Abstract
Background
Sparganium (Typhaceae) is a widespread temperate genus of ecologically important aquatic plants. Previous reconstructions of the phylogenetic relationships among Sparganium species are incompletely resolved partly because they were based on molecular markers comprising < 7,000 bp. Here, we sequenced and assembled the complete chloroplast genomes from 19 Sparganium samples representing 15 putative species and three putative subspecies in order to explore chloroplast genome evolution in this genus, clarify taxonomic lineages, estimate the divergence times of Sparganium species, and reconstruct aspects of the biogeographic history of the genus.
Results
The 19 chloroplast genomes shared a conserved genome structure, gene content, and gene order. Our phylogenomic analysis presented a well-resolved phylogeny with robust support for most clades. Non-monophyly was revealed in three species: S. erectum, S. eurycarpum, and S. stoloniferum. Divergence time estimates suggest that the two subgenera of Sparganium split from each other ca. 30.67 Ma in the middle Oligocene. The subgenus Xanthosparganium diversified in the late Oligocene and Miocene, while the subgenus Sparganium diversified in the late Pliocene and Pleistocene. Ancestral area reconstruction suggested that the two subgenera may have originated in East Eurasia and North America.
Conclusion
The non-monophyletic nature of three putative species underscores the necessity of taxonomic revision for Sparganium: S. stoloniferum subsp. choui may be more appropriately identified as S. choui, and subspecies of S. erectum may be in fact distinct species. The estimated diversification times of the two subgenera correspond to their species and nucleotide diversities. The likely ancestral area for most of subgenus Xanthosparganium was East Eurasia and North America from where it dispersed into West Eurasia and Australia. Most of subgenus Sparganium likely originated in North America and then dispersed into Eurasia. Our study demonstrates some of the ways in which complete chloroplast genome sequences can provide new insights into the evolution, phylogeny, and biogeography of the genus Sparganium.
Similar content being viewed by others
Background
Sparganium L. is an aquatic perennial genus comprising approximately 14–19 species [1,2,3,4]. It mainly occurs in temperate and cool regions of the Northern Hemisphere with only two species extending from eastern Asia southward into Australia and/or New Zealand [1, 2]. Sparganium species often dominate wetlands and play important ecological roles in aquatic communities [1, 2]. The tuberous rhizome of S. stoloniferum is widely used as a gynecological drug in traditional Chinese medicine [5].
Species delimitation is difficult in Sparganium due to phenotypic plasticity and interspecific hybridization. The comprehensive Sparganium monographs by Cook and Nicholls [1, 2] recognized 14 species and six subspecies, divided between two subgenera: Subgenus Xanthosparganium included seven species and one subspecies with translucent perianth segments. Subgenus Sparganium contained seven species and five subspecies with dark brown to black perianth segments [1, 2]. Sulman et al. [6] realigned the subgenera to conform to two clades in a phylogeny of Sparganium that was based on two chloroplast DNA fragments and two nuclear genes. The revised subgenus Sparganium included S. erectum and S. eurycarpum, both of which have bilocular ovary and endocarps with longitudinal ridges. The revised subgenus Xanthosparganium included the remaining 12 species, all of which have unilocular ovary and endocarps without longitudinal ridges [1, 2, 6]. Cook and Nicholls [1] treated S. acaule as a subspecies (S. emersum subsp. acaule), but it was resurrected as a species by Ito et al. [7] due to the non-monophyletic nature of S. emersum sensu lato in a phylogeny of Sparganium that was based on six cpDNA regions plus one nuclear gene. Nevertheless, the phylogenetic relationships among Sparganium species remain incompletely resolved, partly because they have so far been based on limited genetic data (< 4,000 bp [6] or < 7,000 bp [7]).
Sparganium is an early-diverging lineage in Poales [8] with abundant fossils from the Paleocene [6, 9, 10]. These fossils show that the ancestral species from the two subgenera were distinct by the Oligocene, and by the Miocene the endocarps of described fossil species are very difficult to distinguish from extant species [1, 11]. However, previous divergence time analyses using molecular dating based on DNA sequences of four gene regions suggested a mid-Miocene crown origin and Pliocene diversification of Sparganium [6], which is inconsistent with the fossil record. Therefore, it is necessary to re-estimate the divergence time of Sparganium using a more extensive molecular genetic dataset.
In plants, no universal barcode consistently discriminates among plant species and reveals phylogenetic relationships. Researchers are therefore increasingly generating whole-plastid genome sequences to study taxonomy and biogeography [12, 13]. For example, complete chloroplast genome sequences have provided higher resolutions of phylogenetic relationships among plants compared to phylogenetic reconstructions that were based on just a few chloroplast fragments [13,14,15]. Here, we sequenced and assembled chloroplast genomes of 19 Sparganium samples, which were provisionally identified as 15 species and three subspecies. Our aims were to 1) investigate chloroplast genome evolution in Sparganium; 2) clarify the evolutionary relationships among Sparganium species; and 3) estimate the divergence times and infer the ancestral areas of Sparganium.
Results
Feature of chloroplast genomes
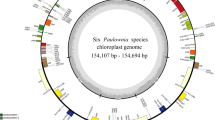
The chloroplast genomes of all Sparganium samples were successfully assembled from genome skimming data. Their sizes ranged from 161,487 to 162,331 bp with a typical quadripartite structure (Fig. 1), including two Inverted Repeat (IR) regions (26,882–27,037 bp), one Large Single Copy (LSC) region (88,026–89,281 bp) and one Small Single Copy (SSC) region (18,684–19,120 bp) (Table 1). Each of the 19 chloroplast genomes that we reconstructed encoded 114 unique genes comprising 80 protein coding genes (PCGs), 30 tRNA genes, and four rRNA genes. The gene arrangement was identical in each genome. The overall GC content of the genomes (36.7–36.9%) was conserved across species (Table 1).
Comparison of border regions and sequence identity
Potential IR expansion or contraction was assessed by comparing the LSC/IR and SSC/IR junctions across species. The locations of LSC/IRa (JLA) and LSC/IRb (JLB) junctions were the same for all Sparganium species: the JLA was consistently located at the psbA-rps19 intergenic spacer (IGS), and the JLB was consistently located at the rpl22-rps19 intergenic spacer (Fig. 2). The distance between psbA and JLA was 91 bp for all species of subgenus Sparganium, and 86 bp for all species of subgenus Xanthosparganium except S. hyperboreum (93 bp). The distance between rpl22 and JLB was 17 bp for all species except S. glomeratum (12 bp). The locations of SSC/IRa (JSA) and SSC/IRb (JSB) were also conserved across species, with the IR region consistently expanding into the ycf1 gene at the JSA or JSB junctions (Fig. 2).
The sequence identity analysis revealed a high similarity (97.53–99.6%) among chloroplast genomes of Sparganium species, especially for species within the subgenus Sparganium that were invariant at > 99% of nucleotide sequences (Table S1). The LSC and SSC regions were more divergent than the IR region (Figs. S1 and S2). In addition, four IGS regions (trnS-trnG, ndhF-rpl32, accD-psaI and petA-psbJ), and two PCGs (ndhF and ndhE) showed relatively high nucleotide diversity with Pi values greater than 0.02 (Fig. S2), making these candidate molecular markers for future phylogenetic and phylogeographic studies.
Phylogenetic analysis
The aligned length of 80 PCGs was 70,612 bp with 3,507 informative sites. Identical topologies were revealed using the maximum likelihood (ML) and Bayesian inference (BI) methods (Fig. 3). Sparganium comprises a strongly supported monophyletic group (BS = 100, PP = 1) divided into two unambiguous clades corresponding to subgenus Sparganium (BS = 100, PP = 1) and subgenus Xanthosparganium (BS = 100, PP = 1). None of the three species in the subgenus Sparganium were monophyletic. Sparganium stoloniferum subsp. choui was placed as sister to the remaining species or subspecies, which formed a clade with high support (BS = 72, PP = 1). Two strongly supported clades, S. stoloniferum + S. erectum subsp. neglectum + S. eurycarpum_ON (BS = 99, PP = 1) and S. erectum subsp. microcarpum + S. erectum (BS = 98, PP = 1), were clustered with high support (BS = 64, PP = 1) and resolved as the sister group of S. eurycarpum_NS. In the subgenus Xanthosparganium, four sister species pairs were revealed, and the backbone was well resolved with robust support for most of the nodes. The topology showed a hierarchical branching structure and the root branching order was S. natans + S. hyperboreum, S. androcladum, S. gramineum + S. fluctuans, S. subglobosum, S. glomeratum + S. acaule, S. fallax, S. japonicum, and S. emersum + S. angustifolium.
Phylogenomic analysis and divergence time dating of Sparganium. The phylogenetic tree was reconstructed from sequences of 80 protein coding genes. Asterisks indicate bootstrap support = 100/posterior probability = 1.00. Triangles indicate the fossil calibration nodes and numbers close to nodes refer to the mean divergent time estimates. Blue bars indicate 95% highest posterior distributions
Divergence time estimation and ancestral area reconstruction
The stem age of Sparganium was estimated to be 74.4 Ma (95% highest posterior densities (HPD):71.28–79.81 Ma). Two subgenera split from each other at approximately 30.67 Ma (95% HPD: 19.58–43.52 Ma) in the middle Oligocene (Fig. 3). The subgenus Xanthosparganium began to diversify approximately 23.97 Ma (95% HPD: 15.54–34.45 Ma) in the late Oligocene, with the majority of species diversification occurring in the Miocene other than the divergence between S. emersum and S. angustifolium, which did not occur until the Pliocene. The subgenus Sparganium did not differentiate until approximately 4.37 Ma (95% HPD: 2.3–7.58 Ma) in the Pliocene, and most of the species/subspecies divergence within this subgenus Sparganium occurred during the Pleistocene.
Based on BioGeoBEARS analysis, the BAYAREALIKE model was supported as the best-fit model (AICc_wt = 0.79) for ancestral area reconstruction. East Eurasia and North America were suggested as high probability ancestral areas for the genus and the two subgenera (Fig. 4). In subgenus Xanthosparganium, the ancestral area of all nodes was East Eurasia and North America except for nodes 22 (S. emersum + S. angustifolium) and 31 (S. natans + S. hyperboreum), which included West Eurasia as a likely ancestral area. In subgenus Sparganium, the ancestral area of all nodes was North America except for node 35 (S. erectum subsp. microcarpum + S. erectum), for which West Eurasia was identified as the most likely ancestral area. Expansion from ancestral areas involved 30 dispersal events across 15 nodes (all except 35, 37, and 38) along with five vicariance events across five nodes (28, 33, 34, 36, and 38; Table S2).
Discussion
Chloroplast genome evolution
The evolution of chloroplast genomes often entails gene inversions, translocations, losses or rearrangements [16, 17]. However, the 19 Sparganium chloroplast genomes were highly conserved, with each having the same numbers and arrangements of genes. Expansion or contraction of the IR is common in chloroplast genomes and plays an important role in the size variation of chloroplast genomes in angiosperms [18]. Among Sparganium chloroplast genomes, the lengths of the IR region were comparable (26,882 bp-27,037 bp), as were the locations of JLA, JLB, JSA and JSB, thus further illustrating the conservative nature of Sparganium chloroplast genomes.
Four chloroplast DNA genes (matK, rbcL, rpoB and rpoC1) and four IGS regions (trnL-trnF, petA-psbJ, psbM-trnD and trnC-petN) were used in two previous studies on the phylogeny of Sparganium [6, 7]. No full resolution of phylogenetic reconstruction was achieved due to the low nucleotide diversity of these regions (except for petA-psbJ, Fig. S2). The relatively variable regions identified in this study (Fig. S2), including trnS-trnG, ndhF-rpl32, accD-psaI, petA-psbJ, ndhF and ndhE, should provide more informative molecular markers for future phylogenetic or phylogeographic studies in this genus.
Phylogenetic relationship
Our phylogenomic analysis revealed two strongly supported clades, corresponding to the subgenera Xanthosparganium and Sparganium that were proposed in a previous study based on partial cpDNA and nuclear sequences plus stigma and endocarp features [6]. Members of the subgenus Xanthosparganium species are floating or emergent and have unilocular ovary and endocarps without longitudinal ridges [1, 6]. The membership that we identified for this subgenus agrees with that identified in earlier studies [6, 7], however, our phylogenetic reconstruction had no polytomies and high support for most clades (Fig. 3), and thus more clearly elucidates the relationships among the 12 species within this subgenus (Fig. 3). Four previously identified sister species pairs, S. natans + S. hyperboreum, S. gramineum + S. fluctuans [6], S. glomeratum + S. acaule, and S. emersum + S. angustifolium [7], were confirmed by our phylogenomic analysis with 96–100% support. The morphological similarities between S. natans + S. hyperboreum and between S. gramineum + S. fluctuans have been described by Cook and Nicolls [1]. Sparganium glomeratum + S. acaule share several features including congested female heads, 1–3 male heads, and the lowest inflorescence bract that is longer than the flowering stem [7]. Sparganium emersum + S. angustifolium differ in leaf shape and male heads [1] although they are often confused when S. emersum is in its floating form; others have suggested that these be considered a single, complex species [19, 20].
Species within the subgenus Sparganium species are emergent and have bilocular ovary and endocarps with longitudinal ridges [1, 6]. There has been some debate about taxonomic demarcations in this subgenus, which in our study included taxa that we initially identified as S. stoloniferum, S. stoloniferum subsp. choui, S. eurycarpum (one sample from Ontario, Canada and one sample from Nova Scotia, Canada), S. erectum, S. erectum subsp. neglectum, and S. erectum subsp. microcarpum. Sparganium stoloniferum and S. stoloniferum subsp. choui did not form a monophyletic group (Fig. 3). Subspecies S. stoloniferum subsp. choui is sometimes considered to be a distinct species, S. choui [3, 21], because compared with S. stoloniferum it has a short panicle, only one male head per branch, a short anther length, and a small fruit [21]. The non-monophyly of S. stoloniferum revealed by our whole-genome phylogeny, combined with morphological differences, suggest that S. choui may be more appropriately considered a species than a subspecies. Sparganium eurycarpum was also polyphyletic, and the sample from Ontario groups with S. erectum subsp. neglectum have high support. Related to this, four genetic groups identified on the basis of AFLPs, genome sizes, and fruit morphology, corresponded to four subspecies of S. erectum (subsp. erectum, subsp. microcarpum, subsp. neglectum and subsp. oocarpum) that were distributed across 64 populations in the Czech Republic [2, 22]. In addition to their non-monophyly in phylogenetic tree (Fig. 3), it may therefore be more appropriate to assign species-status to the subspecies of S. erectum. However, the recency with which subgenus Sparganium diversified, and the associated low levels of sequence divergence, mean that further investigation is needed to clarify some of the taxonomic groups within subgenus Sparganium. Future studies should also compare phylogenies based on nuclear genes with those based on whole chloroplast genomes to test the possibility that historical and/or more recent hybridization, which has been reported among multiple Sparganium species and subspecies [1; 7; 20], is obscuring the phylogenetic inferences in this subgenus.
Biogeographical reconstruction
Accurate estimates of divergence times are needed before we can fully understand biogeographic histories. The stem age of Sparganium or the crown age of Typhaceae was estimated to be 74.4 Ma (Fig. 3), which is similar to the previously reported age of 72 Ma [6] and agrees with the earliest known fossils of Typha from the late Cretaceous [23]. Our time-calibrated tree indicated a crown age of Sparganium at 30.67 Ma, which is much older than the 13 Ma reported in Sulman et al. [6] although the two studies used the same calibration points. However, Sulman et al. [6] based their estimate on < 4,000 bp, and divergence estimates should be more reliable when based on whole chloroplast genomes. The crown age of Sparganium in the Oligocene and its main diversification in the Miocene (Fig. 3) are also consistent with the finding that the endocarps of fossil species from the Miocene onwards are very difficult to distinguish from extant species [1]. The subgenus Xanthosparganium began to diversify in the late Oligocene (an estimated 24 Ma) while the subgenus Sparganium did not begin to differentiate until the late Pliocene (an estimated 4.34 Ma). These dates are consistent with the species diversity of the two subgenera: the subgenus Xanthosparganium contains many species with diverse life forms including boreal and temperate species as well as emerged and floating-leaved species. In contrast, the subgenus Sparganium contains fewer species, but all of which are robust, erect, and temperature species with similar morphological characteristics [1, 2]. In addition, as noted earlier, nucleotide diversity across the chloroplast genome was lower in subgenus Sparganium than in subgenus Xanthosparganium.
The BioGeoBEARS analysis suggested East Eurasia and North America as the ancestral areas for Sparganium and its two subgenera (Fig. 4), which is consistent with the abundant Sparganium fossil records from these regions [1, 2, 6]. In the subgenus Xanthosparganium, the ancestral area for most nodes was East Eurasia and North America, indicating subsequent dispersal from either East Eurasia or North America to West Eurasia. There is no obvious geographical barrier between East and West Eurasia, and many plants, such as Aesculus [24], Sibbaldia [25] and Oxyria digyna [26], spread from Asia to Europe through this route. Alternatively, an important route for plant migration between North America and Europe is the North Atlantic Land Bridge (NALB) [27, 28]. The lifespan of the NALB has been debated, with previous studies suggesting that the NALB was an effective dispersal route between North America and Europe until the Eocene or possibly early Miocene [29,30,31,32,33,34], but a recent study suggested that this land bridge did not close until the late Miocene (8–10 Ma) [35]. Therefore, the subgenus Xanthosparganium, in which most nodes differentiated during the Miocene, could have plausibly spread from North America to Europe through the NALB. Another possible dispersal route was from East Eurasia to the Indo-Pacific and Australia. During the late Oligocene, the Wallace District was uplifted by the collision of the Sunda Shelf with the Sahul Shelf, which led to the formation of a land bridge between Asia and Australia in the Miocene [36, 37]. The two Asia/Australia species S. fallax and S. subglobosum, both of which diverged in the middle Miocene, could have dispersed from Asia to Australia via the land bridge. Alternatively, long distance endozoochoric dispersal could have introduced Sparganium to more distant locations such as Australia, because the seeds of Sparganium plants can be eaten by migratory birds and spread over long distances [1, 38, 39]. A vicariance event suggested by BioGeoBEARS analysis occurs at the node of the Eurasian S. gramineum and the North American S. fluctuans with their divergence at 6.79 Ma (Figs. 3, 4, Table S2) involving the Beringian Land Bridge (BLB). The BLB served as an important route for the exchange of temperate plants between eastern Asia and western North America from the early Paleocene to Pliocene [40,41,42], and the final closure of the BLB occurred at 5.5–5.4 Ma [43]. Therefore, the ancestral area of East Eurasia + North America was likely sundered by the closing of the BLB, thus giving rise to S. gramineum and S. fluctuans.
Our time estimation (4.37 Ma) of the crown node of the subgenus Sparganium closely matches to the final closure of the BLB, thus indicating the crown diversification of the subgenus Sparganium associated with the vicariance event was likely invoked by the closure of the BLB. Within the subgenus Sparganium, other than the node leading to S. erectum/S. erectum subsp. microcarpum, the most likely ancestral area was North America. This subgenus likely dispersed from North America to Asia or Europe possibly via the NALB and BLB land bridges. The final closure of the BLB at 5.5–5.4 Ma [43] is far earlier than the relatively recent species divergence, with the earlier closure of the NALB, suggesting the spread from North America to Asia or Europe through long distance dispersal by birds. Both dispersal and vicariance events occurred in three out of five nodes within the subgenus Sparganium (Table S2), thus indicating that long distance dispersal and vicariance played an important role in the North American-European/Asian diversification of the subgenus Sparganium. Future phylogeographic investigations based on plants sampled from a wider geographical area could provide further insight into the biogeographical history of Sparganium.
Conclusion
IN this study, we assembled 19 chloroplast genomes from Sparganium samples that each represented a distinct evolutionary lineage. The chloroplast genomes of Sparganium species have conserved genome structure, gene content, and gene order. Our phylogenomic analysis presented a well-resolved phylogeny of Sparganium species, although there remains some uncertainty surrounding taxonomic classification within the recently diversified subgenus Sparganium. We also reappraised the divergence time and historical biogeography of Sparganium: Sparganium diversified into two subgenera in the Oligocene. The subgenus Xanthosparganium began to diversify in the late Oligocen and then dispersed from eastern Eurasia and North America into western Eurasia and Australia. The subgenus Sparganium diversified in the late Pliocene and mostly expanded its range from North America into Eurasia. In summary, our study provides new insights into the chloroplast genome evolution, phylogeny, and biogeography of the genus Sparganium.
Methods
Plant sampling and DNA extraction
A total of 19 Sparganium samples comprising 15 putative species (including two samples of S. eurycarpum, one from Nova Scotia and one from Ontario, Canada) and three subspecies were collected (Table 1). Voucher specimens were kept in the herbaria of IBIW, Wuhan University and Trent University with specific voucher numbers (Table 1). Eugeny A. Belyakov and Xinwei Xu performed formal identification of the samples. Our species delimitations follow accepted names of Sparganium from Plants of the World Online (POWO), which incorporates the latest published taxonomy. The pictures of each species were presented in Figure S3. Total genomic DNA was extracted from silica-dried leaves using a DNA Secure Plant Kit (Tiangen Biotech, Beijing, China) following the manufacturer’s protocol.
Genome skimming, chloroplast genome assembly and annotation
Library preparation and genomic sequencing on the Illumina Hiseq 2500 platform were conducted by Benagen (Benagen Inc., Wuhan, China). Approximately 10G paired-end reads (150 bp) were produced for each sample. The chloroplast genomes were assembled de novo using SPAdes v3.9.0 [44] after raw reads were trimmed and filtered using Fastp v0.20.0 [45] with default parameters. Gene annotation was conducted using Geseq [46] with the chloroplast genome of Typha latifolia (GU195652.1) [47] as a reference. The circular map of chloroplast genomes was created using OGDRAW v1.3.1 [48]. The 19 Sparganium chloroplast genomes were deposited in GenBank (see Table 1 for accession numbers). Two of them, S. fallax and S. stoloniferum subsp. choui, were reported in our two previous studies [49, 50].
Comparative analysis of chloroplast genomes
The online program IRscope [51] was used to visualize the junction sites of the chloroplast genomes. Sequences of chloroplast genomes were aligned using MAFFT v7.221 [52]. The sequence and structural variations of Sparganium chloroplast genomes were identified using mVISTA [53] with the chloroplast genome of T. latifolia as a reference. Nucleotide diversity (Pi) was assessed using DnaSP v6.0 [54]. The sliding window method was used with a window length of 800 bp and step size of 200 bp.
Phylogenetic analysis
The chloroplast genomes of T. latifolia, T. orientalis (MN602748.1) [55], Ananas comosus (KR336549.1) [56], and Tillandsia usneoides (KY293680) [57] were downloaded from GenBank as outgroups. The PCGs were extracted from each of the chloroplast genomes and used in the phylogenetic analyses. Sequences of PCGs were aligned using MAFFT v7.221 [52]. The best-fit model of nucleotide substitution was estimated by ModelFinder [58]. Maximum likelihood (ML) and Bayesian inference (BI) methods were used for phylogenetic inference. The ML analysis was performed using RAxML v8.2.12 [59] and 1000 repetitions were performed to summarize the ML bootstrap support. BI implemented in MrBayes v3.2.7 [60] was conducted using two independent runs of 10 million generations, and each run employed four Markov chains, with sampling at every 1,000 generations. Chain convergence was checked using Tracer v1.7.1 [61], and posterior probabilities (PP) were generated from trees after excluding a burn-in of the initial 25% of the trees.
Molecular dating and ancestral area reconstruction
Two calibration points were used for divergence time estimation conducted in BEAST v1.7.4 [62]. One was the stem age of Typha—a minimum age of 70 Ma based on fossil evidence as used in previous studies [6, 63, 64]. The detailed setting was a lognormal prior with an offset of 70, a mean of 1.5, and a standard deviation of 0.5. The other was the stem age of Typhaceae that has a uniform distribution ranging from 90–105 Ma obtained from Givnish et al. [8] and used in a previous study [6]. Markov chain Monte Carlo (MCMC) analyses of 2 × 109 generations were implemented, and every 1,000 generations were sampled. The initial 25% generations were discarded as burn-in, and the effective sample size (ESS) for the convergence of each parameter was checked using Tracer v1.7.1 [61].
Ancestral area reconstruction was conducted using the BioGeoBEARS package [65] implemented in RASP v4.0 [66]. Six geographical areas were defined based on the distribution of Sparganium: (A) North America, (B) Indo-Pacific, (C) West Eurasia, (D) East Eurasia, (E) Africa, and (F) Australia. The input trees for BioGeoBEARS analysis were obtained from BEAST analysis. The best-fit biogeographic model was determined according to the Akaike Information Criterion cumulative weight (AICc_wt).
Availability of data and materials
All chloroplast genomes in this study are available from the National Center for Biotechnology Information (NCBI) (accession numbers: MW789349, MW810982- MW810984, MW817014-MW817019, MW829761-MW829768, and MW874418).
References
Cook CDK, Nicholls MS. A monographic study of the genus Sparganium (Sparganiaceae). Part 1. Subgenus Xanthosparganium Holmberg Bot Helv. 1986;96:213–67.
Cook CDK, Nicholls MS. A monographic study of the genus Sparganium (Sparganiaceae). Part 2. Subgenus Sparganium Bot Helv. 1987;97:1–44.
Sun K, Simpson DA. Typhaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vol. 23. Beijing and St Louis: Science Press and Missouri Botanical Garden Press; 2010. p. 158–63.
Kaul RB. Sparganiaceae. In: Flora of North America Editorial Committee, editor. Flora of North America North of Mexico, vol.22. New York: Oxford University Press; 1997.
Jia J, Li X, Ren X, Liu X, Wang Y, Dong Y, et al. Sparganii Rhizoma: A review of traditional clinical application, processing, phytochemistry, pharmacology, and toxicity. J Ethnopharmacol. 2021;268:113571.
Sulman JD, Drew BT, Drummond C, Hayasaka E, Sytsma KJ. Systematics, biogeography, and character evolution of Sparganium (Typhaceae): diversification of a widespread, aquatic lineage. Am J Bot. 2013;100:2023–39.
Ito Y, Tanaka N, Kim C, Kaul R, Albach D. Phylogeny of Sparganium (Typhaceae) revisited: non-monophyletic nature of S. emersum sensu lato and resurrection of S. acaule. Plant Syst Evol. 2016;302:129–35.
Givnish TJ, Barfuss MHJ, Van Ee B, Riina R, Schulte K, Horres R, et al. Phylogeny, adaptive radiation, and historical biogeography in Bromeliaceae: insights from an eight-locus plastid phylogeny. Am J Bot. 2011;98:872–95.
Hickey LJ. Stratigraphy and paleobotany of the Golden Valley Formation (early Tertiary) of western North Dakota. Geol Soc Am Memoir. 1977;150:1–181.
McIver EE, Basinger JF. Early Tertiary floral evolution in the Canadian High Arctic. Ann Missouri Bot Gard. 1999;86:523–45.
Dorofeev PI. On taxonomy of Tertiary Sparganium. Sovietskaya palaeokarpologya. Moscow: Publishing House Nauka; 1979. ((in Russian)).
Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL. Plant DNA barcoding: from gene to genome. Biol Rev. 2015;90:157–66.
Williams AV, Miller JT, Small I, Nevill PG, Boykin LM. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol Phylogenet Evol. 2016;96:1–8.
Gichira AW, Chen LY, Li ZZ, Hu GW, Saina JK, Gituru RW, et al. Plastid phylogenomics and insights into the inter-mountain dispersal of the eastern African giant senecios (Dendrosenecio, Asteraceae). Mol Phylogenet Evol. 2021;164:107271.
Li ZZ, Lehtonen S, Martins K, Gichira AW, Wu S, Li W, et al. Phylogenomics of the aquatic plant genus Ottelia (Hydrocharitaceae): implications for historical biogeography. Mol Phylogenet Evol. 2020;152:106939.
Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–74.
Mower JP, Vickrey TL. Structural diversity among plastid genomes of land plants. Adv Bot Res. 2018;85:263–92.
Qian J, Song J, Gao H, Zhu Y, Xu J, Pang X, et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE. 2013;8:e57607.
Brayshaw TC. Pondweeds and bur-reeds, and their relatives, of British Columbia. Occas Pap British Columbia Prov Mus. 1985;26:1–167.
Larson GE. Aquatic and wetland vascular plants of the northern Great Plains. General Technical Report RM-238. Fort Collins: US Department of Agriculture, Forest Service, Rocky Mountain Forest and Range Experiment Station; 1993.
Yu D. New plants of Sparganium in Northeastern China. Bull Bot Res. 1992;12:255–61.
Píšová S, Fér T. Intraspecific differentiation of Sparganium erectum in the Czech Republic: molecular, genome size and morphometric analysis. Preslia. 2020;92:137–65.
Knobloch E, Mai DH. Monographie der Früchte und Samen in der Kreide Mitteleuropas. Rozpravy Ústředního Ústavu Geologického. 1986;47:1–219.
Xiang QY, Soltis DE. Dispersal-vicariance analyses of intercontinental disjuncts: historical biogeographical implications for angiosperms in the Northern Hemisphere. Int J Plant Sci. 2001;162(Suppl 6):S29–39.
Allen GA, Marr KL, McCormick LJ, Hebda RJ. Geographical origins, migration patterns and refugia of Sibbaldia procumbens, an arctic–alpine plant with a fragmented range. J Biogeogr. 2015;42:1665–76.
Wang Q, Liu J, Allen GA, Ma Y, Yue W, Marr KL, Abbott RJ. Arctic plant origins and early formation of circumarctic distributions: a case study of the mountain sorrel. Oxyria digyna New Phytol. 2015;209:343–53.
Tiffney BH. Perspectives on the origin of the floristic similarity between Eastern Asia and Eastern North America. J Arnold Arbor. 1985;66:73–94.
Tiffney BH. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J Arnold Arbor. 1985;66:243–73.
Manchester SR. Biogeographical relationships of North American Tertiary floras. Ann Missouri Bot Gard. 1999;86:472–522.
Tiffney BH. Geographic and climatic influence on the Cretaceous and Tertiary history of Euramerican floristic similarity. Acta Universitatis Carolinae -Geologica. 2000;44:5–16.
Milne RI, Abbott RJ. The origin and evolution of Tertiary relict floras. Adv Bot Res. 2002;38:281–314.
Harris AJ, Wen J, Xiang QY. Inferring the biogeographic origins of inter-continental disjunct endemics using a Bayes-DIVA approach. J Syst Evol. 2013;51:117–33.
Tiffney BH, Manchester SR. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. Int J Plant Sci. 2001;162(Suppl 6):S3–17.
Milne RI. Northern Hemisphere plant disjunctions: a window on Tertiary land bridges and climate change? Ann Bot. 2006;98:465–72.
Denk T. The biogeographic history of Iceland - the North Atlantic Land Bridge revisited. In: Denk T, Grimsson F, Zetter R, SíMonarson LA, editors. Late Cainozoic Floras of Iceland. Dordrecht: Springer; 2011. p. 647–68.
Hall R. Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer-based reconstructions, model and animations. J Asian Earth Sci. 2002;20:353–431.
Van Welzen PC, Parnell JAN, Slik JWF. Wallace’s Line and plant distributions: Two or three phytogeographical areas and where to group Java? Biol J Linn Soc. 2011;103:531–45.
Pollux BJA, Santamaria L, Ouborg NJ. Differences in endozoochorous dispersal between aquatic plant species, with reference to plant population persistence in rivers. Freshw Biol. 2005;50:232–42.
Pollux BJA, Jong MD, Steegh A, Ouborg NJ, Groenendael JMV, Klaassen M. The effect of seed morphology on the dispersal of aquatic macrophytes by the common carp (Cyprinus carpio). Freshw Biol. 2006;51:2063–71.
Marincovich L, Gladenkov AY. Evidence for an early opening of the Bering Strait. Nature. 1999;397:149–51.
Vila R, Bell CD, Macniven R, Goldman-Huertas B, Ree RH, Marshall CR, et al. Phylogeny and palaeoecology of Polyommatus blue butterflies show Beringia was a climate-regulated gateway to the New World. Proc R Soc B. 2011;278:2737–44.
Graham A. The role of land bridges, ancient environments, and migrations in the assembly of the North American flora. J Syst Evol. 2018;56:405–29.
Gladenkov AY, Oleinik AE, Marincovich L, Barinov KB. A refined age for the earliest opening of Bering Strait. Palaeogeogr Palaeoclimatol Palaeoecol. 2002;183:321–8.
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90.
Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, et al. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45(W1):W6-11.
Guisinger MM. Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol. 2010;70:149–66.
Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47(W1):W59-64.
Zhang Q, Wang Y, Xu X. The complete chloroplast genome of Sparganium fallax (Typhaceae). Mitochondrial DNA Part B. 2021;6:2097–8.
Huang L, Zhang Q, Xu X. Characterization of the complete chloroplast genome of an aquatic plant, Sparganium stoloniferum subsp. choui (Typhaceae). Mitochondrial DNA Part B. 2021;6:3078–9.
Amiryousefi A, Hyvönen J, Poczai P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34:3030–1.
Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019;20:1160–6.
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, et al. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–7.
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34:3299–302.
Liu Z-D, Zhou X-L, Ma H-Y, Tian Y-Q, Shen S-K. Characterization of the complete chloroplast genome sequence of wetland macrophyte Typha orientalis (Typhaceae). Mitochondrial DNA Part B. 2020;5:136–7.
Redwan RM, Saidin A, Kumar SV. Complete chloroplast genome sequence of MD-2 pineapple and its comparative analysis among nine other plants from the subclass Commelinidae. BMC Plant Biol. 2015;15:196.
Poczai P, Hyvönen J. The complete chloroplast genome sequence of the CAM epiphyte Spanish moss (Tillandsia usneoides, Bromeliaceae) and its comparative analysis. PLoS ONE. 2017;12:e0187199.
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9.
Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67:901–4.
Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73.
Bremer K. Early Cretaceous lineages of monocot flowering plants. Proc Natl Acad Sci U S A. 2000;97:4707–11.
Zhou B, Tu T, Kong F, Wen J, Xu X. Revised phylogeny and historical biogeography of the cosmopolitan aquatic plant genus Typha (Typhaceae). Sci Rep. 2018;8:8813.
Matzke NJ. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in Island Clades. Syst Biol. 2014;63:951–70.
Yu Y, Blair C, He X. RASP 4: Ancestral state reconstruction tool for multiple genes and characters. Mol Biol Evol. 2020;37:604–6.
Acknowledgements
We gratefully thank Kathryn Tisshaw and Haihao Yu for their help in collecting samples.
Funding
This study was supported by the China Scholarship Council (contract 201606275169) and Special Project of Basic Work of Science and Technology, Ministry of Science and Technology, China (No. 2013FY112300).
Author information
Authors and Affiliations
Contributions
X.X., E.A.B. and J.F. designed the research; X.X., J.F., A.G.L. and E.A.B. carried out the field collections; Q.Z. and Y.Z. carried out the experiments and performed the data analysis; Q.Z. drafted the manuscript. X.X., E.A.B. and J.F. revised the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Experimental research and field studies on plants complies with relevant institutional, national, and international guidelines and legislation. All plant species are not rare species for protection. We have the permission to collect the plant samples by the local government. Our work reported here complies with the IUCN Policy Statement on Research Involving Species at Risk of Extinction.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S1. Alignment of chloroplast genomes of Sparganium species. The chloroplast genome of Typha latifolia was used as a reference.
Additional file 2:
Figure S2. Nucleotide diversity hotspot regions in chloroplast genomes of Sparganium species.
Additional file 3:
Figure S3. Pictures of Sparganium species.
Additional file 4:
Table S1. Sequence similarity among the chloroplast genomes of Sparganium species.
Additional file 5:
Table S2. Ancestral area reconstruction using the BioGeoBEARS analysis based on the chronogram inferred using BEAST. Node number is indicated in Figure 4. Letter codes represent biogeographic regions: A - North America, B - Indo-Pacific, C - West Eurasia, D - East Eurasia, E - Africa, F - Australia. Supports for specific reconstructions are indicated by the values to the right of the letter code.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, Q., Belyakov, E.A., Lapirov, A.G. et al. A reappraisal of the phylogeny and historical biogeography of Sparganium (Typhaceae) using complete chloroplast genomes. BMC Plant Biol 22, 588 (2022). https://doi.org/10.1186/s12870-022-03981-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03981-3