004;1033:42–51.
Spiekerkoetter U, Huener G, Baykal T, Demirkol M, Duran M, Wanders R, et al. Silent and symptomatic primary carnitine deficiency within the same family due toidentical mutations in the organic cation/carnitine transporter OCTN2. J Inherit Metab Dis. 2003;26:613–5.
Kilic M, Özgül RK, Coşkun T, Yücel D, Karaca M, Sivri HS, et al. Identification of mutations and evaluation of Cardiomyopathy in Turkish patients with primary Carnitine Deficiency. JIMD Reports - Case and Research Reports, 2011/3. Berlin, Heidelberg: Springer Berlin Heidelberg; 2012. pp. 17–23.
Yamak A, Bitar F, Karam P, Nemer G. Exclusive cardiac dysfunction in familial primary carnitine deficiency cases: a genotype–phenotype correlation. Clin Genet. 2007;72(1):59–62.
Schulze A, Lindner M, Kohlmüller D, Olgemöller K, Mayatepek E, Hoffmann GF. Expanded newborn screening for inborn errors of metabolism by electrospray ionization-tandem mass spectrometry: results, outcome, and implications. Pediatrics. 2003;111(6 Pt 1):1399–406.
Longo N, di Amat C, Pasquali M, editors. Disorders of carnitine transport and the carnitine cycle. American Journal of Medical Genetics Part C: seminars in Medical Genetics. Wiley Online Library; 2006.
Antunes MO, Scudeler TL. Hypertrophic cardiomyopathy. Int J Cardiol Heart Vasculature. 2020;27:100503.
Ino T, Sherwood WG, Benson LN, Wilson GJ, Freedom RM, Rowe RD. Cardiac manifestations in disorders of fat and carnitine metabolism in infancy. J Am Coll Cardiol. 1988;11(6):1301–8.
García-Vielma C, Lazalde-Córdova LG, Arzola-Hernández JC, González-Aceves EN, López-Zertuche H, Guzmán-Delgado NE, et al. Identification of variants in genes associated with hypertrophic cardiomyopathy in Mexican patients. Molecular genetics and genomics: MGG; 2023.
Deswal S, Bijarnia-Mahay S, Manocha V, Hara K, Shigematsu Y, Saxena R, et al. Primary Carnitine Deficiency - A Rare Treatable cause of cardiomyopathy and massive hepatomegaly. Indian J Pediatr. 2017;84(1):83–5.
Li FY, El-Hattab AW, Bawle EV, Boles RG, Schmitt ES, Scaglia F, et al. Molecular spectrum of SLC22A5 (OCTN2) gene mutations detected in 143 subjects evaluated for systemic carnitine deficiency. Hum Mutat. 2010;31(8):E1632–E51.
Dobrowolski SF, McKinney JT, Amat C, Giak Sim K, Wilcken B, Longo N. Validation of dye-binding/high‐resolution thermal denaturation for the identification of mutations in the SLC22A5 gene. Human Mutation. 2005;25(3):306 – 13.
Wang Y, Korman SH, Ye J, Gargus JJ, Gutman A, Taroni F, et al. Phenotype and genotype variation in primary carnitine deficiency. Genet Sci. 2001;3(6):387–92.
El-Hattab AW, Li F-Y, Shen J, Powell BR, Bawle EV, Adams DJ, et al. Maternal systemic primary carnitine deficiency uncovered by newborn screening: clinical, biochemical, and molecular aspects. Genet Sci. 2010;12(1):19–24.
Frigeni M, Balakrishnan B, Yin X, Calderon FR, Mao R, Pasquali M, et al. Functional and molecular studies in primary carnitine deficiency. Hum Mutat. 2017;38(12):1684–99.
Lee N-C, Tang NL-S, Chien Y-H, Chen C-A, Lin S-J, Chiu P-C, et al. Diagnoses of newborns and mothers with carnitine uptake defects through newborn screening. Mol Genet Metab. 2010;100(1):46–50.
Rose EC, di San Filippo CA, Ndukwe Erlingsson UC, Ardon O, Pasquali M, Longo N. Genotype–phenotype correlation in primary carnitine deficiency. Hum Mutat. 2012;33(1):118–23.
Rahbeeni Z, Vaz F, Al-Hussein K, Bucknall M, Ruiter J, Wanders R, et al. Identification of two novel mutations in OCTN2 from two Saudi patients with systemic carnitine deficiency. J Inherit Metab Dis. 2002;25:363–9.
Lamhonwah AM, Olpin SE, Pollitt RJ, Vianey-Saban C, Divry P, Guffon N, et al. Novel OCTN2 mutations: no genotype–phenotype correlations: early carnitine therapy prevents cardiomyopathy. Am J Med Genet. 2002;111(3):271–84.
Rasmussen J, Nielsen OW, Janzen N, Duno M, Køber L, Steuerwald U, et al. Carnitine levels in 26,462 individuals from the nationwide screening program for primary carnitine deficiency in the Faroe Islands. J Inherit Metab Dis. 2014;37:215–22.
Schimmenti LA, Crombez EA, Schwahn BC, Heese BA, Wood TC, Schroer RJ, et al. Expanded newborn screening identifies maternal primary carnitine deficiency. Mol Genet Metab. 2007;90(4):441–5.
Mutlu-Albayrak H, Bene J, Oflaz MB, Tanyalçın T, Çaksen H, Melegh B. Identification of SLC22A5 gene mutation in a family with carnitine uptake defect. Case Reports in Genetics. 2015;2015.
Acknowledgements
The authors wish to acknowledge the kind contribution of the family described herein. The Cardiogenetics Research Center, Rajaie Cardiovascular Medical and Research Center, Tehran, Iran, funded this research.
Funding
The authors received no specific funding for this research.
Author information
Authors and Affiliations
Contributions
SK designed the project and performed WES. AG evaluated the patients clinically and prepared the first version of manuscript. NN and SA performed wet lab. SG conducted docking and modeling studies. AS evaluated patients imaging and echocardiographic studies. MM confirmed the clinical finding of the patient and made complementary revision of the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study complies with the Declaration of Helsinki. Ethical approval was obtained from the Ethics Committees of Rajaie Cardiovascular Medical and Research Center, Iran University of Medical Sciences, Tehran, Iran (IR.RHC.REC.1400.005). Informed consent must have been obtained from a parent and/or legal guardian.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jolfayi, A.G., Naderi, N., Ghasemi, S. et al. A novel pathogenic variant in the carnitine transporter gene, SLC22A5, in association with metabolic carnitine deficiency and cardiomyopathy features. BMC Cardiovasc Disord 24, 1 (2024). https://doi.org/10.1186/s12872-023-03676-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12872-023-03676-z