Abstract
Background
The utilization of heterosis based on three-line system is an effective strategy in crop breeding. However, cloning and mechanism elucidation of restorer genes for cytoplasmic male sterility (CMS) in upland cotton have yet been realized.
Results
This research is based on CMS line 2074A with the cytoplasm from Gossypium harknessii (D2-2) and restorer line R186. The offspring of 2074A × R186 were used to conduct genetic analysis. The fertility mechanism of 2074A can be speculated to be governed by multiple genes, since neither the single gene model nor the double genes model could be used. The bulked segregant analysis (BSA) for (2074A × R186) F2 determined the genetic interval of restorer genes on a region of 4.30 Mb on chromosome D05 that contains 77 annotated genes. Four genes were identified as candidates for fertility restoration using the RNA-seq data of 2074A, 2074B, and R186. There are a number of large effect variants in the four genes between 2074A and R186 that could cause amino acid changes. Evolutionary analysis and identity analysis revealed that GH_D05G3183, GH_D05G3384, and GH_D05G3490 have high identity with their homologs in D2-2, respectively. Tissue differential expression analysis revealed that the genes GH_D05G3183, GH_D05G3384, and GH_D05G3490 were highly expressed in the buds of the line R186. The predicted results demonstrated that GH_D05G3183, GH_D05G3384 and GH_D05G3490 might interact with GH_A02G1295 to regulate orf610a in mitochondria.
Conclusion
Our study uncovered candidate genes for fertility restoration in the restorer line R186 and predicted the possible mechanism for restoring the male fertility in 2074A. This research provided valuable insight into the nucleoplasmic interactions.
Similar content being viewed by others
Background
Plant male sterility mainly includes three types: cytoplasmic male sterility (CMS), cytoplasmic nuclear interaction male sterility, and nuclear male sterility (GMS) [1]. CMS is the most convenient method to produce the population of male sterility in the commercial application of hybrids [2]. CMS gene is reported to be associated with chimeric mitochondrial orf [3]. Sterility of plants with CMS cytoplasm results from the influence of CMS proteins on mitochondria such as toxic effect [4, 5], burst of reactive oxygen species (ROS) [6], inducing the abnormal programmed cell death (PCD) [7] and retrograde regulating nuclear genes [8]. CMS-A2 [9], CMS-B1 [9], CMS-D2-2 [10], CMS-D8 [11] and CMS-AD1 [12] constitute main types of cotton CMS. CMS-D2-2 and CMS-D8 are the most valuable for production and are popular in cotton three-line hybrid breeding among many CMS types of cotton. The cytoplasm of Gossypium harknessii Brandegee (D2-2) was transferred into AD1 nuclear background, which led to the production of CMS-D2-2 [10]. Li [13, 14] constructed fosmid library of mitochondrial genome from D2-2 male sterile line 2074A and identified 28 ORFs specific to CMS 2074A by mitochondrial genome analysis.
Restorer of fertility (Rf) genes rescue the fertility of CMS cytoplasm plants by interacting with CMS gene products and alleviating or eliminating the adverse effects on mitochondria of CMS gene products at DNA [15], transcription [8], post-transcription [16], translation [17,18,19], post-translation [20], and metabolic [21] levels. The mechanisms of fertility restoration are diverse and the types of Rf genes are also not fixed. More than half of cloned restorer genes encode pentatricopeptide repeat (PPR) proteins [22]. There are also some reports that aldehyde dehydrogenase [21], acyl-carrier protein synthase (ACPS)-like domain containing protein [23], glycine-rich protein [24], peptidase-like protein [20], bHLH transcription factor [25] and transcription factors of the plant DREB1 family [8] are designated as Rf genes and can also restore the fertility of plants with CMS genes.
Although some reports found that multiple genes can rescue sterility of CMS-D2-2 cotton [26], Weaver [27] and Zhang [28] confirmed fertility of cotton plant with CMS-D2-2 cytoplasm is conditioned by a single dominance locus at sporophytical level. The gene located at the single dominant locus is designated as Rf1. CMS-D8 was bred by combining cytoplasm of Gossypium trilobum (DC) Skovst (D8) and AD1 nucleus [11]. A single dominant locus containing Rf2 gene can gametophytically dominate fertility of cotton plant with CMS-D8 cytoplasm [29]. The sterility of CMS-D8 cotton can be remedied by Rf1 and Rf2 while fertility restoration of CMS-D2-2 cotton only depends on the function of the Rf1 gene [30].
The tightly linked Rf1 and Rf2 are located in the 0.93 cM interval of same chromosome [28]. Each of the Rf genes is not linked to any morphological markers with known chromosomal locations in cotton [31]. A variety of molecular markers are used for genetic mapping of Rf genes in cotton, such as RAPD [32,33,34], SSR [32, 34], STS [34, 35], AFLP [36], SNP [37, 38] and InDel [39, 40]. Liu [32] mapped the genetic interval of Rf1 gene on the long arm of chromosome 4 (A subgenome). The mapping interval of Rf1 locus was delimited to 100 kb and was between 081-05 K and 052-01N BAC clones [34]. Wang [30] found that Rf1 and Rf2 were delimited to a 1.4 cM genetic distance on chromosome D05 with assistant of four SSR markers. The locus of Rf gene was located on 1.35 Mb of chrD05 by the technology of BSA with SLAF-seq [37]. Feng [38] determined the location of Rf2 on a 1.48 Mb interval of chromosome D05, based on BSA with high-throughput SNP genotyping. While much efforts has been devoted to the genetic mapping of Rf loci, valuable Rf candidates have rarely been identified in cotton. Upgrading of sequencing technology, reduction of costs, and continuous release of high-quality cotton reference genomes [41,42,43,44] provide better opportunities for the localization and cloning of cotton Rf genes.
In this research, the key Rf candidates that dominate the fertility of CMS-D2-2 cotton were mapped by BSA and analyzed for genetics, expression, sequence similarity, and evolution. First, strong restorer lines were screened with a fertility survey of F1 hybrids produced by crossing the CMS-D2-2 line with the restorer line. Genetic analysis of the F2 and BC1F1 populations originating from the CMS-D2-2 line and the strong restorer lines proved that the fertility of CMS-D2-2 cotton is controlled neither by a single locus nor by two loci. Then, the genetic interval of the Rf genes was determined using the BSA technology. Next, RNA-seq and sequence variation information supported the identification of four Rf candidate genes. The homologous proteins of four candidate proteins in Gossypium herbaceum (A1) [42], Gossypium arboreum (A2) [42], G. hirsutum (AD1) [43] and all D genome cotton [45,46,47] were used for evolutionary and sequence similarity analysis. GH_D05G3183, GH_D05G3384, and GH_D05G3490 were identified as candidate fertility restorer genes by evolutionary and sequence similarity analysis. Real-time quantitative PCR (qRT-PCR) proved that GH_D05G3183, GH_D05G3384, and GH_D05G3490 genes were highly expressed in R186 buds. Protein interaction analysis revealed that GH_D05G3183, GH_D05G3384, and GH_D05G3490 may interact with GH_A02G1295 to regulate orf160a and thus restore male fertility in cotton. From the above evidence, we can conclude that GH_D05G3183, GH_D05G3384, and GH_D05G3490 genes are likely Rf genes of CMS-D2-2. This research lays the foundation for the subsequent identification of restoration genes, the elucidation of restoration mechanisms, and the breeding of strong restorer lines with excellent agronomic traits.
Results
Genetic analysis of Rf loci
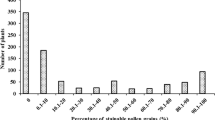
Rf loci can rescue the fertility of 2074A with CMS-D2-2 cytoplasm sporophytically [28]. The fertility surveys of (2074A × R186) F2 in three environments and the BC1F1 population (2074A × (2074A × R186) in two environments were conducted (Additional file 3, Supplemental Table 1). First, we assumed that the Rf locus is single-gene dominant inheritance. The chi-square test results of F2 and BC1F1 population derived from R186 could not unanimously prove this hypothesis (Table 1). Then, we assumed that the fertility of 2074A was controlled by two dominant loci. The ratios of F2 genotypes of double genes interaction including no interaction, dominant complementary effect, inhibiting effect, epistatic recessiveness, epistatic dominance, duplicate effect and additive effect are 9:3:3:1, 9:7, 13:3, 9:3:4, 12:3:1, 15:1 and 9:6:1, respectively. Because the fertility data distribution trend of two F2 population is more similar to the model of dominant complementary effect, epistatic recessiveness and duplicate effect, the fertility survey results of two F2 and two BC1F1 population were tested by chi-square, based on these double genes interaction model (Additional file 3, Supplemental Tables 2–4). However, the genetic model of Rf genes from R186 failed to fit any model of dominant complementary effect, epistatic recessiveness, and duplicate effect. The analytical results of this research proved that the sterility in 2074A with CMS-D2-2 cytoplasm can be remedied by multiple genes, neither single nor double.
Evaluation of whole genome resequencing data and BSA mapping of Rf genes
Two parent lines 2074A and R186, together with extremely fertile and extremely sterile bulks of (2074A × R186) F2 were sequenced on the Illumina HiSeq platform for BSA. A total of 850,956,496 reads and 252.6 Gb data were obtained (Additional file 4, Supplemental Table 5). The average GC content, Q30, genome coverage and coverage depth were 36.10%, 93.16%, 94.38% and 24.47 × in the sequencing results, respectively. A total of 173,351,714 reads and 51.40 Gb data were generated for 2074A with average GC content of 36.21%, a Q30 value of 92.70%, genome coverage of 94.86% and coverage depth of 20.13 × (Additional file 4, Supplemental Table 5). The sequencing results showed that R186 possessed 174,143,920 reads, average GC content of 36.32%, Q30 of 93.01%, coverage depth of 20.34 × and genome coverage of 93.62% (Additional file 4, Supplemental Table 5). On the other hand, 240,953,554 and 262,507,308 reads were gained for extremely fertile and extremely sterile bulks of (2074A × R186) F2, respectively, with Q30 values of 93.62% and 93.31%, average GC content of 35.62% and 36.26%, coverage depth of 27.51 × and 29.91 × , and genome coverage of 94.07% and 94.95% (Additional file 4, Supplemental Table 5). Those reads were aligned to the reference AD1 genome [43]. A total of 805,401 SNPs and 80,717 InDels were obtained from the two bulks of (2074A × R186) F2 (Additional file 4, Supplemental Table 6). D05 chromosome has the highest number and density of SNPs and InDels among the variants detected in two bulks from (2074A × R186) F2 (Fig. 1a, Table 2). Both A/G and C/T type SNPs accounted for the highest proportions among the SNPs detected in two bulks from (2074A × R186) F2 (Fig. 1b). The InDels with length of 1 bp accounted for the largest proportion among the InDels detected in two bulks (Fig. 1c).
The algorithms of Δ(SNP-index) and euclidean distance (ED) algorithms were used to locate intervals of Rf genes in BSA. The Rf genes from R186 were delimited on 34,943,848–35,280,626 bp, 37,694,536–38,093,258 bp, 38,227,690–38,918,070 bp, 43,648,410–43,742,747 bp, 44,220,658–44,835,843 bp, 45,653,643–45,811,858 bp, 46,818,876–47,801,436 bp, 49,315,266–50,052,334 bp, 51,046,570–51,244,308 bp and 51,557,591–51,715,302 bp of chromosome D05 using Δ(SNP-index) algorithm (Fig. 2a, Additional file 5, Supplemental Table 7). ED algorithm determined the position of Rf genes on 34,937,629–35,327,222 bp, 37,656,846–38,088,236 bp, 38,266,076–38,923,764 bp, 43,619,667–43,777,276 bp, 44,162,548–44,834,728 bp, 45,654,955–45,821,873 bp, 46,834,875–47,798,159 bp, 49,311,406–50,065,365 bp, 51,034,546–51,244,603 bp and 51,563,873–51,719,761 bp of chromosome D05 (Fig. 2b, Additional file 5, Supplemental Table 8). Eventually, the Rf genes from R186 were mapped in the 4.30 Mb interval of chromosome D05, based on the Δ(SNP-index) and ED algorithms, including intervals 34,943,848–35,280,626 bp, 37,694,536–38,088,236 bp, 38,266,076–38,918,070 bp, 43,648,410–43,742,747 bp, 44,220,658–44,834,728 bp, 45,654,955–45,811,858 bp, 46,834,875–47,798,159 bp, 49,315,266–50,052,334 bp, 51,046,570–51,244,308 bp and 51,563,873–51,715,302 bp on chromosome D05, which contain a total of 77 genes (Table 3).
Go annotation of genes in the association interval
Fourty-two genes from seventy-seven genes located in BSA interval were identified through gene ontology (GO) analysis (Fig. 3a). Twenty-eight, thirty-seven, and nine genes, respectively, are involved in biological process, molecular function, and cellular component. The significantly enriched molecular function mainly includes binding, molecular function regulator, catalytic activity, structural molecule activity and transporter activity (Fig. 3a). The purine ribonucleoside triphosphate binding genes were speculated to be involved in male fertility, which includes GH_D05G3176, GH_D05G3177, GH_D05G3263, GH_D05G3269, GH_D05G3270, GH_D05G3273, GH_D05G3328, GH_D05G3386, GH_D05G3461, GH_D05G3467, GH_D05G3468, GH_D05G3469 and GH_D05G3491 genes (Fig. 3b). The Rf proteins, which regulate male fertility at the post-transcriptional level, usually modulate the stability of the abortive gene mRNA by binding to it. The purine ribonucleoside binding and ribonucleoside binding are molecular functions that need to be focused, including GH_D05G3176, GH_D05G3177, GH_D05G3263, GH_D05G3269, GH_D05G3270, GH_D05G3273, GH_D05G3328, GH_D05G3386, GH_D05G3461, GH_D05G3467, GH_D05G3468, GH_D05G3469 and GH_D05G3491 (Fig. 3b). In addition, the relationship between other types of molecular function and male fertility needs to be clarified in follow-up studies.
Determination of Rf candidate genes by expression analysis
More than half of the reported fertility restoration genes for cytoplasmic male sterility belong to the PPR genes. Only one PPR gene, GH_D05G3465 gene, was identified in the 4.30 Mb interval, based on the reference AD1 genome [43]. However, the identity between GH_D05G3465 and homologous protein in D2-2 was only 22.96% (Additional file 6, Supplemental Table 9). No significant difference in GH_D05G3465 gene expression levels was detected in buds of 2074A, 2074B and R186 (Additional file 6, Supplemental Table 9). Therefore, GH_D05G3465 was hardly considered as Rf candidate gene.
In order to identify Rf candidate genes in the mapping interval, RNA-seq of 2074A, R186, E5903, R144 and R245 buds with diameters of 0–1.5 mm (the earlier stage of pollen abortion) and 1.5–9.0 mm (stage of pollen abortion) were performed. The total DEGs of R186_2 vs 2074A_2 and R186_2 vs 2074B_2 were 12,166 and 21,483 while the up-regulated DEGs in restorer lines were 8191 and 10,798 in R186_2 vs 2074A_2 and R186_2 vs 2074B_2, respectively (Fig. 4a-b). The VENN diagram revealed that the GH_D05G3183, GH_D05G3265, GH_D05G3384, GH_D05G3388 and GH_D05G3490 genes from the BSA interval had significantly higher expression levels in R186 than 2074A and 2074B (Fig. 4c). Figure 4d showed that the expression levels of the genes GH_D05G3183, GH_D05G3265, GH_D05G3384 and GH_D05G3490 were highest in the abortive stage buds of R186. The heatmap analysis found that the expression of GH_D05G3183, GH_D05G3265, GH_D05G3384 and GH_D05G3490 genes in buds with stage of pollen abortion of E5903, R144 and R245 were higher than 2074A and 2074B (Fig. 4e).
Identification of CMS-Rf candidate genes based on RNA-seq. a Volcanic map of R186_2 vs. 2074A_2. b Volcanic map of R186_2 vs. 2074B_2. c Determination of Rf candidate genes in BSA interval by venn diagram, based on gene expression in 2074A, 2074B and R186. d The heatmap for five candidate genes in the buds of 2074A, 2074B, and R186 at the early abortion stage and abortion stage. e The heatmap for four candidate genes in the buds of 2074A, 2074B, E5903, R144, and R245 at the early abortion stage and abortion stage. R186_2, the R186 buds with diameters of 1.5–9.0 mm (the stage of pollen abortion). 2074A_2, the 2074A buds with diameters of 1.5–9.0 mm (the stage of pollen abortion). 2074B_2, the 2074B buds with diameters of 1.5–9.0 mm (the stage of pollen abortion)
Sequence, evolutionary and tissue differential expression analysis of candidate genes
Four candidate genes were screened by BSA and RNA-seq analysis, including GH_D05G3183, GH_D05G3265, GH_D05G3384 and GH_D05G3490. Sequence analysis revealed that there are two SNPs in the exon of GH_D05G3183, including SNP_D05_35158622 and SNP_D05_35160174. Nucleotides of 2074A (sterile bulk) and R186 (fertile bulk) at SNP_D05_35158622 are T and A, respectively, which changes the amino acid from K to M (Table 4). Nucleotides of 2074A (sterile bulk) and R186 (fertile bulk) at SNP_D05_35160174 are G and C, respectively, which changes the amino acid from F to L (Table 4). Furthermore, there are two, seven and two SNPs that can cause changes in amino acids in exons of GH_D05G3265, GH_D05G3384 and GH_D05G3490 genes, respectively (Table 4). Nucleotides of 2074A and sterile bulk at those large effect variants (LEVs) are the same as the reference genome while R186 and fertile bulk are alternative nucleotides (Table 4). The index of all LEVs for GH_D05G3183, GH_D05G3265, GH_D05G3384 and GH_D05G3490 genes in fertile bulk are 1 while those in sterile bulk are 0 (Table 4).
The phylogenetic analysis revealed that the evolutionary relationship between GH_D05G3183, GH_D05G3384 and their homologues from D2-2 are all relatively close while the evolutionary relationship between GH_D05G3265, GH_D05G3490 and their homologues from D2-2 are relatively distant (Fig. 5). The analysis of sequence identity found that the identity between GH_D05G3183, GH_D05G3384, GH_D05G3490 and their homologues from D2-2 are high while the identity between GH_D05G3265 and its homologue from D2-2 is only 33.76% (Fig. 5). GH_D05G3183, GH_D05G3384 and GH_D05G3490 genes were further identified as candidate genes for fertility restoration based on phylogenetic and sequence identity analysis.
The expression characteristics of the genes GH_D05G3183, GH_D05G3384 and GH_D05G3490 in multiple organs of the restorer line R186 were further analyzed by qRT-PCR. GH_D05G3183 gene expression in buds was 6.17, 8.55 and 1.79 times higher than in root, stem and leaf, respectively (Fig. 6a). GH_D05G3384 gene expression in bud was 27.57 times higher than in roots while GH_D05G3384 gene expression was not detected in stem and leaf (Fig. 6b). Although GH_D05G3490 gene expression was higher in leaf than in bud, its expression was higher in bud than in root and stem (Fig. 6c).
Eventually, GH_D05G3183, GH_D05G3384, and GH_D05G3490 were identified as Rf candidate genes. The GH_D05G3183 gene, which encodes purple acid phosphatase 3, was annotated with acid phosphatase activity. The GH_D05G3384 gene encodes the putative protein NRT1/PTR FAMILY 2.14 with transmembrane transporter activity. DNA-directed RNA polymerases II, IV and V subunit 8B encoded by the GH_D05G3490 gene has DNA-directed RNA polymerase activity (Table 5).
Discussion
Elite CMS lines and restorer lines are the important parts of three-line breeding
As the male parent of the hybrid F1, the restoring power and agronomic traits of the restorer line have a significant impact on the hybrid F1. Screening of restorer lines with strong restoring power can contribute to the utilization of heterosis. R186 (Additional file 1, Supplemental Fig. 1b), a strong restorer line in this research, was selected by analyzing the fertility of 16 (2074A × R) F1 and possesses the strongest restoring power among 16 (2074A × R) F1 (Additional file 7, Supplemental Table 10). Although F2 derived from female parent upland cotton CMS line 3096 and male parent restorer line 866 had been used to locate fertility restorer genes by BSA [37], type of abortion cytoplasm of sterile lines between 3096 and 2074A are completely different. The CMS line (A) with CMS-D8 cytoplasm in Feng [38] and the CMS line (6001A) with Gossypium thurberi cytoplasm [26] are also different from 2074A in this research. In addition to the sterile line, another important difference is the restorer line. R186 is completely different from 866R [37], restorer line (R) [38] and the restorer line (7R13) [26].
D2-2 CMS fertility restorer genes are located on chromosome D05
Many types of molecular markers, such as SNP [37, 38], InDel [39, 40], RAPD [32,33,34], SSR [32, 34], STS [34, 35], and AFLP [36] are effective tools for genetic mapping of fertility restoration genes. As sequencing cost has come down and technology has been upgraded, SNP and InDel have been the most popular molecular markers. In this research, the genetic location of Rf genes from R186 was determined by BSA, based on 805,401 SNPs and 80,717 InDels (Additional file 4, Supplemental Table 6), respectively. The long arm of chromosome 4 (A subgenome) was determined to be the genetic location of Rf1 [32]. However, the Rf genes were considered to be located on chromosome D, since the abortive cytoplasm was derived from D2-2. Wang [30] delimited Rf1 and Rf2 on chromosome D05 with the help of four SSR markers. Zhao [37] found that the fertility of upland cotton CMS line 3096 was dominated by the locus of Rf genes located on 1.35 Mb of chrD05. The Rf genes dominating sterile line (A) with CMS-D8 cytoplasm were mapped on a 1.48 Mb interval of chromosome D05 [38]. The Rf genes for CMS line (6001A) derived from the crossing progenies of G. thurberi (D1) and AD1 were located in the interval of 2.05 Mb (53,632,812–55,682,586 bp) [26]. In this research, the Rf genes from R186 was delimited in a 4.30 Mb interval of chromosome D05. From our BSA results, it can be determined that the Rf genes for 2074A were mapped on chromosome D05, which was consistent with the location of the Rf genes for CMS lines with AD1, D1 and D8 abortive cytoplasm.
The fertility of 2074A is controlled by multiple nuclear genes
The fertility of most reported cytoplasmic male sterile lines is controlled by single gene in plants [8, 24, 48–52,] or double [16, 18, 53,54,55,56]. However, the fertility of CMS-Charrua (C) maize and Triticum timopheevii (T)-type CMS wheat are dominated by multiple genes [25, 57]. 2074A, a cotton cytoplasmic male sterile line, possesses the nucleus of AD1 and the abortive cytoplasm of D2-2. Weaver [27] and Zhang [28] found that there is one restorer gene in D2R while Gao [26] believed that sterility of CMS-D2-2 cotton can be rescued by multiple genes. In this research, chi-square analysis results showed that multiple nuclear genes dominate the fertility of 2074A. The discovery is consistent with the views of Gao [26]. Zhang [58] revealed that orf610a can lead to excessive accumulation of reactive oxygen species, reduction in ATP content and inhibition of cellular growth of yeast and abnormal development of male reproductive organs in Arabidopsis. The qRT-PCR result revealed that the expression of orf610a in R186 buds with abortive cytoplasm was extremely significantly lower than that of 2074A, suggesting that the Rf genes may restore male fertility by reducing the expression level of orf610a in mitochondria (Additional file 2, Supplemental Fig. 2a). The cytoplasm of 2074B is normal, so the expression of orf610a in 2074B was almost undetectable (Additional file 2, Supplemental Fig. 2a). To investigate the potential mechanism of the three fertility restoration genes affecting male fertility, the interaction between the three fertility restoration genes and orf610a was analyzed by STRING V11.5 [59]. Prediction of protein interaction showed that GH_D05G3183, GH_D05G3384 and GH_D05G3490 may co-regulate orf610a in mitochondria by interacting with GH_A02G1295, thereby regulating male fertility of 2074A (Additional file 2, Supplemental Fig. 2b). We speculated that the genes GH_D05G3183, GH_D05G3384 and GH_D05G3490 co-regulated the fertility restoration of D2-2 abortion. Follow-up works will focus on simultaneous silencing of those genes to validate the relationship between GH_D05G3183, GH_D05G3384 and GH_D05G3490 genes and fertility restoration.
The relationship between the PPR genes and 2074A fertility restoration
PPR genes, one of the largest gene families in land plants, function by targeting mitochondria or chloroplast, binding, editing, and processing organelle transcripts [60]. As Rf genes, PPR genes can restore the fertility of CMS-Boro II (BT) rice [53], CMS-Honglian (HL) rice [16, 55], CMS-wild-abortive (WA) rice [54], Triticum timopheevii (T)-type CMS wheat [57], Ogura (ogu) CMS oilseed rape [49, 50], Polima (pol) CMS oilseed rape [56], Kosena (kos) CMS radish [18], CMS-NJCMS1A soybean [52], Shahdara (Sha)-CMS Arabidopsis [51] and CMS-pcf petunia [48] at multiple different levels of regulation. In this research, only one PPR gene, GH_D05G3465, is localized in a 4.30 Mb interval on chromosome D05. However, the identity between GH_D05G3465 protein and its homologous proteins in D2-2 is low (Additional file 6, Supplemental Table 9). The expression of GH_D05G3465 gene were not significantly different in 2074A, 2074B and R186 (Additional file 6, Supplemental Table 9). Although more than half of the reported Rf proteins belong to the PPR family, there are also many non-PPR Rf proteins in plants such as ACPS-like domain containing protein [23], glycine-rich protein [24], aldehyde dehydrogenase [21], bHLH transcription factor [25], transcription factors of the plant DREB1 family [8] and peptidase-like protein [20]. It can be determined that the types of CMS restorer genes are diverse. Based on the results of this research, valuable PPR fertility restoration candidate genes could not be discovered by BSA. Rf gene of 2074A may not be limited to PPR genes.
The characteristics of CMS fertility restoration genes
The primary sequences of the proteins encoded by Rf genes are usually different between the sterile line and the restorer line [25]. Most Rf genes restore the fertility of CMS lines by positive regulation and usually have high expression levels in the restorer lines but low expression in the sterile lines [16, 18, 20, 21, 24, 25, 48,49,50, 53,54,55,56,57]. The expression level of the Rf genes in the restorer line should be significantly higher than that in the maintainer line, since the maintainer line does not contain the Rf genes. In this research, GH_D05G3183, GH_D05G3265, GH_D05G3384 and GH_D05G3490 genes with LEVs were most highly expressed in abortive buds of R186 and were screened as Rf candidate genes from 77 genes located in the 4.30 Mb interval of chromosome D05 with the assistance of R186, 2074B and 2074A bud transcriptome data (Fig. 4). Since the abortive cytoplasm of 2074A originated from D2-2, the Rf genes should exist in D2-2 nuclear genome. The evolutionary relationship or identity of Rf proteins from AD1 and D2-2 should be high. In the present study, GH_D05G3183, GH_D05G3265, GH_D05G3384 and GH_D05G3490 genes were identified as Rf candidate genes while GH_D05G3265 gene was ruled out as Rf candidate gene, based on phylogenetic and sequence identity analysis (Fig. 5). Restorer genes are usually highly expressed in the stamens [8, 56]. In this work, GH_D05G3183, GH_D05G3384 and GH_D05G3490 genes were retained by qRT-PCR as a result of the high expression level of the 3 genes in buds (Fig. 6). The mechanism of GH_D05G3183, GH_D05G3384 and GH_D05G3490 genes affecting the male fertility of 2074A and the mining of other Rf genes that control 2074A male fertility based on the whole genome sequencing of R186 will be the focus of follow-up research.
Conclusions
In the present study, genetic analysis revealed that male fertility in 2074A could be regulated by multiple Rf genes. The Rf loci were localized in a 4.3 Mb interval of chromosome D05. The genes GH_D05G3183, GH_D05G3384 and GH_D05G3490 were identified as Rf candidate genes based on RNA-seq, sequence and evolutionary analyses. Protein interaction analysis revealed that GH_D05G3183, GH_D05G3384 and GH_D05G3490 might restore male fertility in 2074A by co-regulating orf610a in mitochondria. Our study laid a foundation for exploring the Rf genes in D2-2 cytoplasmic male sterility and clarifying the mechanism of the Rf genes.
Methods
Plant materials and growth conditions
2074A, a cotton CMS line, possesses G. harknessii Brandegee CMS-D2-2 cytoplasm originated from DES-HAMS 277 and AD1 nucleus with no Rf genes (Additional file 1, Supplemental Fig. 1a) [10, 13]. R186 (Additional file 1, Supplemental Fig. 1b) was selected as strong restorer line through the male fertility identification of F1 produced by the hybrid of CMS-D2-2 line and restorer lines from 16 restorer lines (Additional file 7, Supplemental Table 10) in the summer of 2019 at Hejian Guoxin Cotton Base (Cangzhou City, China) (38°38′N, 116°13′E).
The F2 population derived from the 2074A and R186 hybrids, together with the BC1F1 population derived from the 2074A and R186 parents, was used to perform a genetic analysis of the restoring genes in cotton. (2074A × R186) F2 was planted in the winter of 2019 at Sanya Base of Cotton Research Institute of Chinese Academy of Agricultural Sciences (Sanya, China) (18°34′N, 109°65′E) and in the summer of 2020, 2021 at Hejian Guoxin Cotton Base. 2074A × (2074A × R186) was planted in the winter of 2019 at Sanya Base and in the summer of 2020 at Hejian Guoxin Cotton Base.
(2074A × R186) F2:3 was planted in the summer of 2020 at Hejian Guoxin Cotton Base. The F2:3 population, together with (2074A × R186) F2 planted in the winter of 2019 at Sanya Base, was used for fertility survey and BSA sampling. 2074A and R186 were planted in the summer of 2019 at Hejian Guoxin Cotton Base and used for RNA-seq sampling.
Fertility investigation and plant sampling
The morphological standard of flower fertility is divided into three levels, such as fully fertile (full pollen), partially fertile (less pollen) and completely sterile (no pollen). There are also three types for fertility of plant individuals including fully fertile individuals (all flowers of individual are fertile), partially fertile individuals (individual possesses fertile and sterile flowers) and completely sterile individuals (all flowers of individual are sterile).
The fresh leaves of 19 extremely fertile individuals and 30 extremely sterile individuals from (2074A × R186) F2 population were collected for DNA extraction and BSA, based on the fertility survey results of (2074A × R186) F2 and (2074A × R186) F2:3 population. The fertility of the F2:3 lines from the extremely fertile F2 individuals must be fully fertile. The DNA of the extremely fertile and extremely sterile individuals were used to construct of the BSA extremely fertile and sterile bulks, respectively. Fresh leaves of 2074A and R186 were also used to extract DNA and were taken as the parents for BSA.
The buds of 2074A, R186, E5903 (restorer line), R144 (restorer line) and R245 (restorer line) with diameters of 0–1.5 mm (the earlier stage of pollen abortion) and 1.5–9 mm (stage of pollen abortion) [61] were collected in three biological replicates for RNA-seq.
Whole genome sequencing and BSA
The DNA of cotton leaves including extremely fertile and extremely sterile individuals of (2074A × R186) F2, 2074A and R186 was isolated by cetyltrimethylammonium bromide (CTAB) method [62]. When the qualified sample DNA was prepared, the library was constructed in strict accordance with the protocol provided by the kit of NEBNext® Ultra™ II DNA Library Prep Kit for Illumina® (NEB). Sequencing could be performed on the Illumina HiSeq platform when the library quality met the requirements. The library quality was tested as follows: First, Qubit3.0 was used for preliminary quantification. Then, the insert size of the library was detected using Agilent 2100. The next experiment could only be performed after the insert size met the expectations and no connector contamination was present. Last, a qualified library whose effective concentration was more than 2 nM was obtained by accurately quantifying the effective concentration of the library using the German ANALYTIKJENA (Jena) QTOWER real-time fluorescent quantitative PCR instrument (German). The library was pooled, and paired-end 150 bp (PE150) sequencing was performed on the Illumina HiSeq platform. The raw data obtained by sequencing was transformed into clean data by a three-step filter. (i) Linker sequence contained in reads was removed using ‘cutadapt’ software (1.13); (ii) Low-quality bases in reads were eliminated by ‘trimmomatic’ software (0.36); (iii) The length of reads must be greater than 50 bp. The MEM algorithm of ‘BWA’ software (0.7.15-r1140) was used to align clean reads to the reference AD1 genome [43] and the result file was output in SAM format. The SAM format file was converted to BAM format with ‘samtools’ software (1.3.1). The final BAM file could be used for statistics of coverage and depth and variant calling after reads in the BAM file being sorted by SortSam of Picard tool (1.91). The HaplotypeCaller module in the GATK (3.7) software package was used to generate gvcf files for each sample, and then variants detection (SNPs and InDels) of all samples were performed using the GenotypeGVCFs module. The variation information output by GATK was stored in a file in vcf format, which contains all the variations present between the sample and the reference AD1 genome. In order to analyze the variants between samples, the original mutations were screened based on the following criterion: (i) The sequencing depth of the parent is not less than 5; (ii) The sequencing depth of the bulks is not less than 10; (iii) The parents are all homozygous and there are polymorphisms among the parents; (iv) The SNP-index value of bulks cannot be more than 0.8 or less than 0.2 at the same time. ANNOVAR software (2016Feb1) [63] was used to annotate variants and predict the effect of variants on gene function. There are two algorithms suitable for the location of Rf locus in BSA, including Δ(SNP-index) and ED. The ED algorithm, also called MMAPPR, calculates the frequency distance of each mutant between different bulks, and uses the distance difference to reflect the linkage strength between marker and target interval [64]. DeepBSA software was used to calculate the Δ(SNP-index) of each mutation site and evaluate ED between mutation sites based on default parameters [65]. The LEVs in the candidate interval were focused on. LEVs are mutations that cause changes in the protein sequence, including non-synonymous SNP, frameshift InDel, non-frameshift InDel, stop-gain SNP/InDel stop-loss SNP/InDel, and splicing.
RNA-seq analysis
The total RNA was extracted from buds by CTAB-ammonium acetate method with slight modifications [66]. The detection of RNA samples mainly includes four methods: (i) The contamination and degradation of RNA were monitored on 1% agarose gels; (ii) RNA purity (OD260/280) was checked by the NanoPhotometer® spectrophotometer (IMPLEN, CA, USA); (iii) The concentration of RNA was determined by Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, CA, USA); (iv) The RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used to assess RNA integrity. A total amount of 3 µg RNA each sample was prepared as input material for the RNA-seq. Sequencing libraries were constructed with the assistance of NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following manufacturer’s recommendations. After the library was constructed, Qubit2.0 was used for preliminary quantification, and the library was diluted to 1 ng·ul−1. The inserting size of library was then evaluated using the Agilent Bioanalyzer 2100 system. The Q-PCR method was used to accurately quantify the effective concentration of the library (the effective concentration of the library > 2 nM) to ensure the quality of the library after insert size of library meeting expectations. Sequencing was performed on an Illumina Hiseq platform when different libraries were pooled according to the requirements of library effective concentration and the target off-machine data volume. Raw data obtained from HiSeq sequencing was transformed into clean data by a three-step data processing. (i) Removing reads with adapters; (ii) Removing reads with more than 10% uncertain bases; (iii) Removing low-quality reads. At the same time, the Q20, Q30, and GC content of the clean data were calculated. High-quality clean data was the basis for all the downstream analyses. STAR was used to align paired-end clean reads to the reference cotton genome [43]. The length of the gene and reads count mapped to this gene were the foundation of calculating expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs sequenced (FPKM) of per gene [67]. DESeq R package (1.18.0) was used to perform differential expression analysis of genes, and the threshold for significantly differential expression of genes is P-value of 0.05 and |log2(Fold change)|≥ 1.
Application of public transcriptome data
RNA-seq data of the maintainer line 2074B were referred to Nie [68].
Functional enrichment analysis
Functional enrichment analysis of candidate genes was performed at online website GO (http://www.geneontology.org/).
qRT-PCR analysis
RNA from root, stem, leaves and the buds with diameters of 1.5–9 mm of R186 was extracted by the CTAB method mentioned above [66], which was used for expression verification of candidate genes.
PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) was used to complete the reverse transcription of RNA. The experiment of qRT-PCR was executed with the assistance of PrimeScript™ RT reagent Kit (Perfect Real Time). The primers used in the experiments are shown in Additional file 8, and Supplemental Table 11. Three replicates were set for each sample and GhUBQ7 (GenBank accession number: DQ116441) was the internal reference gene in all qRT-PCR experiments. 2−ΔΔCt method was adopted to calculate the relative expression level of each gene [69].
Homology, evolutionary and protein interaction analysis
The homologous proteins sequence of the candidate proteins in A1, A2, AD1 and all D genome cotton were searched and downloaded from the CottonGen (https://www.cottongen.org) [70]. The identity between homologous proteins of different cotton species was calculated using the DNAMAN software. Evolutionary analysis between homologous proteins was performed with the assistance of the MEGA 7.0.26 software.
Protein interaction analysis of candidate proteins with reported mitochondrial abortive orf610a in cotton [58] were predicted based on their homologous proteins in Gossypium raimondii L. using STRING V11.5 [59]. The minimum required interaction score was set to 0.150.
Statement
Complying with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora, we confirm that the plant materials used in the present study does not involve any species at risk of extinction. All methods performed are in accordance with the relevant institutional, national, and international guidelines and legislation.
The cytoplasmic male sterile lines DES-HAMS277, DES-HAMS16 and the restorer lines DES-HAF277 and DES-HAF16 are originated from Gossypium harknessii and were released since 1970s [10]. All these lines were introduced into China in 1980 by Dr. Tianjue Zuo, and the seeds were divided into two parts. One was sent to the Institute of Cash Crops, Hubei Academy of Agricultural Sciences, Wuhan 430,064, Hubei, China, and the other to the Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang 455,000, Henan, China. Using these original allo-cytoplasm lines serial new lines such as 2074A, R186 were developed in our lab and issued to China Agricultural University since 2005 [13, 14, 68, 71]. We confirm that all the introduced processes have been authorized.
Availability of data and materials
Sequencing data have been uploaded to GSA of the NGDC website (https://ngdc.cncb.ac.cn/sso/login?service=https://ngdc.cncb.ac.cn/gsa/login). The accession numbers of GSA are CRA007637 and CRA007640, respectively: CRA007637 is for BSA data (https://ngdc.cncb.ac.cn/gsa/s/E7jGRJ7p), and CRA007640 is for RNA-seq data (https://ngdc.cncb.ac.cn/gsa/s/64zE2c3u). R186, 2074A, (2074A × R186) F2 in the manuscript are accordingly recorded using their line numbers: 19A1114, 19A1117, 19A1102, respectively.
Abbreviations
- CMS:
-
Cytoplasmic male sterility
- BSA:
-
Bulked segregant analysis
- GMS:
-
Genic male sterility
- ROS:
-
Reactive oxygen species
- Rf:
-
Restorer of fertility
- PPR:
-
Pentatricopeptide repeat
- ED:
-
Euclidean distance
- LEVs:
-
Large effect variants
- FPKM:
-
Expected number of Fragments Per Kilobase of transcript sequence per Millions base pairs
- GO:
-
Gene ontology
- qRT-PCR:
-
Real-time quantitative PCR
References
Wan XY, Wu SW, Li ZW, Dong ZY, An XL, Ma B, et al. Maize genic male-sterility genes and their applications in hybrid breeding: progress and perspectives. Mol Plant. 2019;12(3):321–42.
Chase CD. Genetically engineered cytoplasmic male sterility. Trends Plant Sci. 2006;11(1):7–9.
Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3(5):175–80.
Chen LT, Liu YG. Male sterility and fertility restoration in crops. Annu Rev Plant Biol. 2014;65:579–606.
Dewey RE, Timothy DH, Levings CS. A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc Natl Acad Sci USA. 1987;84(15):5374–8.
de Souza A, Wang JZ, Dehesh K. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu Rev Plant Biol. 2017;68:85–108.
Luo DP, Xu H, Liu ZL, Guo JX, Li HY, Chen LT, et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat Genet. 2013;45(5):573–7.
Xiao SL, Zang J, Pei YR, Liu J, Liu J, Song W, et al. Activation of mitochondrial orf355 gene expression by a nuclear-encoded DREB transcription factor causes cytoplasmic male sterility in maize. Mol Plant. 2020;13(9):1270–83.
Meyer VG, Meyer JR. Cytoplasmically controlled male sterility in cotton. Crop Sci. 1965;5:444–8.
Meyer VG. Male sterility from Gossypium harknessii. Heredity. 1975;66(1):23–7.
Zhang J, Turley RB, Stewart JM. Comparative analysis of gene expression between CMS-D8 restored plants and normal non-restoring fertile plants in cotton by differential display. Plant Cell Rep. 2008;27(3):553–61.
Jia ZC. Breeding of cotton male sterile line 104–7A and three-line matching. China Cotton. 1990;17(6):11.
Li SS, Chen ZW, Zhao N, Wang YM, Nie HS, Hua JP. The comparison of four mitochondrial genomes reveals cytoplasmic male sterility candidate genes in cotton. BMC Genomics. 2018;19(1):775.
Li SS, Liu GZ, Chen ZW, Wang YM, Li PB, Hua JP. Construction and initial analysis of five Fosmid libraries of mitochondrial genomes of cotton (Gossypium). Chinese Sci Bull. 2013;58(36):4608–15.
Mackenzie SA, Chase CD. Fertility restoration is associated with loss of a portion of the mitochondrial genome in cytoplasmic male-sterile common bean. Plant Cell. 1990;2(9):905–12.
Hu J, Wang K, Huang WC, Liu G, Gao Y, Wang JM, et al. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell. 2012;24(1):109–22.
Koizuka N, Imai R, Iwabuchi M, Sakai T, Imamura J. Genetic analysis of fertility restoration and accumulation of ORF125 mitochondrial protein in the kosena radish (Raphanus sativus cv. Kosena) and a Brassica napus restorer line. Theor Appl Genet. 2000;100:949–55.
Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, et al. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 2003;34(4):407–15.
Wang CD, Lezhneva L, Arnal N, Quadrado M, Mireau H. The radish Ogura fertility restorer impedes translation elongation along its cognate CMS-causing mRNA. Proc Natl Acad Sci USA. 2021;118(35): e2105274118.
Kitazaki K, Arakawa T, Matsunaga M, Yui-Kurino R, Matsuhira H, Mikami T, et al. Post-translational mechanisms are associated with fertility restoration of cytoplasmic male sterility in sugar beet (Beta vulgaris). Plant J. 2015;83(2):290–9.
Liu F, Cui X, Horner HT, Weiner H, Schnable PS. Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell. 2001;13(5):1063–78.
Kim YJ, Zhang DB. Molecular control of male fertility for crop hybrid breeding. Trends Plant Sci. 2018;23(1):53–65.
Fujii S, Toriyama K. Suppressed expression of Retrograde-Regulated Male Sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc Natl Acad Sci USA. 2009;106(23):9513–8.
Itabashi E, Iwata N, Fujii S, Kazama T, Toriyama K. The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J. 2011;65(3):359–67.
Jaqueth JS, Hou ZL, Zheng PZ, Ren RH, Nagel BA, Cutter G, et al. Fertility restoration of maize CMS-C altered by a single amino acid substitution within the Rf4 bHLH transcription factor. Plant J. 2020;101(1):101–11.
Gao B, Ren GF, Wen TW, Li HP, Zhang XL, Lin ZX. A super PPR cluster for restoring fertility revealed by genetic mapping, homocap-seq and de novo assembly in cotton. Theor Appl Genet. 2022;135(2):637–52.
Weaver DB, Weaver JB. Inheritance of pollen fertility restoration in cytoplasmic male-sterile upland cotton. Crop Sci. 1977;17(4):497–9.
Zhang JF, Stewart JM. Inheritance and genetic relationships of the D8 and D2–2 restorer genes for cotton cytoplasmic male sterility. Crop Sci. 2001;41(2):289–94.
Zhang JF, Stewart JM. CMS-D8 restoration in cotton is conditioned by one dominant gene. Crop Sci. 2001;41(2):283–8.
Wang F, Yue B, Hu JG, Stewart JM, Zhang JF. A target region amplified polymorphism marker for fertility restorer gene Rf1 and chromosomal localization of Rf1 and Rf2 in cotton. Crop Sci. 2009;49(5):1602–8.
Zhang JF, Stewart JM, Wang TH. Linkage analysis between gametophytic restorer Rf2 gene and genetic markers in cotton. Crop Sci. 2005;45(1):147–56.
Liu L, Guo W, Zhu X, Zhang T. Inheritance and fine mapping of fertility restoration for cytoplasmic male sterility in Gossypium hirsutum L. Theor Appl Genet. 2003;106(3):461–9.
Zhang JF, Stewart JM. Identification of molecular markers linked to the fertility restorer genes for CMS-D8 in cotton. Crop Sci. 2004;44(4):1209–17.
Yin JM, Guo WZ, Yang LM, Liu LW, Zhang TT. Physical mapping of the Rf1 fertility-restoring gene to a 100 kb region in cotton. Theor Appl Genet. 2006;112(7):1318–25.
Feng CD, Stewart JM, Zhang JF. STS markers linked to the Rf1 fertility restorer gene of cotton. Theor Appl Genet. 2005;110(2):237–43.
Wang F, Stewart JM, Zhang JF. Molecular markers linked to the Rf2 fertility restorer gene in cotton. Genome. 2007;50(9):818–24.
Zhao CP, Zhao GY, Geng Z, Wang ZX, Wang KH, Liu S, et al. Physical mapping and candidate gene prediction of fertility restorer gene of cytoplasmic male sterility in cotton. BMC Genomics. 2018;19(1):6.
Feng JJ, Zhang XX, Zhang M, Guo LP, Qi TX, Tang HN, et al. Physical mapping and InDel marker development for the restorer gene Rf2 in cytoplasmic male sterile CMS-D8 cotton. BMC Genomics. 2021;22(1):24.
Wu JY, Zhang M, Zhang XX, Guo LP, Qi TX, Wang HL, et al. Development of InDel markers for the restorer gene Rf1 and assessment of their utility for marker-assisted selection in cotton. Euphytica. 2017;213:251.
Feng JJ, Zhu HY, Zhang M, Zhang XX, Guo LP, Qi TX, et al. Development and utilization of an InDel marker linked to the fertility restorer genes of CMS-D8 and CMS-D2 in cotton. Mol Biol Rep. 2020;47(2):1275–82.
Zhang TZ, Hu Y, Jiang WK, Fang L, Guan XY, Chen JD, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–7.
Huang G, Wu ZG, Percy RG, Bai MZ, Li Y, Frelichowski JE, et al. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton A-genome evolution. Nat Genet. 2020;52(5):516–24.
Hu Y, Chen JD, Fang L, Zhang ZY, Ma W, Niu YC, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–48.
Peng RH, Xu YC, Tian SL, Unver T, Liu Z, Zhou ZL, et al. Evolutionary divergence of duplicated genomes in newly described allotetraploid cottons. Proc Natl Acad Sci USA. 2022;119(39): e2208496119.
Grover CE, Arick MA, Thrash A, Conover JL, Sanders WS, Peterson DG, et al. Insights into the evolution of the new world diploid cottons (Gossypium, subgenus Houzingenia) based on genome sequencing. Genome Biol Evol. 2019;11(1):53–71.
Udall JA, Long E, Hanson C, Yuan D, Ramaraj T, Conover JL, et al. De Novo genome sequence assemblies of Gossypium raimondii and Gossypium turneri. G3 (Bethesda). 2019;9(10):3079–85.
Wang MJ, Li JY, Wang PC, Liu F, Liu ZP, Zhao GN, et al. Comparative genome analyses highlight transposon-mediated genome expansion and the evolutionary architecture of 3D genomic folding in cotton. Mol Biol Evol. 2021;38(9):3621–36.
Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA. 2002;99(16):10887–92.
Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, et al. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J. 2003;35(2):262–72.
Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, et al. Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 2003;4(6):588–94.
Durand S, Ricou A, Simon M, Dehaene N, Budar F, Camiller C. A restorer-of-fertility-like pentatricopeptide repeat protein promotes cytoplasmic male sterility in Arabidopsis thaliana. Plant J. 2021;105(1):124–35.
Wang TL, He TT, Ding XL, Zhang QQ, Yang LS, Nie ZX, et al. Confirmation of GmPPR576 as a fertility restorer gene of cytoplasmic male sterility in soybean. J Exp Bot. 2021;72(22):7729–42.
Wang ZH, Zou YJ, Li XY, Zhang QY, Chen LT, Wu H, et al. Cytoplasmic male sterility of rice with Boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell. 2006;18(3):676–87.
Tang HW, Luo DP, Zhou DG, Zhang QY, Tian DS, Zheng XM, et al. The rice restorer Rf4 for wild-abortive cytoplasmic male sterility encodes a mitochondrial-localized PPR protein that functions in reduction of WA352 transcripts. Mol Plant. 2014;7(9):1497–500.
Huang WC, Yu CC, Hu J, Wang LL, Dan ZW, Zhou W, et al. Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc Natl Acad Sci USA. 2015;112(48):14984–9.
Liu Z, Yang ZH, Wang X, Li KD, An H, Liu J, et al. A mitochondria-targeted PPR protein restores pol cytoplasmic male sterility by reducing orf224 transcript levels in Oilseed Rape. Mol Plant. 2016;9(7):1082–4.
Melonek J, Duarte J, Martin J, Beuf L, Murigneux A, Varenne P, et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat Commun. 2021;12(1):1036.
YJ Zhang H Yang M Zhang XX Zhang LP Guo TX Qi et al 2022 The cotton mitochondrial chimeric gene orf610a causes male sterility by disturbing the dynamic balance of ATP synthesis and ROS burst Crop J. https://doi.org/10.1016/j.cj.2022.02.008
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13.
Barkan A, Small I. Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol. 2014;65:415–42.
Zhao HY, Huang JL. Study on microspore abortion of male sterile cotton Yamian A and Yamian B. Scientia Agricultura Sinica. 2012;45:4130–40.
Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP and PCR analysis. Plant Mol Biol Rep. 1993;11:122–7.
Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from nextgeneration sequencing data. Nucleic Acids Res. 2010;38: e164.
Hill JT, Demarest BL, Bisgrove BW, Gorsi B, Su YC, Yost HJ. MMAPPR: mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013;23(4):687–97.
Li Z, Chen XX, Shi SQ, Zhang HW, Wang X, Chen H, Li WF, et al. DeepBSA: A deep-learning algorithm improves bulked segregant analysis for dissecting complex traits. Mol Plant. 2022;15(9):1418–27.
Zhao L, Ding Q, Zeng J, Wang FR, Zhang J, Fan SJ, et al. An improved CTAB-ammonium acetate method for total RNA isolation from cotton. Phytochem Anal. 2012;23(6):647–50.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5.
Nie HS, Wang YM, Su Y, Hua JP. Exploration of miRNAs and target genes of cytoplasmic male sterility line in cotton during flower bud development. Funct Integr Genomics. 2018;18(4):457–76.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25(4):402–8.
Yu J, Jung S, Cheng CH, Ficklin SP, Lee T, Zheng P, et al. CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 2014;42:D1229–36.
Nie HS, Cheng C, Hua JP. Mitochondrial proteomic analysis reveals that proteins relate to oxidoreductase activity play a central role in pollen fertility in cotton. J Proteomics. 2020;227: 103938.
Acknowledgements
Not applicable
Funding
This research was supported in part by National Natural Science Foundation of China (31671741) and National Key Research and Development Program for Crop Breeding (2016YFD0101407) to J HUA.
Author information
Authors and Affiliations
Contributions
CC investigated and collected data, analyzed the results, and prepared the manuscript. HSN, HJL, DA, BL, KYJ, YNC, MZ, BXZ and AHG attended the data collection and discussion. JPH conceived and designed the research direction, guided the entire research process and revised the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplemental figure 1
Additional file 2.
Supplemental figure 2
Additional file 3.
Supplemental table 1-4
Additional file 4.
Supplemental table 5-6
Additional file 5.
Supplemental table 7-8
Additional file 6.
Supplemental table 9
Additional file 7.
Supplemental table 10
Additional file 8.
Supplemental table 11
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cheng, C., Nie, H., Li, H. et al. Identification of fertility restoration candidate genes from a restorer line R186 for Gossypium harknessii cytoplasmic male sterile cotton. BMC Plant Biol 23, 175 (2023). https://doi.org/10.1186/s12870-023-04185-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-023-04185-z