Abstract
Background
The proper identification of feather grasses in nature is often limited due to phenotypic variability and high morphological similarity between many species. Among plausible factors influencing this issue are hybridisation and introgression recently detected in the genus. Nonetheless, to date, only a bounded set of taxa have been investigated using integrative taxonomy combining morphological and molecular data. Here, we report the first large-scale study on five feather grass species across several hybrid zones in Russia and Central Asia. In total, 302 specimens were sampled in the field and classified based on the current descriptions of these taxa. They were then genotyped with high density genome-wide markers and measured based on a set of morphological characters to delimitate species and assess levels of hybridisation and introgression. Moreover, we tested species for past introgression and estimated divergence times between them.
Results
Our findings demonstrated that 250 specimens represent five distinct species: S. baicalensis, S. capillata, S. glareosa, S. grandis and S. krylovii. The remaining 52 individuals provided evidence for extensive hybridisation between S. capillata and S. baicalensis, S. capillata and S. krylovii, S. baicalensis and S. krylovii, as well as to a lesser extent between S. grandis and S. krylovii, S. grandis and S. baicalensis. We detected past reticulation events between S. baicalensis, S. krylovii, S. grandis and inferred that diversification within species S. capillata, S. baicalensis, S. krylovii and S. grandis started ca. 130–96 kya. In addition, the assessment of genetic population structure revealed signs of contemporary gene flow between populations across species from the section Leiostipa, despite significant geographical distances between some of them. Lastly, we concluded that only 5 out of 52 hybrid taxa were properly identified solely based on morphology.
Conclusions
Our results support the hypothesis that hybridisation is an important mechanism driving evolution in Stipa. As an outcome, this phenomenon complicates identification of hybrid taxa in the field using morphological characters alone. Thus, integrative taxonomy seems to be the only reliable way to properly resolve the phylogenetic issue of Stipa. Moreover, we believe that feather grasses may be a suitable genus to study hybridisation and introgression events in nature.
Similar content being viewed by others
Background
The proper delimitation of species plays an important role in taxonomy as well as in studies related to speciation, biogeography and ecology, leading to effective conservation and management of biodiversity. In the last two decades, traditional approaches relying mostly on morphological features have been supplemented by molecular data that boosted the discovery of new species. Although one estimate of the number of plant species is around 298,000 [1], recently it has been shown that the plant kingdom is comprised of at least 374,000 taxa [2]. Nowadays, many systematicists emphasise the need to apply multidisciplinary data, so-called integrative approaches or integrative taxonomy [3]. For instance, information from a variety of disciplines, e.g., morphology, biochemistry, cytogenetics and ‘omics studies, increases the reliability and validity in identifying taxa [4,5,6].
To date, among the molecular methods, DNA barcoding has been a widely utilised tool to identify taxa at different levels aiming not only to facilitate revisionary taxonomy, but also to broaden our understanding of molecular phylogenetics and population-level variations [7,8,9]. Among standard plant markers, chloroplast regions rbcL and matK and the nuclear internal transcribed spacer (ITS) locus have been proposed for DNA barcoding of land plants [10, 11]. Additionally, several non-coding plastid regions have been suggested as supplementary loci where further resolution is required [10].
Stipa is one of the largest genera in the grass subfamily Pooideae (Poaceae), currently comprising nearly 150 cool-season species with C3 photosynthesis common in Eurasia and North Africa [12, 13]. Based on ITS and the plastid trnK region, the genus has been proven to be monophyletic [14, 15]. Nonetheless, the traditional barcodes are not able to validate the sectional subdivision in Stipa proposed, e.g., by Tzvelev [16, 17] or Freitag [18]. Recently, the nuclear intergenic spacer (IGS) region [19] and marker sets derived from whole chloroplast genomes of 19 species [20] were proposed for phylogenetic studies of feather grasses. Although these markers are more phylogenetically informative in comparison to the previously used barcodes, they are still unable to discriminate all taxa, causing unresolved nodes in the reconstructed trees [19, 20].
One of the plausible explanations for this unresolved branching in Stipa is that many feather grasses are of hybrid origin [16, 21,22,23]. Presently, hybridisation is considered to be widespread among at least 25% of plant taxa, mostly the youngest species [24]. This phenomenon is often accompanied by introgression via repeated backcrossing to one or both parental species that can lead to diversification and speciation [25, 26]. In grasses, hybrid speciation has been explicitly studied at the genome level, e.g., in Triticum [27] and Brachypodium [28]. Nonetheless, previously many hybrids and introgressed individuals were characterised exclusively based on morphology that limited their successful identification in nature [29, 30]. In addition, hybridisation may lead new organisms not only to intermediate traits of parental species but also to extreme, or transgressive, phenotypes [31] that complicate their proper taxonomic treatment. In feather grasses, the hypothesis of hybrid origin of some species was initially tested using multivariate morphological analyses [23, 32] and, more recently, applying molecular markers among genetically closely related species in the Stipa heptapotamica complex [33] as well as within two genetically distant species, S. krylovii and S. bungeana [34]. Furthermore, due to the usage of integrative taxonomic approaches, it was shown that some Stipa taxa, previously assigned to S. richteriana and S. grandis, appeared to be cryptic species [33, 35]. Thus, taking into account that ca. 30% of feather grasses could be of hybrid origin [13] and that cryptic species are present, integrative taxonomy seems to be the only reliable way to properly resolve the phylogenetic issue of Stipa.
Importantly, the advent of next generation sequencing technologies, primarily Illumina, and the continuously reducing sequencing cost have facilitated the implementation of genomic data in studies of non-model organisms. For instance, high-throughput techniques based on restriction enzymes, e.g., RADseq [36] and genotyping-by-sequencing [37], which have been foremost used in agricultural species [38], are currently widely applied in phylogenetics and studies related to hybridisation in many wild plant genera with little or no previous genomic information [39,40,41]. Recently, a promising result has also been demonstrated in Stipa where the usage of the DArTseq technique resulted in an increased number of markers that was several 100-fold higher than in the previous genomic studies [34].
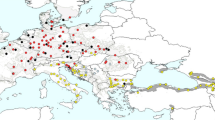
During field studies on steppe communities, we observed high morphological variability in populations of genetically related plants. In particular, we noticed that some specimens of feather grasses representing S. capillata, S. grandis and S. krylovii seemingly share mixed morphological characteristics between these species, while taxon S. baicalensis is frequently observed within populations of the aforesaid taxa and resembles an intermediate phenotype between S. grandis and S. krylovii or S. capillata and S. krylovii. The above-mentioned species have wide distribution ranges (Fig. 1 and Supplementary Table S1). Specifically, S. capillata is the most widespread taxon within the genus, grows on the dry grasslands and is common in Siberia, Western Asia and is also present in a limited number of refugia in Europe and North Africa. Two species, S. baicalensis and S. grandis, share similar ranges in southwestern Siberia, in the Baikal region, in the south part of Zabaykalsky Krai, in Mongolia and in northeastern China; S. baicalensis is also present in the south part of the Russian Far East, whereas S. grandis occurs in Central China and Tibet. Finally, S. krylovii grows in Siberia, Mongolia, China, Northern Nepal, Southern Tajikistan, Eastern Kazakhstan and Eastern Kyrgyzstan [13, 42].
The general distribution map of (a) S. baicalensis (yellow), S. capillata (red), S. grandis (green), S. krylovii (blue) and sampling locations (b) in East Kazakhstan and southwestern Siberia (Russia), (c) in southeastern Siberia and (d) in Eastern Kyrgyzstan. The dashed lines indicate hypothetical borders. The coloured circles depict species found in the numbered locations. The exact coordinates of the locations are presented in the Supplementary Table S1
We hypothesise that the observed variability in S. baicalensis, S. capillata, S. grandis and S. krylovii is due to the presence of interspecific hybrids and that may lead to species misidentification based on the current descriptions of these taxa [16, 43,44,45]. Thus, in the current study, we aim to use an integrative taxonomy approach to (1) delimitate species and test if S. baicalensis is a hybrid between S. grandis × S. krylovii or S. capillata × S. krylovii; (2) assess levels of hybridisation and introgression (if present) between the examined taxa and populations at the molecular level; (3) estimate divergence times between the studied taxa; (4) obtain insight into the extent of hybridisation between these species at the morphological level and (5) assess whether morphological characters can be used to identify hybrids in the field.
Results
DNA-based species delimitation
The DArTseq technique was applied to obtain a total of 8660 SNP markers to infer the genetic structure of 302 Stipa specimens. Firstly, analyses of genetic clustering with an unweighted pair group method using arithmetic average (UPGMA) and fastSTRUCTURE revealed five major clades corresponding to morphospecies S. glareosa, S. capillata, S. grandis, S. krylovii and S. baicalensis (Fig. 2). According to the fastSTRUCTURE analysis, the first and the fourth clades consisted exclusively of pure specimens of S. glareosa and S. krylovii, respectively. The remaining clades beside pure specimens of S. capillata, S. grandis and S. baicalensis included hybrid individuals. In particular, the second clade comprised pure specimens of S. capillata and the admixed individuals S. capillata × S. baicalensis and S. capillata × S. krylovii. The third cluster consisted of pure specimens of S. grandis and hybrids S. grandis × S. krylovii and S. grandis × S. baicalensis. The fifth clade included pure specimens of S. baicalensis and the admixed individuals S. baicalensis × S. krylovii.
The UPGMA dendrogram (at the top) aligned with the best supported fastSTRUCTURE model K = 5 (on the bottom). The genetic distance was calculated using the Jaccard Similarity Coefficient (y-axis, top). Individuals are represented by coloured bars according to the proportion of membership (y-axis, bottom) of a genotype to the respective cluster
In total, fastSTRUCTURE inferred 52 individuals with an admixture of two genetic clusters including an exception of S. capillata × S. baicalensis (0454631) that had a minor proportion (0.02) of a third cluster representing S. krylovii. Among pure individuals only one specimen of S. krylovii (0454646) had an insignificant admixed proportion (0.03) of S. grandis. Noteworthy, the vast majority of admixed individuals (49 or 94%) had a proportion of membership in the range from 0.46 to 0.54 indicating F1 hybrids or later generations of hybrids that have no backcrossing to the parental species. The remaining admixed samples represented: (1) one individual (0477009) was formed by a 0.78–0.22 admixture between S. baicalensis and S. krylovii evidencing a first-generation backcross (F1× S. baicalensis), (2) one individual (000948) was shared between S. grandis (0.88) and S. krylovii (0.12) indicating a second-generation backcross (first-generation backcross × S. grandis), (3) one individual (000956) was admixed between S. grandis (0.64) and S. krylovii (0.36) that may suggest a first-generation backcross (F1× S. grandis) or a more complex backcross to S. grandis via different intermediate combinations.
A consistent result was also found with a principal coordinates analysis (PCoA). The first three axes explained 29.6, 19.9 and 19.2% of the total genetic divergence within the studied taxa, respectively. According to the PCoA, pure individuals were grouped into five markedly differentiated groups correspondingly to their taxonomic classifications (Fig. 3; an interactive version of the three-dimensional plot can be accessed via https://plot.ly/~eugenebayahmetov/40/). The remaining hybrids F1 had intermediate positions between their parental species. Two hybrid individuals S. grandis × S. krylovii (000948 and 000956) were grouped closer to S. grandis reflecting a higher proportion of membership with the first taxon established earlier with fastSTRUCTURE. Similarly, an admixed individual S. baicalensis × S. krylovii (0477009) with the proportion of 0.78 and 0.22 was closer to S. baicalensis.
Hybrid generation identification
The NewHybrids analysis revealed a more complex pattern of hybridisation than it was inferred with fastSTRUCTURE. Among 16 admixed specimens of S. baicalensis × S. krylovii, previously assigned as F1, six individuals with posterior probabilities (PP) in a range of 0.84 and 1.00 were identified being F2 (F1× F1) hybrids suggesting that F1 hybrids are able to reproduce further. One specimen, S. baicalensis × S. krylovii (0477009), was proven to be a first-generation backcross (F1× S. baicalensis) having PP of 0.81. In addition, five mixed individuals had PP between two categories (F1 and F2 hybrids) in a range of 0.22–0.78 suggesting uncertainty in the assignment (Fig. 4a and Supplementary Table S2). These mixed assignment individuals may represent a more advanced hybrid generation than can be detected by NewHybrids. Within 14 hybrids of S. capillata × S. krylovii, previously assigned to F1 hybrids, we detected six individuals of F1 (PP of 0.87–1.00) and one specimen was identified as an F2 hybrid with PP of 0.83. The remaining seven individuals had mixed assignments in a range of 0.39–0.73 for the F1 class and 0.27–0.61 for the F2 class, respectively (Fig. 4b). The analysis also demonstrated that 13 out 14 hybrids of S. capillata × S. baicalensis were F1 (PP of 0.86–1.00) and one individual remained unclassified sharing PP (0.54 and 0.46) between F1 and F2 categories (Fig. 4c). Among S. grandis × S. krylovii only one F1 hybrid was detected (PP of 0.91), two individuals were assigned to the F2 class (PP of 0.83 and 1.00) and two specimens were classified as first generation backcrosses (F1× S. grandis) having PP of 0.88 and 0.99. Although the specimen 000948 was inferred to be an F1 backcross, it is more plausible that it represents rather a second-generation backcross established by fastSTRUCTURE due to NewHybrids being unable to detect more advanced backcrosses than F1. Additionally, one individual had mixed assignments between the F1 (PP of 0.27) and F2 (PP of 0.72) classes (Fig. 4d). Finally, the only hybrid detected for S. grandis × S. baicalensis appeared to be a first generation hybrid with PP of 1.00 (Fig. 4e).
The assignment of Stipa taxa into four hybrid classes according to the posterior probabilities (y-axis) inferred in NewHybrids. a S. baicalensis × S. krylovii, (b) S. capillata × S. krylovii, (c) S. capillata × S. baicalensis, (d) S. grandis × S. krylovii, (e) S. grandis × S. baicalensis. Hybrid classes are coloured by black (F1 hybrid), grey (F2), cyan (backcross to the first parental species, BC to parent 1) and pink (backcross to the second parental species, BC to parent 2)
Testing for introgression
A total of 6894 SNP markers were used to test for reticulation events between the studied taxa. Due to five different admixed combinations detected by fastSTRUCTURE and NewHybrids, we tested all possible four-species combinations regardless of their phylogenetic positions (Fig. 2). The results of the f4 statistic suggest no gene flow between S. capillata and the remaining species because of negligible deviations from the expected 50/50 ratio of BABA/ABBA patterns and the lowest Z-scores of any tests (Table 1). This finding disagrees with the presence of contemporary hybrids S. capillata × S. krylovii and S. capillata × S. baicalensis inferred with fastSTRUCTURE and NewHybrids. Nonetheless, it can be explained by the fact that all identified admixed individuals were excluded from this analysis. On the other hand, introgression events were suggested between S. grandis and S. baicalensis (combinations 5 and 11), S. grandis and S. krylovii (combinations 4, 6, 7 and 8), S. krylovii and S. baicalensis (combinations 9 and 12). Additionally, when S. grandis, S. krylovii and S. baicalensis were analysed together (combinations 4, 7 and 10) the ratio of BABA/ABBA patterns were either almost equal (combination 10) or relatively lower (combinations 4 and 7) compared to the other tests that indicated gene flow among these species. One potential explanation is that these species are involved in introgression at the same rate, which theoretically cancel out each other.
Population differentiation
A total of 3483 SNP markers were used to investigate the genetic differentiation in populations of S. baicalensis, 6288 SNPs in S. capillata, 4635 SNPs in S. grandis and 6912 SNPs in S. krylovii (Supplementary Fig. S1). The pairwise Fst values demonstrated strong differentiation among four populations of S. baicalensis, while the results of STRUCTURE and PCoA revealed two and three genetic clusters, respectively (Fig. 5a and Supplementary Fig. S2), where populations 1 and 4 are merged (Fst of 0.32, Supplementary Table S3) regardless of the fact that the distance between them is more than 1000 km. Additionally, the second most likely K according to STRUCTURE was K = 3 indicating that this number of clusters is also a likely option. Relatively strong differentiation was also shown for populations of S. capillata with an exception of populations 5 and 6 with a moderate Fst value of 0.13, suggesting potential gene flow. According to the PCoA, almost all individuals were grouped together excluding population 3 and a few specimens of populations 2 and 5. Nonetheless, the first two axes of PCoA explained only 18% of the total genetic divergence within the specimens. On the other hand, STRUCTURE supported K = 4 as the best fitting model, while two and nine clusters were also among the probable options (Fig. 5b and Supplementary Fig. S2). Within S. grandis, evidence for weak differentiation was shown for geographically close populations 5 and 6 (Fst of 0.10) as well as for 7 and 8 (Fst of 0.08), while the first two axes of the PCoA explained 23.3% of the variation and revealed the close genetic relationship between geographically distant populations 1 and 2 (Fst of 0.42). In addition, based on the second axis of the PCoA, populations 3 and 4 were distant to each other as well as to the remaining populations (Fig. 5c). Further, the STRUCTURE analysis suggested that K = 2 was the most probable number of separate clusters within S. grandis (Supplementary Fig. S2), where one pure cluster represented individuals from populations 7 and 8, while 9 out of 13 specimens of population 3 and 4 constituted the second pure group. The rest of S. grandis specimens were admixed between these pure clusters. Lastly, based on the PCoA individuals of S. krylovii were clustered into the two main groups representing population 8 from Kyrgyzstan and the remaining populations from Russia (Fig. 5d). Although few specimens of population 1 were genetically more distant to the other individuals, the second axis of PCoA explained only 5.4% of the total genetic divergence. The pairwise Fst values also supported the division within S. krylovii into two groups indicating the strong differentiation of population 8 (Fst in a range of 0.37–0.46) and a moderate or near to moderate differentiation among the remaining populations (Fst in a range of 0.06–0.19). Similarly, the STRUCTURE analysis revealed two genetic clusters (Supplementary Fig. S2) that described the population structure the best: the first one represented population 8 from Kyrgyzstan, while the second cluster comprised populations from Russia.
Divergence-time estimation
The SNAPP phylogeny based on 2717 SNP markers among pure individuals revealed largely the same topology as the UPGMA dendrogram, except for the pair S. grandis and S. krylovii that were grouped together, while S. baicalensis was a sister taxon (Fig. 6 and Supplementary Fig. S3). The result suggests that the potential split between S. capillata and three species, namely, S. grandis, S. krylovii and S. baicalensis, took place approximately 1.07 Mya with the 95% Highest Posterior Density interval (HPD) of 1.51–0.71 Mya. The most recent common ancestor for S. grandis, S. krylovii and S. baicalensis was inferred to be 0.79 Mya (95% HPD: 1.12–0.53 Mya), whereas the lowest divergence time of 0.73 Mya was registered for S. grandis and S. krylovii (95% HPD: 1.02–0.48 Mya). The chronogram also indicates that diversification within S. capillata, S. krylovii and S. baicalensis started at relatively the same time, ca. 130–114 kya (95% HPD: 181–74 kya), while S. grandis was established to be the youngest species (96 kya, 95% HPD: 137–63 kya). Lastly, although the topologies within the species had large uncertainty, some nodes had comparatively high Bayesian posterior probabilities (BPP ≥ 0.80; Supplementary Fig. S3). In particular, individuals of S. capillata from Kyrgyzstan started to differentiate 97 kya (95% HPD: 137–58 kya), while the well-supported split (BPP of 0.92) between specimens from localities 5 (Russia, the Republic of Khakassia) and 19 (Russia, the Republic of Buryatia) took place 86 kya (95% HPD: 122–53 kya). The most recent common ancestor for individuals of S. krylovii from Kyrgyzstan was inferred to be 62 kya (95% HPD: 94–35 kya), whereas specimens from localities 22 (Mongolia) and 24 (Russia, Zabaykalsky Krai) had the potential split 68 kya (95% HPD: 99–40 kya). Interestingly, specimens of S. baicalensis from localities 11 and 13 (both from Russia, the West shore of Lake Baikal) and individuals from the remaining localities had nearly the same divergence times of 97 kya (95% HPD: 140–60 kya) and 93 kya (95% HPD: 132–58 kya), respectively. Additionally, the potential split between populations of S. grandis from the Republic of Khakassia (locality 5) and the Republic of Buryatia (locality 18) took place 65 kya (95% HPD: 91–39 kya), while the well-supported split (BPP of 0.95) between specimens from localities 23 and 24 (both from Russia, Zabaykalsky Krai) took place 58 kya (95% HPD: 86–34 kya).
Phylogeny and divergence date estimates inferred by SNAPP. Blue coloured trees represent the most probable topology. Numbers at each node represent mean ages of divergence time estimates and the 95% HPD intervals (in the brackets). The black rectangles on the nodes indicate the 95% HPD intervals of the estimated posterior distributions of the divergence times. The red circle indicates the presumed divergence time split set as a reference. The Bayesian posterior probabilities were 1.00 for the nodes with the shown 95% HPD intervals. The scale shows divergence time in Mya
Morpho-molecular analysis
As non-parametric Spearman correlation coefficients did not demonstrate any strong correlation (> 0.90) between the measured variables (Supplementary Fig. S4), we retained all morphological characters (Table 3) for a factor analysis of mixed data (FAMD) and analyses of notch plots. Subsequently, to investigate if the observed phenotypes are congruent with molecular data, we supplemented the result of the FAMD analysis with the genetic clusters inferred by fastSTRUCTURE for the best K = 5. As a result, the FAMD revealed five markedly differentiated groups (Fig. 7) of operational taxonomic units (OTUs) in accordance with the detected clusters using the SNP data (Figs. 2, 3 and 4). The first four dimensions explained 31.0, 18.9, 10.9 and 8.1% of the total variability, respectively. The first three axes are mainly composed by the contribution of 11 quantitative and four qualitative variables (Supplementary Table S4).
The factor analysis of mixed data performed on 17 quantitative and six qualitative characters of the five examined species of Stipa. a Plot of the two principal axes. b Plot of the three principal axes. The pie charts represent the proportions of membership established by fastSTRUCTURE for the best K = 5
Importantly, due to having the genetic clusters assigned by fastSTRUCTURE, the two-dimensional plot revealed the slight overlapping of OTUs belonging to the pure species S. baicalensis and S. krylovii, whereas OTUs representing the admixed specimens S. baicalensis × S. krylovii were present in both clouds of the parental taxa (Fig. 7a). Furthermore, hybrids S. capillata × S. baicalensis were also present in both clouds of the pure species. On the other hand, all the admixed individuals S. capillata × S. krylovii were grouped together with only one parental species, S. capillata. Interestingly, hybrids between S. grandis and S. krylovii were mainly grouped together with OTUs of S. baicalensis with an exception of the first- and the second-generation backcrosses. The only hybrid detected between S. grandis and S. baicalensis was clustered together with the pure individuals of the former taxon. A more clear dispersal of the pure species can be seen in the three-dimensional plot, where differences between the studied species are explained by the third principal axis (Fig. 7b; an interactive version of the plot available at https://plot.ly/~eugenebayahmetov/42/). Additionally, all combinations of two axes plots for the four dimensions are present in Supplementary Fig. S5.
The result of FAMD and notch plots of the quantitative variables demonstrated that the studied species can be differentiated morphologically mainly based on the length of the lower segment of the awn (Col1L), the distance from the end of the dorsal line of hairs to the top of the lemma (DDL), the length of the anthecium (AL), the length of the middle segment of the awn (Col2L), the length of the callus (CL), the length of ligules of the internal vegetative shoots (LigIV), the length of ligules of the middle cauline leaves (LigC) and the length of hairs on the top of the lemma (LHTA). In addition, the length of hairs on the lower segment of the awn (HLCol1), the length of hairs on the middle segment of the awn (HLCol2) and the length of the callus base (CBL) can aid to distinguish S. glareosa from the remaining taxa (Supplementary Fig. S6). For instance, the notch plot of Col1L showed significant differences between means and strong evidence of differing medians within the pure species except the pair S. baicalensis and S. capillata, while the hybrid individuals had mostly intermediate positions between the parental species except specimens of S. capillata × S. baicalensis that were significantly different from the samples of the pure taxa. The differences across all quantitative variables between individuals of the pure species and the hybrids can be better evaluated in the interactive box plots presented in Supplementary File 1.
Among the qualitative variables, the main contribution to the axes of the FAMD had the abaxial surface of vegetative leaves (AbSVL), the type of hairs on the top of the anthecium (HTTA), the type of the awn geniculation (AG) and the presence of hairs below nodes (PHBN) (Supplementary Table S4). For instance, vegetative leaves with prickles were common in S. glareosa (all samples), S. capillata (61 out of 66 samples), S. capillata × S. krylovii (12 out of 14 samples), S. capillata × S. baicalensis (10 out of 14 samples), less frequent in S. grandis (2 out of 54 samples), S. krylovii (5 out of 81 samples), S. baicalensis × S. krylovii (2 out of 14 samples) and totally absent in S. baicalensis, S. grandis × S. krylovii and S. grandis × S. baicalensis (Supplementary Fig. S7).
Thus, based on the results of the molecular analyses and the FAMD combining both phenotypic and SNP data, we were able to differentiate the pure species and the hybrid individuals at the morphological level. We established that using the traditional identification keys [16, 43,44,45] 71 out of 302 specimens had been misidentified, mostly due to their hybrid nature (Supplementary Table S5). In particular, 47 samples previously identified as pure species appeared to be hybrids. The remaining 24 specimens earlier were classified either as hybrids (15 samples) or misleadingly assigned to S. baicalensis (9 samples). Interestingly, in the latter case, all individuals were previously reported from the northeastern part of Kazakhstan [46].
In general, S. baicalensis was the most problematic species for taxonomic identification comprising 54 doubtful samples. Specifically, the above-mentioned specimens from Kazakhstan genetically were proven to be pure S. capillata, 10 specimens appeared to be hybrids S. capillata × S. baicalensis, 8 were S. baicalensis × S. krylovii and 10 were hybrids between S. capillata and S. krylovii. On the other hand, specimens previously identified as hybrids S. baicalensis × S. krylovii (3 samples) and S. grandis × S. krylovii (2 samples) were genetically assigned to the pure species S. baicalensis. Oppositely, specimens morphologically identified as S. krylovii (8 samples), S. grandis (1 sample) and S. capillata (1 sample) appeared to be hybrids S. baicalensis × S. krylovii (6 samples) and S. capillata × S. baicalensis (2 samples), S. grandis × S. baicalensis and S. capillata × S. baicalensis, respectively. Additionally, one specimen S. capillata × S. grandis was proven to be S. capillata × S. baicalensis. At last, one individual S. capillata × S. baicalensis was genetically verified to be the pure species S. capillata.
The remaining doubtful samples morphologically were either misleadingly assigned as the pure species or as taxa of hybrid nature. For instance, 4 specimens of S. capillata were hybrids between S. capillata and S. krylovii, while specimens of S. grandis (2 samples) and S. krylovii (2 samples) were shown to be genetically admixed as S. grandis × S. krylovii. In opposite, 9 individuals identified as S. capillata × S. krylovii appeared to be the pure species S. capillata. Interestingly, only 5 out of 52 hybrid taxa were properly identified based on morphology including S. baicalensis × S. krylovii (3 samples) and S. grandis × S. krylovii (2 samples). The only one taxon that had not any questionable individuals was S. glareosa representing the section Smirnovia.
Discussion
The current understanding of taxonomy and species limits in Stipa is still largely based on morphological characters. Our study highlights the necessity of using molecular tools to properly identify taxa and detect processes underlying speciation. This is of particular relevance in hybrid zones where ongoing hybridisation and introgression may lead admixed individuals to phenotypes similar to one of the parental species, complicating identification of such taxa using morphological characters alone. Furthermore, integrative studies suggest that apparently intermediate phenotypes between two species are not necessarily hybrids [47]. On the other hand, as indicated in the present work, although some interspecific hybrids have intermediate characters between parental taxa, their phenotypic traits can also overlap with non-parental species leading to misidentification. Given the circumstances, here we utilised an integrative approach combining genome-wide data and morphology to delimitate species and ascertain the extent of hybridisation in feather grasses.
The study clearly illustrates that a molecular-based analysis, e.g., such as fastSTRUCTURE, combined with a factor analysis of mixed data, utilising both quantitative and qualitative variables, can largely resolve the problem with species identification in the face of ongoing hybridisation and introgression. Primarily, such an approach allows to visualise and easily trace if the observed phenotypes are congruent with molecular data. Besides that, this approach may aid in selecting a set of traits that can be useful for species identification in the field. Our findings clearly demonstrate that the studied individuals can be clustered into five species groups representing S. capillata, S. baicalensis, S. glareosa, S. grandis and S. krylovii. Thus, here we found no evidence that S. baicalensis is of hybrid origin from S. grandis × S. krylovii or S. capillata × S. krylovii but is instead a genetically distinct species. The general branching of the phylogenetic trees is in good agreement with the current taxonomic classification. In particular, a representative of the section Smirnovia, S. glareosa, was genetically distant to the remaining species from the section Leiostipa. Nevertheless, our result contradicts a previous research, where S. capillata, S. krylovii and S. baicalensis represented one clade and S. grandis was a sister taxon [19], while here, such a sister taxon was S. capillata. The current result is likely more accurate due to applying several thousand SNPs across the genome in comparison to only one nuclear locus in the above-mentioned study. Additionally, we demonstrated that the potential split between S. capillata and the remaining representatives of Leiostipa took place approximately 1.07 Mya, which is similar to our previous estimate of 1.73 Mya based on the nucleolar organising regions [48]. Here, we also reported for the first time that diversification within species S. capillata, S. baicalensis, S. krylovii and S. grandis started ca. 130–96 kya (95% HPD: 181–63 kya). These ages may correspond to the potential window of time between the Last Interglacial period, which began around 130 kya, and the Last Glacial Period (LGP), which started about 110 kya. Thus, the observed pattern is similar to dispersing events reported for different taxa across the plant kingdom [49, 50] suggesting climatic changes as a feasible factor in the current distribution of feather grasses. Of note, due to the divergence times that were inferred in SNAPP, which uses the multi-species coalescent model ignoring possible introgression, the confidence may be exaggerated. On the other hand, although introgression does cause biased errors in coalescent-based species tree inference [51,52,53], it should not affect the estimates in the present study, since all admixed individuals were excluded from the analysis.
Importantly, the results of molecular analyses were congruent and provided evidence for the existence of hybridisation between pairs S. capillata × S. baicalensis, S. capillata × S. krylovii, S. grandis × S. krylovii, S. grandis × S. baicalensis and S. baicalensis × S. krylovii. The presence of F2 or more advanced hybrid generations suggests that F1 individuals are able to reproduce further. This observation is in agreement with our previous findings on hybridisation within the genus [32, 33], where a direct approach proved that a hybrid taxon S. heptapotamica produces fertile pollen grains and is capable of backcrossing to primarily one parental species [33]. Indeed, here we detected backcrosses S. baicalensis × S. krylovii and S. grandis × S. krylovii to their former parental species. Moreover, the analysis of BABA/ABBA patterns among species revealed signs of past introgression between S. baicalensis and S. krylovii, S. baicalensis and S. grandis, S. krylovii and S. grandis. Taking into account the diversification times of these species, we can hypothesise that if such a gene flow occurred it was relatively recent in evolutionary terms and seemingly still present between S. baicalensis and S. krylovii, as well as in pair S. grandis and S. krylovii. Nevertheless, we treat our BABA/ABBA analysis with caution due to such a test originally being applied in human studies whose genome is available at the chromosome level and the number of SNPs was remarkably higher than used here. Thus, we intend to reassess the past gene flow within these taxa when a more continuous genome will be obtained. Additionally, although here we did not detect any backcrosses between S. baicalensis and S. grandis, it cannot be excluded that such a combination exists in nature, especially since both species are mostly common in Mongolia and China; however specimens from there were not presented in the study. Lastly, our study revealed only unidirectional backcrossing of hybrid taxa either to S. baicalensis or S. grandis, but not to S. krylovii. Similarly, due to the relatively small sample size and the limited number of localities used here, we cannot draw any reliable conclusions concerning possible barriers to gene flow to the latter taxon.
According to our results, the populations’ expansion seemingly started during the LGP. Nonetheless, the assessment of genetic structures revealed signs of contemporary gene flow between populations across all species, despite significant geographical distances between some of them. For instance, populations of S. baicalensis, S. capillata and S. grandis from the eastern part of Khakassia and the southwestern area of Buryatia represented either one genetic cluster or were admixed between two. Among potential explanations are shifting their distributions in response to climate change, or seasonal migration of wild animals and livestock grazing. While we currently do not possess enough data to verify the first assumption, seeds of feather grasses usually are spread naturally by wind or water. On the other hand, they also can be frequently dispersed by the wool of mammals including, e.g., sheep, goats and horses [16, 18]. Although this study was not intended to explore population differences in detail, we believe that our findings regarding gene flow merit further studies in order to better understand the intraspecific variation and relationships among populations as well as to discuss potential consequences of such events.
Our results also illustrated a complex association between species at the morphological level. There are usually few phenotypic characters differentiating hybridising species and these characters are often functionally or developmentally correlated [54]. In the present case, although the studied species had a set of distinctive characters, the current identification keys do not provide a solution on distinguishing admixed individuals in the section Leiostipa. As a result, only 5 out of 52 hybrid taxa were properly identified based on morphology. Furthermore, the results demonstrated that the most problematic taxon is S. baicalensis, which was frequently misidentified either with S. capillata or a cross S. capillata × S. krylovii, while a few individuals were misleadingly assigned to S. grandis × S. krylovii. Additionally, several hybrids between S. baicalensis and S. capillata were determined as pure S. krylovii. Therefore, we believe that the identification keys should be revised in order to properly delimitate pure species and propose a taxonomic treatment for the hybrid taxa identified in the study. Moreover, for a more comprehensive morphological assessment we suggest using scanning electron microscopy that can assist in finding unique ultrastructures among pure and admixed individuals.
Although here we highlight that morphological characters alone cannot be used to properly identify hybrids and backcrossed individuals in the field, it is a common issue in plants rather than an exception in feather grasses. For instance, in a study on three species of willows it was shown that based on phenotype only 5% of specimens were classified as introgressed individuals, which was much less than the 19% detected using SNP data [55]. Another research on tropical trees demonstrated that even limited genomic sampling, when combined with morphology and geography, can greatly improve estimates of species diversity for clades where hybridisation contributes to taxonomic difficulties [56]. Recently, an investigation on several pine species using SNP data derived from the DArTseq pipeline revealed that one species, previously considered of hybrid origin, is a genetically distinct species and provided insights into the challenges of solely using morphological traits when identifying taxa with cryptic hybridisation and variable morphology [57].
In grasses, hybridisation and introgression phenomena are still mainly studied in crop species, e.g., rice [58], wheat [59] and sugarcane [60]. To date, we have detected hybrids not only between genetically closely related species [33], but also among genetically distant Stipa taxa [13, 34]. The results present here and our previous findings helps us to shift toward thinking of the Stipa phylogeny as reticulate webs rather than a strictly bifurcating tree. Nonetheless, studying hybridisation in feather grasses is not only of particular interest to plant taxonomists. The presence of parental species, multiple generation hybrids and backcrosses in different proportions in a hybrid zone may indicate renewed sympatry providing important data for studying species boundaries and patterns of speciation [61]. From this point of view, Stipa may be a suitable genus to study these phenomena. Despite the increasing interest in feather grasses at the molecular level [19, 20, 48, 62,63,64,65], there is still a lack of substantial knowledge regarding, e.g., chromosome numbers of admixed and pure taxa in hybrid zones, fertility of pollen grains in F1 and later generation hybrids and backcrosses and genomic information related to specific loci contributing to reproductive barriers. We believe that only an integrative approach combining the aforesaid data can properly interpret evolutionary patterns and processes in feather grasses.
Conclusions
In the current study we revealed a complex taxonomic issue in feather grasses with variable morphology exhibited due to extensive hybridisation. Based on SNPs derived from genome-wide genotyping we detected five genetic groups representing separate morphospecies and showed that S. baicalensis is a genetically distinct species instead of a taxon of hybrid origin as it was previously hypothesised. We demonstrated the presence of F1 hybrids between S. capillata × S. baicalensis, S. capillata × S. krylovii, S. grandis × S. krylovii, S. grandis × S. baicalensis, S. baicalensis × S. krylovii and F2 individuals in S. capillata × S. krylovii, S. grandis × S. krylovii, S. baicalensis × S. krylovii indicating low levels of reproductive isolation in these species. We also discovered a few backcrosses S. baicalensis × S. krylovii and S. grandis × S. krylovii to their former parental species suggesting possible introgression among the taxa.
Furthermore, we detected reticulation events between S. baicalensis and S. krylovii, S. baicalensis and S. grandis, S. krylovii and S. grandis. On the other hand, we revealed signs of contemporary gene flow between populations of the species from the section Leiostipa. Another important outcome of the research is divergence date estimates inferred at the species and population levels. Here we deduce that diversification within the studied species started ca. 130–96 kya and hypothesise that climatic changes during the LGP were a driving force behind the current distribution of feather grasses. Importantly, here we also emphasise the usefulness of applying integrative approaches combining molecular and morphological data to delimitate species and detect hybridisation and introgression events in feather grasses. Finally, we conclude that Stipa may be a suitable genus to study these phenomena.
Methods
Plant material
In total, 302 fully developed Stipa samples were used for molecular and morphological studies. We gathered individuals representing the section Leiostipa (S. baicalensis, S. capillata, S. grandis and S. krylovii) from localities where these taxa grow together as well as from areas where they grow separately from each other (Fig. 1, Supplementary Table S1). Additionally, we included two populations of S. baicalensis previously reported from the northeastern part of Kazakhstan [46] and 13 specimens of S. glareosa belonging to the section Smirnovia that were found in localities 10, 12, 13 and 16. All voucher specimens used in the study are preserved at TK and KRA. All maps were visualised using ArcGIS Pro 2.7.1 (ESRI, Redlands, USA). The species distribution ranges were established based on the revision of herbarium specimens preserved at AA, ALTB, B, BM, BRNU, COLO, E, FR, FRU, G, GAT, GFW, GOET, IFP, K, KAS, KFTA, KHOR, KRA, KRAM, KUZ, L, LE, LECB, M, MO, MSB, MW, NY, P, PE, PR, PRC, TAD, TASH, TK, UPS, W, WA, WU and Z.
DNA extraction, amplification and DArT sequencing
This section was performed according to the previously reported procedures [34]. In brief, genomic DNA was isolated from dried leaf tissues using a Genomic Mini AX Plant Kit (A&A Biotechnology, Poland) and sent to Diversity Arrays Technology Pty Ltd. (Canberra, Australia) for the following genome complexity reduction using restriction enzymes and high-throughput polymorphism detection [66].
All DNA samples were processed in digestion/ligation reactions as described previously [66], but replacing a single PstI-compatible adaptor with two different adaptors corresponding to two different restriction enzyme overhangs. The PstI-compatible adapter was designed to include Illumina flowcell attachment sequence, sequencing primer sequence and “staggered”, varying length barcode region, similar to the sequence previously reported [37]. The reverse adapter contained a flowcell attachment region and MseI-compatible overhang sequence. Only “mixed fragments” (PstI-MseI) were effectively amplified by PCR using an initial denaturation step of 94 °C for 1 min, followed by 30 cycles with the following temperature profile: denaturation at 94 °C for 20 s, annealing at 58 °C for 30 s and extension at 72 °C for 45 s, with an additional final extension at 72 °C for 7 min. After PCR equimolar amounts of amplification products from each sample of the 96-well microtiter plate were bulked and applied to c-Bot (Illumina, USA) bridge PCR followed by sequencing on Hiseq2500 (Illumina, USA). The single read sequencing was performed for 77 cycles.
Sequences generated from each lane were processed using proprietary DArT analytical pipelines. In the primary pipeline, the fastq files were first processed to filter away poor quality sequences, applying more stringent selection criteria to the barcode region compared to the rest of the sequence. In that way the assignments of the sequences to specific samples carried in the “barcode split” step were reliable. Approximately 2.5 mln sequences per barcode/sample were identified and used in marker calling.
DArTseq data analysis
For the downstream analyses, we applied co-dominant single nucleotide polymorphism (SNP) markers processed in the R-package dartR v.1.5.5 [67] with the following parameters: (1) a scoring reproducibility of 100%, (2) SNP loci with read depth < 5 or > 50 were removed, (3) at least 95% loci called (the respective DNA fragment had been identified in greater than 95% of all individuals), (4) monomorphic loci were removed, (5) SNPs that shared secondaries (had more than one sequence tag represented in the dataset) were randomly filtered out to keep only one random sequence tag.
DNA-based species delimitation
Five approaches were used to analyse the genetic structure at the species level: (1) Unweighted Pair Group Method with Arithmetic Mean (UPGMA), (2) fastSTRUCTURE analysis, (3) Principal Coordinates Analysis (PCoA), (4) NewHybrids analysis, (5) calculation of the f4 statistic. In addition, to assess the genetic differentiation at the population level within S. baicalensis, S. capillata, S. grandis and S. krylovii we performed PCoA and STRUCTURE analyses and calculated FST. Furthermore, we used SNAPP to estimate divergence times within the studied species and populations.
Firstly, a UPGMA cluster analysis based on Jaccard’s distance matrix was performed using R-packages dartR and visualised with stats v.3.6.2 [68]. Next, the genetic structure was investigated using fastSTRUCTURE v.1.0, which implements the Bayesian clustering algorithm STRUCTURE, assuming Hardy-Weinberg equilibrium between alleles, in a fast and resource-efficient manner [69]. A number of clusters (K-values) ranging from 1 to 10 were tested using the default parameters with ten replicate runs per dataset. The most likely K-value was estimated with the best choice function implemented in fastSTRUCTURE. The output matrices for the best K-values were reordered and plotted using the R package pophelperShiny v.2.1.0 [70]. We applied the threshold of 0.10 < q < 0.90 as the most widely utilised measure for the assessment of hybridisation [71, 72] with q-values > 0.9 being pure species and 0.45 < q < 0.55 being F1 hybrids, while first- and second-generation backcrosses with one parent were considered at q-values 0.25 and 0.125, respectively [73]. Then, a PCoA on a Euclidean distance matrix was performed using the R-package dartR and visualised with ggplot2 v.3.3.0 [74] to show the first two components and plotly v.4.9.2 [75] to illustrate the first three components.
Hybrid generation identification
Next, to assign individuals to a genetic category (pure, hybrid F1 or F2, backcross hybrid) based on their multilocus genotypes, we performed analyses using a Bayesian model-based clustering method implemented in NewHybrids v.1.1 [76] via the R package dartR. The program utilises Markov chain Monte Carlo (MCMC) simulations to compute the posterior probability of an individual belonging to pre-defined categories comparing two parental genotypes at a time. Thus, we used five data sets representing parental species and their hybrids defined by the UPGMA, fastSTRUCTURE and PCoA outputs from the previous steps. Specifically, we had the following datasets: (1) S. baicalensis, S. krylovii and the admixed individuals between the parental species; (2) S. baicalensis, S. capillata and the admixed individuals; (3) S. grandis, S. krylovii and the admixed individuals; (4) S. capillata, S. krylovii and the admixed individuals; (5) S. baicalensis, S. grandis and the admixed individuals. Due to limitations of NewHybrids, only 200 loci were used per run. In the beginning, the first 200 loci ranked on information content (option “AvgPIC”) were chosen via dartR. Then, to ensure the convergence of the algorithm, we used ten different 200-SNP subsets selected in random (option “random”). The Jeffreys prior was used for θ and π and a burn-in of 10,000 MCMC generations followed by 10,000 post burn-in sweeps. The resulting posterior probabilities were calculated based on 11 runs and a probability threshold > 0.8 was set for the assignment into a genetic category. The calculated posterior probabilities for the assigned categories were visualised using the R package pophelperShiny.
Testing for introgression
Further, to calculate the f4 statistic we retained only pure individuals determined via the UPGMA, fastSTRUCTURE and PCoA analyses. All filtering steps were conducted using the R-package dartR with the above-mentioned sequence. Next, the processed data was converted into the EIGENSTRAT format using the R-package dartR. The subsequent calculation of f4 statistics was performed in ADMIXTOOLS v. 7.0.1 [77] using the R package admixr v.0.9.1 [78]. In brief, the f4 statistic [79] is similar to the D statistic [80, 81] and measures the average correlation in allele frequency differences between four populations, e.g., A, B, C and D [82]. If a divergent outgroup is provided as population A, we can test for gene flow between B and C (if the statistic is negative and Z-score < − 3) or B and D (if the statistic is positive and Z-score > 3). Due to such a test originally being applied to continuous genome-wide data, it is important to mention some limitations of the analysis while working with non-model organisms. If a draft genome is available, it is possible to specify positions of SNPs along contigs or scaffolds by a special parameter “blockname” implemented in ADMIXTOOLS. Nonetheless, the package currently does not support more than 600 contigs/scaffolds [83] decreasing the potential number of used SNPs for very fragmented genomes. That was the case of Stipa, where only one draft genome comprising 5931 contigs is currently available [48]. Additionally, SNPs positions and genetic distances are used for a block jackknife method to test for a significant deviation from the null expectation of the f4 statistic. Thus, in the absence of a reference genome or the presence of a very fragmented genome the f4/D statistics should be treated with caution. Here we consider signatures of past gene flow events only if BABA or ABBA patterns are greater than 50. All possible four-species combinations were tested, while one species, S. glareosa, was selected as an outgroup (A) for all runs.
Population differentiation
To perform PCoA and STRUCTURE analyses and calculate FST at the population level we retained only pure species and kept populations with more than three individuals per population. To maintain as many populations as possible, we merged several individuals growing relatively close to each other (Table 2 and Supplementary Table S1). All filtering steps were conducted using the R-package dartR with the above-mentioned sequence. PCoA analyses were also performed according to the aforesaid flow. Next, we used STRUCTURE v.2.3.4 [84] instead of fastSTRUCTURE due to the former software being markedly superior to the latter one under weak population differentiation [85]. To overcome the relatively slow speed of the analysis we applied parallel computing using StrAuto v.1.0 [86]. Five replicate runs were performed for each number of clusters (K) from one to ten with a burn-in of 50,000 iterations followed by 500,000 MCMC iterations. The optimal K value was identified based on Evanno’s method of ΔK statistics [87] as implemented in Structure Harvester [88]. The calculated posterior probabilities for the assigned categories were visualised using the R package pophelperShiny. Then, the fixation index Fst was calculated using the R-package dartR with 1000 bootstraps to obtain p-values. This measure assesses genetic differentiation among populations, where values of 0.00–0.05 indicate low differentiation, 0.05–0.15 indicate moderate differentiation, while Fst > 0.15 indicates high levels of differentiation [89].
Divergence-time estimation
Finally, to estimate divergence times between the studied taxa, we retained only one pure individual per species and population using dartR and followed the above-mentioned sequence of filtering steps with an exception of the called loci (100% instead of 95%). Then, the SNP data was converted to the Nexus format using dartR. Subsequently, we created an XML file using the SNAPP-specific template provided in Stange et al., 2018. SNAPP v.1.5.1 [90] utilises the multi-species coalescent approach and is well-suited for analyses of genome-wide data deducing the species tree and divergence times directly from SNPs [91]. We applied one time calibration, setting a log-normal distribution with a mean of 4.39 Mya and a standard deviation (SD) of 0.18 for the split between S. glareosa (section Smirnovia) and S. capillata (section Leiostipa) as it was inferred in our previous study [48]. The analysis was performed three times independently, 1.25 million MCMC generations for each run using BEAST2 v.2.6.3 [92]. Tracer v.1.7.1 [93] was used to visually check the combined log file regarding Effective Sample Size (ESS) values. As all ESSs exceeded 200, we combined tree files using LogCombiner v.2.6.3 (a part of the BEAST package) with the first 10% discarded as burn-in from each run. The final maximum clade credibility tree was summarised in TreeAnnotator v.2.6.3 (a part of the BEAST package). Lately, we visualised a pattern across all of the posterior trees via DensiTree v.2.01 [94], while FigTree v.1.4.4 [95] was used to inspect the Bayesian posterior probabilities and the 95% credibility intervals of the final tree.
Morphological analysis
A total of 302 specimens were examined under a light microscope SMZ800 (Nikon, Japan) across the 17 most informative quantitative and six qualitative morphological characters commonly used in keys and taxonomic descriptions of Stipa (Table 3). Firstly, the Shapiro-Wilk test was used in the R-package MVN v.5.8 [96] to assess the normality of the distribution of each characteristic. Secondly, the non-parametric Spearman’s correlation test was applied using R-packages stats and Hmisc v.4.3–1 [97] to examine relations between the studied characters. The combined correlogram with the significance test was visualised using the R-package corrplot v.0.84 [98].
Next, a Factor Analysis of Mixed Data (FAMD) [99] was accomplished using the R-package FactoMineR v.2.3 [100] to characterise the variation within and among groups of taxa without a priori taxonomic classification and to extract the variables that best identified them. The number of principal components used in the analysis was chosen based on Cattell’s scree test [101]. R-packages factoextra v.1.0.6 [102] and plotly were used to visualise the first two and the first three principal components, respectively. Subsequently, the plots were supplemented with the result of the fastSTRUCTURE analysis for the best K = 5.
Additionally, to evaluate distributional relationships between each response variable and the studied taxa, notch plots and interactive box plots were created using R-packages ggplot2 v.3.3.0 and plotly v.4.9.2, respectively. The notched box plots display a confidence interval around the median, which is normally based on the median ± 1.57 × interquartile range/square root of n. According to this graphical method for data analysis, if the notches of the two boxes do not overlap, there is “strong evidence” (95% confidence) that their medians differ. In addition, to reveal significant differences between means of particular characters across all examined taxa the nonparametric Kruskal-Wallis test followed by the post-hoc Wilcoxon test with Bonferroni correction were performed using the R-package stats v.3.6.2.
Availability of data and materials
The SNP dataset derived from the DArTseq pipeline in the genlight format is available via Figshare repository, https://doi.org/10.6084/m9.figshare.14461802.
Change history
08 January 2022
A Correction to this paper has been published: https://doi.org/10.1186/s12870-021-03357-z
References
Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. How many species are there on earth and in the ocean? PLoS Biol 2011;9(8):e1001127. https://doi.org/10.1371/journal.pbio.1001127.
Christenhusz MJM, Byng JW. The number of known plants species in the world and its annual increase. Phytotaxa. 2016;261(3):201–217. https://doi.org/10.11646/phytotaxa.261.3.1
Dayrat B. Towards integrative taxonomy. Biol J Linn Soc 2005;83:407–415. https://doi.org/10.1111/j.1095-8312.2005.00503.x.
Raupach MJ, Amann R, Wheeler Q, Roos C. The application of "-omics" technologies for the classification and identification of animals. Org Divers Evol 2016;16:1–12. https://doi.org/10.1007/s13127-015-0234-6.
Massó S, López-Pujol J, Vilatersana R. Reinterpretation of an endangered taxon based on integrative taxonomy: the case of Cynara baetica (Compositae). PLoS One 2018;13(11):e0207094. https://doi.org/10.1371/journal.pone.0207094.
Lovrenčić L, Bonassin L, Boštjančić LL, Podnar M, Jelić M, Klobučar G, et al. New insights into the genetic diversity of the stone crayfish: taxonomic and conservation implications. BMC Evol Biol 2020;20:146. https://doi.org/10.1186/s12862e-020-01709-1.
Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet 2007;23(4):167–172. https://doi.org/10.1016/j.tig.2007.02.001.
Kress WJ. Plant DNA barcodes: applications today and in the future. J Syst Evol 2017;55(44):291–307. https://doi.org/10.1111/jse.12254.
Tizard J, Patel S, Waugh J, Tavares E, Bergmann T, Gill B, et al. DNA barcoding a unique avifauna: an important tool for evolution, systematics and conservation. BMC Evol Biol 2019;19:52. https://doi.org/10.1186/s12862-019-1346-y.
CBOL Plant Working Group. A DNA barcode for land plants. P Natl Acad Sci USA 2009;106(31):12794–12797. https://doi.org/10.1073/pnas.0905845106.
Li DZ, Gao LM, Li HT, Wang H, Ge XJ, Liu JQ, et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. P Natl Acad Sci USA. 2011;108(49):19641–19646. https://doi.org/10.1073/pnas.1104551108.
Nobis M. Taxonomic revision of the central Asiatic Stipa tianschanica complex (Poaceae) with particular reference to the epidermal micromorphology of the lemma. Folia Geobot 2014;49:283–308. https://doi.org/10.1007/s12224-013-9164-2.
Nobis M, Gudkova P, Nowak A, Sawicki J, Nobis A. A revision of the genus Stipa (Poaceae) in middle Asia, including a key to species identification, an annotated checklist and phytogeographic analysis. Ann Mo Bot Gard 2020;105:1–63. https://doi.org/10.3417/2019378.
Hamasha HR, von Hagen KB, Röser M. Stipa (Poaceae) and allies in the Old World: molecular phylogenetics realigns genus circumscription and gives evidence on the origin of American and Australian lineages. Plant Syst Evol 2012;298:351–367. https://doi.org/10.1007/s00606-011-0549-5.
Romaschenko K, Peterson PM, Soreng RJ, Garcia-Jacas N, Futorna O, Susanna A. Systematics and evolution of the needle grasses (Poaceae: Pooideae: Stipeae) based on analysis of multiple chloroplast loci, ITS, and lemma micromorphology. Taxon. 2012;61:18–44. https://doi.org/10.1002/tax.611002.
Tzvelev NN. Zlaki USSR. Leningrad: Nauka Press; 1976.
Tzvelev NN. Notes on the tribe Stipeae Dumort. (Poaceae). Novosti Sist Vyssh Rast. 2012;43:20–9.
Freitag H. The genus Stipa (Gramineae) in southwest and South Asia. Notes Roy Bot Gard Edinburgh. 1985;42:355–489.
Krawczyk K, Nobis M, Nowak A, Szczecińska M, Sawicki J. Phylogenetic implications of nuclear rRNA IGS variation in Stipa L. (Poaceae). Sci Rep. 2017;7:11506. https://doi.org/10.1038/s41598-017-11804-x.
Krawczyk K, Nobis M, Myszczyński K, Klichowska E, Sawicki J. Plastid superbarcodes as a tool for species discrimination in feather grasses (Poaceae: Stipa). Sci Rep 2018;8:1924. https://doi.org/10.1038/s41598-018-20399-w.
Smirnov PA. Stiparum Armeniae minus cognitarum descriptiones. Byull Moskovsk Obshch Isp Prir Otd Biol. 1970;75:113–5.
Kotukhov YA. Synopsis of feather grass (Stipa L.) and false needlegrasses (Ptilagrostis Griseb.) the eastern of Kazakhstan (the Kazakh Altai, Zaisan valley and Prialtayskie ranges). Bot Issl Sib Kazakhst. 2002;8:3–16.
Nobis M. Taxonomic revision of the Stipa lipskyi group (Poaceae: Stipa section Smirnovia) in the Pamir alai and tian-Shan Mountains. Plant Syst Evol 2013;299:1307–1354. https://doi.org/10.1007/s00606-013-0799-5.
Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol 2005;20:229–237. https://doi.org/10.1016/j.tree.2005.02.010.
Hegarty MJ, Hiscock SJ. Hybrid speciation in plants: new insights from molecular studies. New Phytol 2005;165:411–423. https://doi.org/10.1111/j.1469-8137.2004.01253.x.
Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, et al. Hybridization and speciation. J Evol Biol 2013;26:229–246. https://doi.org/10.1111/j.1420-9101.2012.02599.x.
Matsuoka Y, Takumi S, Nasuda S. Genetic mechanisms of allopolyploid speciation through hybrid genome doubling: novel insights from wheat (Triticum and Aegilops) studies. Int Rev Cel Mol Bio 2014;309:199–258. https://doi.org/10.1016/b978-0-12-800255-1.00004-1.
Gordon SP, Contreras-Moreira B, Levy JJ, Djamei A, Czedik-Eysenberg A, Tartaglio VS, et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat Commun 2020;11:3670. https://doi.org/10.1038/s41467-020-17302-5.
Rieseberg LH, Ellstrand NC. What can molecular and morphological markers tell us about plant hybridization. Crit Rev Plant Sci 1993;12:213–241. https://doi.org/10.1080/07352689309701902.
Hardig TM, Brunsfeld SJ, Fritz RS, Morgan M, Orians CM. Morphological and molecular evidence for hybridization and introgression in a willow (Salix) hybrid zone. Mol Ecol 2000;9:9–24. https://doi.org/10.1046/j.1365-294x.2000.00757.x.
Rieseberg L, Archer M, Wayne R. Transgressive segregation, adaptation and speciation. Heredity. 1999;83:363–372. https://doi.org/10.1038/sj.hdy.6886170.
Nobis M, Nowak A, Nobis A, Nowak S, Żabicka J, Żabicki P. Stipa ×fallax (Poaceae: Pooideae: Stipeae), a new natural hybrid from Tajikistan, and a new combination in Stipa drobovii. Phytotaxa. 2017;303:141–154. https://doi.org/10.11646/phytotaxa.303.2.4
Nobis M, Gudkova PD, Baiakhmetov E, Żabicka J, Krawczyk K, Sawicki J. Hybridisation, introgression events and cryptic speciation in Stipa (Poaceae): a case study of the Stipa heptapotamica hybrid-complex. Perspect Plant Ecol Evol Syst 2019;39:125457. https://doi.org/10.1016/j.ppees.2019.05.001.
Baiakhmetov E, Nowak A, Gudkova PD, Nobis M. Morphological and genome-wide evidence for natural hybridisation within the genus Stipa (Poaceae). Sci Rep 2020;10:13803. https://doi.org/10.1038/s41598-020-70582-1.
Nie B, Jiao BH, Ren LF, Gudkova PD, Chen WL, Zhang WH. Integrative taxonomy recognized a new cryptic species within Stipa grandis from loess plateau of China. J Syst Evol 2020. https://doi.org/10.1111/jse.12714.
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One 2008;3:e3376. https://dx.doi.org/10.1371%2Fjournal.pone.0003376
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 2011;6:e19379. https://doi.org/10.1371/journal.pone.0019379.
Poland JA, Rife TW. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012;5:92–102. https://doi.org/10.3835/plantgenome2012.05.0005.
Schilling MP, Gompert Z, Li FW, Windham MD, Wolf PG. Admixture, evolution, and variation in reproductive isolation in the Boechera puberula clade. BMC Evol Biol 2018;18:61. https://doi.org/10.1186/s12862-018-1173-6.
Wagner ND, Gramlich S, Hörandl E. RAD sequencing resolved phylogenetic relationships in European shrub willows (Salix L. subg. Chamaetia and subg. Vetrix) and revealed multiple evolution of dwarf shrubs. Ecol Evol 2018;8(16):8243–8255. https://doi.org/10.1002/ece3.4360.
Hodkinson TR, Perdereau A, Klaas M, Cormican P, Barth S. Genotyping by sequencing and plastome analysis finds high genetic variability and geographical structure in Dactylis glomerata L in Northwest Europe despite lack of ploidy variation Agronomy 2019;9:342. https://doi.org/10.3390/agronomy9070342.
Gudkova PD, Olonova MV, Feoktisov DS. The comparison of ecologo-climatic niches of two species feather grass Stipa sareptana a.K. Becker and S. krylovii Roshev. (Poaceae). Ukr J Ecol 2017;7:263–269. https://doi.org/10.15421/2017_115
Tzvelev NN. Gramineae. In: Grubov VI, editor. Plantae Asiae Centralis (secus materies Instituti Botanici nomine V. L. Komarovii). Leningrad: Nauka; 1968. p. 1–243.
Wu ZL, Phillips SM. Stipa. In: Wu ZY, et al., editors. Flora of China. Beijing: Science Press; 2006. p. 196–203.
Tzvelev NN, Probatova NS. Grasses of Russia. Moscow: KMK Scientific Press; 2019.
Kotukhov YA, Anufrieva OA. Addition to the flora of Kazakhstan. Terra. 2008:48–55.
Curtu AL, Gailing O, Finkeldey R. Evidence for hybridization and introgression within a species-rich oak (Quercus spp.) community. BMC Evol Biol 2007;7:218. https://doi.org/10.1186/1471-2148-7-218.
Baiakhmetov E, Guyomar C, Shelest E, Nobis M, Gudkova PD. The first draft genome of feather grasses using SMRT sequencing and its implications in molecular studies of Stipa. Sci Rep 2021;11:15345. https://doi.org/10.1038/s41598-021-94068-w.
Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, et al. (2017). African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. P Natl Acad Sci USA. 2017;114(20):5213–5218. https://doi.org/10.1073/pnas.1616736114.
Cornejo-Romero A, Vargas-Mendoza CF, Aguilar-Martínez GF, Medina-Sánchez J, Rendón-Aguilar B, Valverde PL, et al. Alternative glacial-interglacial refugia demographic hypotheses tested on Cephalocereus columna-trajani (Cactaceae) in the intertropical Mexican drylands. PLoS One 2017;12(4):e0175905. https://doi.org/10.1371/journal.pone.0175905.
Long C, Kubatko L. The effect of gene flow on coalescent-based species-tree inference. Syst Biol 2018;67(5):770–785. https://doi.org/10.1093/sysbio/syy020.
Jaio X, Flouri T, Rannala B, Yang Z. The impact of cross-species gene flow on species tree estimation. Syst Biol 2020;69(5):830–847. https://doi.org/10.1093/sysbio/syaa001.
Jiao X, Yang Z. Defining species when there is gene flow. Syst Biol 2021;70(1):108–119. https://doi.org/10.1093/sysbio/syaa052.
Rieseberg L, Wendel J. lntrogression and Its Consequences in Plants. In: Harrison RG, editor. Hybrid Zones and the Evolutionary Process. Oxford: Oxford University Press; 1993. p. 70–109.
Fogelqvist J, Verkhozina AV, Katyshev AI, Pucholt P, Dixelius C, Rönnberg-Wästljung AC, et al. Genetic and morphological evidence for introgression between three species of willows. BMC Evol Biol 2015;15:193. https://doi.org/10.1186/s12862-015-0461-7.
Federman S, Donoghue MJ, Daly DC, Eaton DAR. Reconciling species diversity in a tropical plant clade (Canarium, Burseraceae). PLoS One 2018;13(6):e0198882. https://doi.org/10.1371/journal.pone.0198882.
Buck R, Hyasat S, Hossfeld A, Flores-Rentería L. Patterns of hybridization and cryptic introgression among one- and four-needled pinyon pines. Ann Bot 2020;126(3):401–411. https://doi.org/10.1093/aob/mcaa045.
Civán P, Brown TA. Role of genetic introgression during the evolution of cultivated rice (Oryza sativa L.). BMC Evol Biol. 2018;18:57. https://doi.org/10.1186/s12862-018-1180-7.
Cheng H, Liu J, Wen J, Nie X, Xu L, Chen N, et al. Frequent intra- and inter-species introgression shapes the landscape of genetic variation in bread wheat. Genome Biol 2019;20:136. https://doi.org/10.1186/s13059-019-1744-x.
Pachakkil B, Terajima Y, Ohmido N, Ebina M, Irei S, Hayashi H, et al. Cytogenetic and agronomic characterization of intergeneric hybrids between Saccharum spp hybrid and Erianthus arundinaceus Sci Rep 2019;9:1748. https://doi.org/10.1038/s41598-018-38316-6.
Harrison RG, Larson EL. Hybridization, introgression, and the nature of species boundaries, J Hered 2014;105(1):795–809. https://doi.org/10.1093/jhered/esu033.
Yang YQ, Li X, Kong X, Ma L, Hu X, Yang Y. Transcriptome analysis reveals diversified adaptation of Stipa purpurea along a drought gradient on the Tibetan plateau. Funct Integr Genomic 2015;15:295–307. https://doi.org/10.1007/s10142-014-0419-7.
Wan D, Wan Y, Hou X, Ren W, Ding Y, Sa R. De novo assembly and transcriptomic profiling of the grazing response in Stipa grandis. PLoS One 2015;10:e0122641. https://doi.org/10.1371/journal.pone.0122641.
Schubert M, Marcussen T, Meseguer AS, Fjellheim S. The grass subfamily Pooideae: cretaceous–Palaeocene origin and climate-driven Cenozoic diversification. Glob Ecol Biogeogr 2019;28:1168–1182. https://doi.org/10.1111/geb.12923.
Yan D, Ren J, Liu J, Ding Y, Niu J. De novo assembly, annotation, marker discovery, and genetic diversity of the Stipa breviflora Griseb. (Poaceae) response to grazing. PLOS One. 2020;15(12): e0244222. https://doi.org/10.1371/journal.pone.0244222
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, et al. Diversity arrays technology: a generic genome profiling technology on open platforms. Methods Mol Biol 2012;888:67–89. https://doi.org/10.1007/978-1-61779-870-2_5.
Gruber B, Unmack PJ, Berry OF, Georges A dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing Mol Ecol Resour 2018;18:691–699. https://doi.org/10.1111/1755-0998.12745.
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2021. https://www.R-project.org. Accessed 23 Apr 2021.
Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. https://doi.org/10.1534/genetics.114.164350.
Francis RM. POPHELPER: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 2017;17(1):27–32. https://doi.org/10.1111/1755-0998.12509.
Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, Petit RJ, et al. Detection of hybrids in nature: application to oaks (Quercus suber and Q. ilex). Heredity. 2009;102:442–452. https://doi.org/10.1038/hdy.2009.8.
Winkler KA, Pamminger-Lahnsteiner B, Wanzenböck J, Weiss S. Hybridization and restricted gene flow between native and introduced stocks of Alpine whitefish (Coregonus sp.) across multiple environments. Mol Ecol. 2011;20:456–472. https://doi.org/10.1111/j.1365-294X.2010.04961.x.
Beugin MP, Gayet T, Pontier D, Devillard S, Jombart T. A fast likelihood solution to the genetic clustering problem. Methods Ecol Evol 2018;9:1006–1016. https://doi.org/10.1111/2041-210X.12968.
Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer; 2016.
Sievert C, Parmer C, Hocking T, Chamberlain S, Ram K, Corvellec M, et al. plotly: create interactive web graphics via 'plotly.js'. 2021. https://rdrr.io/cran/plotly. Accessed 23 Apr 2021.
Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus genetic data. Genetics. 2002;160(3):1217–29.
Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, et al. Ancient admixture in human history. Genetics. 2012;192(3):1065–1093. https://doi.org/10.1534/genetics.112.145037.
Petr M, Vernot B, Kelso J. Admixr–R package for reproducible analyses using ADMIXTOOLS. Bioinformatics. 2019;35(17):3194–3195. https://doi.org/10.1093/bioinformatics/btz030.
Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. https://doi.org/10.1038/nature08365.
Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. https://doi.org/10.1126/science.1188021.
Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol 2011;28:2239–2252. https://doi.org/10.1093/molbev/msr048.
Lipson M. Applying f4-statistics and admixture graphs: theory and examples. Mol Ecol Resour 2020;20(6):1658–1667. https://doi.org/10.1111/1755-0998.13230.
Taylor RS, Manseau M, Horn RL, Keobouasone S, Golding GB, Wilson PJ. The role of introgression and ecotypic parallelism in delineating intraspecific conservation units. Mol Ecol 2020;29(15):2793–2909. https://doi.org/10.1111/mec.15522.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59.
Stift M, Kolář F, Meirmans PG. STRUCTURE is more robust than other clustering methods in simulated mixed-ploidy populations. Heredity. 2019;123:429–441. https://doi.org/10.1038/s41437-019-0247-6.
Chhatre VE, Emerson KJ. StrAuto: automation and parallelization of STRUCTURE analysis. BMC Bioinformatics 2017;18:192. https://doi.org/10.1186/s12859-017-1593-0.
Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 2005;14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x.
Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 2012;4(2):359–361. https://doi.org/10.1007/s12686-011-9548-7.
Hartl DL, Clark AG. Principles of population genetics. 3rd ed. Sunderland: Sinauer Associates; 1997.
Bryant D, Bouckaert R, Felsenstein J, Rosenberg NA, RoyChoudhury A. Inferring species trees directly from Biallelic genetic markers: bypassing gene trees in a full coalescent analysis. Mol Biol Evol 2012;29(8):1917–1932. https://doi.org/10.1093/molbev/mss086.
Stange M, Sánchez-Villagra MR, Salzburger W, Matschiner M. Bayesian divergence-time estimation with genome-wide single-nucleotide polymorphism data of sea catfishes (Ariidae) supports Miocene closure of the Panamanian isthmus. Syst Biol 2018;67(4):681–699. https://doi.org/10.1093/sysbio/syy006.
Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. Plos Comput Biol. 2019;15(4):e1006650. https://doi.org/10.1371/journal.pcbi.1006650.
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian Phylogenetics using tracer 1.7. Syst Biol 2018;67(5):901–904. https://doi.org/10.1093/sysbio/syy032.
Bouckaert RR. DensiTree: making sense of sets of phylogenetic trees. Bioinformatics. 2010;26(10):1372–1373. https://doi.org/10.1093/bioinformatics/btq110.
Rambaut A. Figtree v1.4.4. 2021. http://tree.bio.ed.ac.uk/software/figtree. Accessed 23 Apr 2021.
Korkmaz S, Goksuluk D, Zararsiz G. Mvn: an r package for assessing multivariate normality. R J 2014;6:151–162. https://doi.org/10.32614/rj-2014-031
Harrell FEJ. Hmisc: Harrell Miscellaneous. 2021. https://CRAN.R-project.org/package=Hmisc. Accessed 23 Apr 2021.
Wei T, Simko V. R package "Corrplot": visualization of a correlation matrix. 2021. https://github.com/taiyun/corrplot. Accessed 23 Apr 2021.
Pagès J. Analyse Factorielle de Donnees Mixtes. Rev Stat Appl. 2004;4:93–111.
Lê S, Josse J, Husson F. FactoMineR: an R package for multivariate analysis. J Stat Softw 2008;25(1):1–18. https://doi.org/10.18637/jss.v025.i01
Cattell RB. The scree test for the number of factors. Multivariate Behav Res. 1966;1:245–76.
Kassambara A. Factoextra: extract and visualize the results of multivariate data analyses. 2021. https://rdrr.io/cran/factoextra. Accessed 23 Apr 2021.
Acknowledgements
We would like to express our gratitude to two anonymous reviewers for providing valuable comments on the manuscript.
Funding
The study was supported by a RSF grant (project no. 19–74-10067), partially by a DS grant of the Jagiellonian University (446.31150.2.2020) and by the National Science Centre, Poland (grant no. 2018/29/B/NZ9/00313). The open-access publication of this article was funded by the BioS Priority Research Area under the program “Excellence Initiative – Research University” at the Jagiellonian University in Krakow.
Author information
Authors and Affiliations
Contributions
E.B., M.N., P.D.G. supervised the study. P.D.G., M.N. planned the field studies, carried out the sampling, revised herbarium materials and performed the taxonomic identification. P.D.G., M.N. established the general distribution of the species and D.R. created the map. D.R., E.B., P.D.G. conducted the morphological measurements. E.B. performed all the analyses and wrote the manuscript. All authors revised the draft, provided comments and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: the author identified an error in Supplementary Materials. Additional File 1, Interactive box plots, is missing. Additionally, revised and high resolution figures should also be captured.
Supplementary Information
Additional file 1.
Interactive box plots.
Additional file 2: Supplementary Table S1
. List of samples used in the study. Table S2. Average posterior probabilities inferred in NewHybrids for first (F1) and second (F2) generation hybrids and backcrosses (F1xP1 and F1xP2). Table S3. Pairwise Fst values for population differentiation across the four studied species. Fst > 0.15 indicating high levels of differentiation are in bold type. Table S4. Contribution (%) by dimension of each character (abbreviations according to Table 3) in FAMD. The first five characters contributing the most are in bold type. Abbreviations of the qualitative variables and their contributions to the principal axes are underlined. Table S5. The assigned species names based on morphological and molecular data. Mismatches are shown in bold type. Supplementary Figure S1 Venn diagram representing polymorphic SNPs among four pure Stipa species. The admixed individuals and S. glareosa, which did not show patterns of hybridisation, were omitted in the metric’s calculation. Supplementary Figure S2 Delta K values calculated by Evanno’s method across four species. (a) S. baicalensis. (b) S. capillata. (c) S. grandis. (d) S. krylovii. Supplementary Figure S3 Phylogeny (at the top) and divergence date estimates at the species level (on the bottom) inferred by SNAPP. The scale shows divergence time in Mya. The red circles indicate nodes with the Bayesian posterior probabilities (BPP) > 0.8. The lower-case letters refer to the embedded table containing data regarding the exact estimates of the divergence times (in kya), BPPs and 95% HPD intervals. Supplementary Figure S4 Correlation matrix of the studied morphological characters (abbreviations according to Table 3). Colour intensity and the size of the circle are proportional to the correlation coefficients (displayed in the circle). Positive correlations are blue while negative are red. All p-values of Pearson correlations were < 0.01. Supplementary Figure S5 Factor analysis of mixed data performed on 17 quantitative and six qualitative characters of the five examined species of Stipa. (a) Plot of the principal axes one and two. (b) Plot of the principal axes one and three. (c) Plot of the principal axes one and four. (d) Plot of the principal axes two and three. (e) Plot of the principal axes two and four. (f) Plot of the principal axes three and four. The pie charts represent the proportions of membership established by fastSTRUCTURE for the best K = 5. Supplementary Figure S6 Notched boxplot demonstrating the mean (white circle), the median (dark black line), 95% confidence interval around the median (notch), inter-quartile ranges (25 to 75%), whiskers (5 and 95%) and minimum and maximum measurements (crosses) of quantitative characters (a-q) for the studied species. Statistical significance was tested by Wilcoxon rank-sum test for post hoc group comparisons with Bonferroni correction, p < 0.001, p < 0.01, p < 0.05, p < 0.1 and p < 1 noted as ‘***’, ‘**’, ‘*’, ‘.’ and no symbol, respectively. Due to the small sample size, p-values cannot be properly estimated for S. grandis × S. krylovii and S. grandis × S. baicalensis. Each dot represents an observation. Supplementary Figure S7 Bar charts displaying frequencies of the qualitative characters. (a) AbSVL. (b) AdSVL. (c) HTTA. (d) AG. (e) CN. (f) PHBN.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Baiakhmetov, E., Ryzhakova, D., Gudkova, P.D. et al. Evidence for extensive hybridisation and past introgression events in feather grasses using genome-wide SNP genotyping. BMC Plant Biol 21, 505 (2021). https://doi.org/10.1186/s12870-021-03287-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-021-03287-w