Abstract
Background
Alfalfa (Medicago sativa L.) is an important forage crop grown worldwide. Alfalfa is called the “queen of forage crops” due to its high forage yield and nutritional characteristics. The aim of this study was to undertake quantitative trait loci (QTL) mapping of yield and yield-related traits in an F1 population of two alfalfa varieties that differ in their yield and yield-related traits.
Results
We constructed a high-density linkage map using single nucleotide polymorphism (SNP) markers generated by restriction-site associated DNA sequencing (RAD-seq). The linkage map contains 4346 SNP and 119 simple sequence repeat (SSR) markers, with 32 linkage groups for each parent. The average marker distances were 3.00 and 1.32 cM, with coverages of 3455 cM and 4381 cM for paternal and maternal linkage maps, respectively. Using these maps and phenotypic data, we identified a total of 21 QTL for yield and yield components, including five for yield, five for plant height, five for branch number, and six for shoot diameter. Among them, six QTL were co-located for more than one trait. Five QTL explained more than 10% of the phenotypic variation.
Conclusions
We used RAD-seq to construct a linkage map for alfalfa that greatly enhanced marker density compared to previous studies. This high-density linkage map of alfalfa is a useful reference for mapping yield-related traits. Identified yield-related loci could be used to validate their usefulness in developing markers for maker-assisted selection in breeding populations to improve yield potential in alfalfa.
Similar content being viewed by others
Background
Biomass yield is the most important trait for alfalfa production and increasing yield is the primary goal for improving alfalfa. Yield is a complex quantitative trait affected by the interaction of genetics and the environment (G × E). Understanding the genetic basis of yield-related traits can help increase yield in alfalfa [1]. However, cultivated alfalfa is autotetraploid and its genetic composition is complex [2]. The outcrossing nature and high heterozygosity of alfalfa increase challenges in genetic analyses. Quantitative trait loci (QTL) have been mapped to study yield-related traits in alfalfa [3,4,5]. However, the use of low-density genetic markers such as SSRs (simple sequence repeats) and RFLPs (restriction fragment length polymorphisms) for mapping genetic loci has limited the resolution of QTL intervals in alfalfa.
SNP markers are abundant and have high coverage in plant genomes compared to RFLP and SSR markers [6]. In alfalfa, high density genetic linkage maps have been constructed using SNP markers [7] and SNPs have also been used for genome-wide association studies (GWAS) [8,9,10,11]. SNP markers can be generated in many ways including RNA-seq, genotyping-by-sequencing (GBS), and restriction associated DNA sequencing (RAD-seq). The application of RAD-seq has been reported in many species, such as pear [12], carnation [13], and eggplant [14]. However, RAD-seq has not been used in alfalfa. Using RAD-seq to develop SNP markers and construct a high density genetic linkage map to increase resolution of QTL mapping and reduce QTL interval length [15] would be valuable for identifying causal loci to enhance traits of interest.
In the present study, we used an F1 population of alfalfa to map QTL for yield-related traits. Genotyping was done using RAD-seq followed by SNP calling. The resulting SNPs were used to construct a high-density linkage map. Using this map, we were able to map yield-related QTL in the alfalfa population. The ultimate objective is to identify QTL and closely linked markers that can be used for molecular breeding to improve alfalfa yield production.
Results
Analysis of phenotypic variation
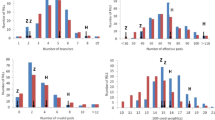
Phenotypic data for yield, plant height, shoot diameter, and branch number were analyzed using best linear unbiased prediction (BLUP). The minimum and maximum BLUP values for yield were − 65.71 and 61.65, respectively. The range of values for F1 plants was wider than that of the parents, reflecting the presence of transgressive segregation (Table 1). The same was true for plant height, shoot diameter, and branch number (Table 1). The kurtosis and skewness of yield and branch number were close to zero. Broad sense heritability (H2) was calculated as described in a previous study [16]. The H2 values were 46.7, 58.4, 34.9, and 32.0% for yield, plant height, shoot diameter, and branch number, respectively (Table 1). Analysis of variance (ANOVA) for yield, plant height, shoot diameter, and branch number were conducted using PROC GLM (SAS Institute, 2010). Genotypic variations were significant for yield, plant height, shoot diameter, and branch number (p < 0.001) when the mixed model of genotype*years*location was used (Table 2). Significant differences were also found for all traits analyzed when genotype*year and genotype*location models were used (Table 2). Correlation analysis showed a significant correlation according to Pearson’s test (P < 0.01) among four yield-related traits (Table 3). The correlation coefficient between yield and plant height is 0.79. The correlation coefficient between yield and shoot diameter is 0.51. The correlation coefficient between yield and branch number is 0.82 (Table 3).
Genetic linkage map
A total of 113,837 SNP markers were obtained after genotype calling with the Universal Network Enable Analysis Kit (UNEAK) pipeline. After removal of markers with missing values > 50%, 82,668 SNP markers were retained. Of those, 2317 were single-dose allele (SDA) markers in P1 and 4553 SDA in P2. For SSR markers, 56 and 84 SDA markers were obtained in P1 and P2, respectively. The SNP markers and SSR markers were combined to construct linkage maps. Thirty-two linkage groups were obtained for each parent (Fig. 1). The chromosome numbers were assigned based on SSR markers with known chromosome positions and the alignment of SNP markers sequences to the physical map of Medicago truncatula.
The P1 linkage map spanned a total of 3455 cM with 1153 mapped markers and an average marker density of 3.00 cM. The highest number of markers were found in linkage group (LG) 1A (106 markers) and lowest in LGs 1C and 1D (16 markers in each LG) (Table 4). The P2 linkage map spanned a total of 4381 cM with 3312 mapped markers and an average marker density of 1.32 cM (Fig. 1a and Table 4). The highest number of markers was found in LG 4B (201 markers) and the lowest in LG 7C (31 markers) (Fig. 1b, Table 4).
QTL mapping
Phenotypic data was collected from five environments: 3 years from Langfang (LF2014, LF2015, LF2016) and 2 years from Taizhou (TZ2014, TZ2015) were used for QTL mapping. Phenotypic data were analyzed using BLUP and the BLUP values were used for QTL mapping. A total of 21 QTL were identified, including five QTL for yield, five QTL for plant height, five QTL for branch number, and six QTL for shoot diameter in two parents (Table 5). Five QTL explained more than 10% of the phenotypic variation (PVE). The PVE ranged from 4.4 to 13.6% for yield related traits. Among those, three pairs of QTL were co-located: qyield-1 and qbranch-2, qyield-2 and qbranch-3, and qheight-5 and qdiameter-6 (Figs. 2 and 3). Sequences and SNP variance of the nearest markers of every QTL were supplied (Table 6).
Yield related QTL on 32 linkage groups from a genetic linkage map of parent 1. Each linkage group is named based on Medicago truncatula, with four group ordered A to D randomly. The name qyield, qheight, qdiameter and qbranch represent the QTL of yield, plant height, shoot diameter and branch number using blup value. Distances among markers are indicated in cM to the left of the linkage groups; names of markers are shown on the right. Only those SNP markers are shown which were in and around the QTL regions. QTLs are depicted as colored vertical bars to the right of the linkage groups. Different colors represent different traits. Those groups have not detected QTL are not been shown
Yield related QTL on 32 linkage groups from a genetic linkage map of parent 2. Each linkage group is named based on Medicago truncatula, with four group orderd A to D randomly. The name qyield, qheight, qdiameter and qbranch represent the QTL of yield, plant height, shoot diameter and branch number using BLUP values. Distances among markers are indicated in cM to the left of the linkage groups; names of markers are shown on the right. Only those SNP markers are shown which were in and around the QTL regions. QTLs are depicted as colored vertical bars to the right of the linkage groups. Different colors represent different traits. Those groups have not detected QTL are not been shown
Yield
QTL analysis identified a total of five QTL for yield. The phenotypic variation explained (PVE) by a single QTL ranged from 7.2 to 13.6%. The highest value of PVE, 13.6%, was found for the QTL qyield-1. It was located on linkage group 3C of P1 at the position from 98.0 to 99.8 cM with a logarithm of odds (LOD) value of 4.3 (Table 5). The QTL qyield-1 overlapped with the reported QTL for forage yield (72.3–102.2 cM) [3] and root dry weight (93–117 cM) [17]. Similarly, the other four QTL (qyield-2, qyield-3, qyield-4, and qyield-5) were also detected within a similar position as QTL for yield. The QTL qyield-2 was in a similar location as other reported QTL for forage yield (43.8–54.5 cM) [3] and root dry weight (39–55 cM) [17]. The QTL qyield-2 identified in the present study was also co-located with the reported QTL for fall plant height QTL [17] and seeds per seedpod QTL [18]. The QTL qyield-3 was located within the reported QTL for shoot dry weight and winter injury [17]. The QTL qyield-4 was located at a similar position as the reported QTL for crown dry weight (8–30 cM) and fall plant height (24–40 cM) [17]. The QTL qyield-5 was in a similar location as reported QTL for forage yield (42.3–48.8 cM) and winter hardiness (47.6–50.1 cM) [19].
Plant height
Five QTL with PVE ranging from 6.1 to 9.8% were identified for plant height. They were mapped to five LGs (1B, 5A, 7D, 8B, 8D). All of these QTL were identified in the high yield parent (P2). However, contrary to QTL qheight-2, qheight-3, and qheight-5, QTL qheight-1 and qheight-4 had negative effects (Add< 0) on plant height (Table 5). There were two QTL (qheight-2 and qheight-3) mapped within similar positions of reported QTL for plant height in alfalfa. The QTL qheight-2 identified in this study was located at a similar position as the reported QTL for fall plant height (48–68 cM), shoot dry weight (50–70 cM), and root dry weight (52–70 cM) [17]. The QTL qheight-3 was located within a QTL for fall plant height identified in a previous study [17]. Additional reports of QTL for fall dormancy (47.3–52 cM) [19] were mapped in the same chromosome region as qheight-3. Furthermore, qheight-4 was located within the genomic location of shoot dry weight (0–17 cM) and root dry weight (0–27 cM) [17].
Shoot diameter
For shoot diameter, six QTL were identified, explaining 7.1 to 13.6% of the phenotypic variance. The LOD value ranged from 3.4 to 6.9. The QTL qdiameter-5 on LG 2D (position 12.5–16.6 cM) had the highest LOD of 6.9 and highest PVE of 13.6% (Table 5). The QTL qdiameter-5 was located in a similar location as previously reported QTL for crown dry weight (8–30 cM) in alfalfa [17]. Similarly, three other QTL (qdiameter-1, qdiameter-2, and qdiameter-3) were located in a similar position as yield-related traits. The QTL qdiameter-1 was in a location similar to QTL for root dry weight (60–70 cM) [17] and drought-stressed forage biomass (59–72 cM) [20]. The QTL qdiameter-2 was located in a similar position as reported QTL for shoot dry weight (0–8 cM) [17]. The QTL qdiameter-3 was in a similar location as reported QTL for shoot dry weight (0–23 cM), crown dry weight, and root dry weight (17–29 cM) [17]. The QTL qdiameter-4 was in a similar location as reported QTL for fall dormancy (21.1–23.7 cM) [19].
Branch number
Among the five QTL identified for branch number, two QTL (qbranch-2 and qbranch-4) had PVE greater than 10%. The QTL qbranch-2 was located on linkage group 3C of P1 at the position from 98.0 to 99.8 cM. The PVE of qbranch-2 was 13.5%. The QTL qbranch-4 was located on linkage group 6A of P1 at the position from 87.6 to 90.5 cM. The PVE of qbranch-4 was 12.1% (Table 5). The QTL qbranch-2 was located in a similar genomic location of reported QTL for root dry weight (93–117 cM) and shoot dry weight (101–127 cM). The QTL qbranch-4 was in a similar location as reported QTL for fall plant height (82–101 cM) [17]. Both of these QTL had positive effects (Add> 0) for branch number.
Discussion
The use of RAD-seq and methods for allele calling
RAD-seq has been used to generate SNP markers in many plant species, including barley [21], pear [12], and grape [22]. It has been applied to construct high-density genetic linkage maps in plant species [22], however, RAD-seq has not been used in alfalfa. Both GBS and RAD-seq use methylation-sensitive enzymes for cutting genome sequences to reduce redundancy. However, the sensitive enzymes and sequencing strategies are different for the two method [23]. GBS has been used for generating SNPs in alfalfa [7, 19]. We are the first to use RAD-seq to generate SNPs to construct high density linkage maps in alfalfa with improved coverage. RAD-seq used in the present study produced more SNP markers (113,837) compared to previous studies using GBS [7, 19].
In the present study, we used the UNEAK pipeline for SNP calling. UNEAK was developed for GBS analysis in species with non-reference genomes. We used the UNEAK pipeline and called SNPs with RAD-seq. The combination of RAD-seq and the UNEAK pipeline provides a comprehensive strategy to obtain an adequate number of markers for QTL mapping. We compared the UNEAK pipeline with the reference pipeline, TASSEL-GBS [24], for genotype calling. The UNEAK pipeline gave better results for genotype calling than the reference pipeline. Using the UNEAK pipeline, we obtained high density SNPs and constructed linkage maps with 32 linkage groups (LGs). In contrast, we were unable to group the variants called using the reference pipeline and could not establish 32 LGs. This was probably because the reference genome of M. truncatula was diploid and did not align well with the tetraploid alfalfa genome, resulting in losing genetic information of alfalfa when aligned to the reference genome. The UNEAK pipeline is a platform using network comparison without alignment to the reference genome, and therefore, it resulted in more markers than the reference pipeline.
Marker density
The marker density is comparable to that of the genetic linkage map in alfalfa constructed in previous studies [7, 19]. This congruence suggests that the genotyping method used in this study is appropriate for a genetic mapping approach in alfalfa. However, the total length of the P1 map (3455 cM) was less than that of the P2 map (4381 cM). Different coverages were also found between parents in the previous studies [7, 19, 25]. Genetic differences in the parents are likely the main reason for different SNP coverage between the parents [26]. Furthermore, sequencing bias may cause a low number of raw reads for P1. Nevertheless, we were able to map 3312 markers to 32 LGs that covered all four sets of genomes in tetraploid alfalfa.
QTL for yield-related traits in alfalfa have been reported in previous studies [5, 17, 27]. However, the genetic maps were constructed using SSR and RFLP markers [5, 27] and the limited coverage provided by SSR and RFLP platforms resulted in large QTL intervals (> 10 cM) with low resolution. The use of single-dose alleles (SDAs) for genetic mapping is feasible in tetraploid species. Adhikari et al. identified QTL for fall dormancy and winter hardiness in alfalfa F1 population using SDAs [19]. This method is a pseudo-testcross strategy, which uses the simplex markers (AAAB x BBBB) of an F1 population for autotetraploid genetic linkage map construction using diploid software like JoinMap [28]. The pseudo-testcross strategy allows us to use thousands of SDA markers to construct linkage map followed by QTL mapping. Using SDA markers and the composite interval mapping method, QTL intervals were greatly reduced (< 3 cM) in the present analysis.
Comparison of QTL associated with yield-related traits in alfalfa
To reduce the effect of the interaction between genetics and the environment (G × E), we used BLUP to estimate phenotypic variation across five environments and identified QTL for each trait analyzed in this study. Among 21 QTL associated with yield and yield-related traits, several QTL were co-located among the yield-related traits. This indicated that phenotyping in multiple environments and adjusted BLUP is a useful way to control environmental variation. Most QTL identified in the present study were co-located with previously reported yield-related QTL [3, 17] and fall dormancy QTL [19]. There were five QTL (qyield-1, qbranch-2, qbranch-4, qdiameter-1, and qdiameter-5) with PVE > 10% and all of them are co-located with reported yield QTL except qbranch-4. These QTL may play a major role in controlling yield. Additional QTL (such as qyield-2 and qheight-2) were co-located with other traits, such as fall dormancy QTL, which may suggest pleiotropic effects of the genes.
We are the first to map QTL for shoot diameter and branch number in alfalfa. Although the heritability was not very high (34.9 and 32.0% for shoot diameter and branch number, respectively), we were able to detect six QTL for shoot diameter and five QTL for branch number. Several QTL for shoot diameter and branch number were co-located with QTL for yield and plant height: qyield-1 was co-located with qbranch-2; qyield-2 with qbranch-3; and qheight-5 with qdiameter-6 (Figs. 2 and 3). Furthermore, most shoot diameter and branch number QTL identified in the present study were co-located with shoot dry weight, crown dry weight, or root dry weight QTL [17]. These QTL may have an indirect effect in controlling yield traits [29].
Given that the average heritability (43%) of yield-related traits was reasonable for genetic analysis, the QTL identified in the present analysis need further validation. Overlapping QTL have been reported using different populations with different genetic backgrounds in different environments [3, 17, 19, 20]. In the present study, six QTL overlapped with three reported QTL. Among those, one of them had high PVE (> 13%). QTL with high PVE may play major roles for yield-related traits. Furthermore, in the present analysis, we were able to narrow down the QTL interval with high PVE (qyield-1 and qbranch-2) to 1.8 cM (98.0–99.8 cM), which will facilitate further investigations such as fine mapping and gene cloning.
Conclusions
In the present study, we constructed high density genetic maps using SNPs generated by RAD-seq in an F1 population derived from two local alfalfa cultivars Cangzhou and Zhongmu No. 1 with low and high yield, respectively. Our results showed that RAD-seq is an appropriate method for generating genetic markers that can be used to construct linkage maps in alfalfa. The QTL detected in this study will help us to understand the genetic basis of yield-related traits. However, these QTL may be not robust in different populations or environments and thus must be validated in breeding populations in future studies. With further investigation, markers closely linked to the major QTL can be used for marker-assisted selection to improve yield in alfalfa.
Methods
Plant materials and experimental design
P1 (paternal parent; Cangzhou) and P2 (maternal parent; Zhongmu No. 1) were individually selected from a panel of, respectively, low forage yield of Cangzhou and high forage yield of Zhongmu No. 1. They were crossed to generate an F1 population consisting of 149 progeny lines. In 2012, the seeds of the F1 population were planted in the greenhouse of the Chinese Academy of Agricultural Sciences (CAAS) in Langfang, Hebei Province, China, under conditions of 16 h day/8 h night, 22 °C and 40% relative humidity. Light intensity (a combination of natural light and incandescent lamps) was approximately 200 μmol m− 2 s− 1. Clones were propagated from individual plants by stem cuttings. During the early branching stage in 2013, the cloned plants were moved from propagation flats to the field of the CAAS research station in Langfang, Hebei Province (39.59°N, 116.59°E). F1 and parent individuals were also transplanted to establish a field trial in Tongzhou, Beijing (39.92°N, 116.65°E).
The field trial was carried out using a randomized complete block design (RCBD) with three replications at each of the two field sites. Every replication had one clone plant for every individual. The plant spacing was 100 cm between rows and 80 cm within rows. The individuals were not similar with each other after transplanting. Alfalfa is a perennial plant and is harvested by cutting; its rapid regrowth after cutting makes alfalfa a high yielding forage crop. To ensure that the plants were uniform before regrowth, they were clipped to a height of 5 cm; plants were clipped twice before regrowth for phenotyping. Weeds were removed manually and there was no cover crop in the field.
Phenotypic data collection and analysis
Phenotypic data were collected at two locations in multiple years. Two and 3 years of phenotypic data were respectively collected in Tongzhou in 2014 and 2015 (TZ2014, TZ2015) and in Langfang in 2014, 2015, and 2016 (LF2014, LF2015 and LF2016). Alfalfa plants were harvested at the early flowering stage (when 10% of plants begin flowering). Yield was measured after harvested plants were dried in a forced-air dryer. Plant height was measured based on the tallest stem at the date of harvesting. Branch number (the number of all stems) was counted at the same time. Diameters of five randomly selected shoots were measured at the shoot bottom after harvesting and the mean value was calculated. Data of yield-related traits from the first harvest was used for our analysis.
Best linear unbiased predictions (BLUPs) were used for statistical analysis of phenotypic data collected from years at different locations using PROC MIXED [30]. Analyses of the interaction of genotype with location and years were conducted using PROC GLM (SAS Institute, 2010). The correlation analysis of yield-related traits were conducted using PROC CORR (SAS Institute, 2010).
The random effect model used for BLUP was as follows:
where Yijkh is the Y for the jth genotype in the ith replication of the kth location in the hth year; m is the grand mean; ri(k) is the effect of the ith replication nested in the kth location; yh is the effect of the hth year; gj is the genetic effect of the jth genotype; gljk is the interaction effect of the jth genotype and kth location; gyjh is the interaction effect of the jth genotype and hth year; glyjkh is the interaction effect of the jth genotype, kth location, and hth year; and eijkh is the residual. All factors were random effects except for the grand mean.
DNA isolation and RAD library construction
DNA was extracted from 100 mg fresh young leaf tissue using the CWBIO Plant Genomic DNA Kit (CoWin Biosciences, Beijing, China), according to the manufacturer’s protocol. DNA was quantified using a Nanodrop 2000 spectrophotometer based on absorbance of 260 nm (Thermo Scientific, Waltham, MA, USA). DNA concentrations were adjusted to 50 ng/μl and subsequently used for RAD library preparation. DNA was digested by the EcoRI restriction enzyme (New England BioLabs, NEB), then ligated to a unique barcode adapter. Samples were pooled together and randomly sheared ultrasonically. The average length of sheared fragments was confined to around 400 bp using Qseq100 DNA Analyzer (Bioptic Inc.). The AMPure XP Beads PCR Purification kit (Beckman Coulter, Inc.) was applied to purify the sheared DNA fragments. The fragment ends were repaired with the Quick Blunting kit (NEB). A 3′-dA overhang was added using the dA-tailing module contained in the kits (NEB), and then ligated to a common adapter. The collected fragments were enriched by PCR amplification and purified using the AMPure XP Beads PCR purification kit (Beckman Coulter, Inc.). Each sample was normalized to 10 nM for sequencing using a Hi-seq X Ten (Illumina) at ORI-GENE (Beijing, China). The RAD sequences were submitted to the NCBI Sequence Read Archive with BioProject ID: PRJNA503672.
Sequence analysis and SNP genotyping
The Tassel 3.0 Universal Network Enable Analysis Kit (UNEAK) pipeline [31] was used for SNP genotype calling. We initially used the Medicago truncatula genome (Mt4.0 v1) as a reference for SNP calling (TASSEL-GBS), but we could not identify linkage groups using the resulting SNP markers. Briefly, a minimum of five was used for the tag number count step for the UQseqToTagCountPlugin command line of the UNEAK build in TASSEL and a 64 bp target length was used for trimming sequences. Marker missing values were less than 50%. Other parameters were set as default.
SSR marker genotyping
In total, 184 SSR markers were also used for genotyping. Primer sequences of SSR markers were obtained from the Samuel Roberts Noble Foundation (Ardmore, OK, USA). Polymerase chain reaction (PCR) was based on the method of Diwan et al. [32]. Briefly, PCR steps were as follows: 94 °C for 3 min, followed by 33 cycles with 30 s at 94 °C, 30 s at 56 °C, and 60 s at 72 °C, followed by an elongation step of 7 min at 72 °C. PCR products of SSR markers were separated using polyacrylamide gel electrophoresis (PAGE) and each allele of an SSR marker was scored as 1 for present, 0 for absent, and 9 for missing.
Linkage mapping and QTL analysis
Single-dose alleles (SDAs) of SNP markers with a ratio of less than 2:1 among F1 progenies were used to construct a genetic linkage map as described by Li et al. [7]. SSR markers with a ratio of 1:1 in F1 progenies were used to construct genetic linkage maps. Those SDA markers and SSR markers were combined together to construct a genetic linkage map using JoinMap 4.0 [28]. First, markers were grouped using a minimum logarithm of odds (LOD) of five (> 5) for P1 and seven (> 7) for P2. Second, for each group, markers were clustered using default parameters. Linkage group numbers were assigned based on SSRs with known chromosome locations. The R package R/qtl was used to display the linkage map [33]. Phenotypic data were analyzed using means across the three replicates for each site/year and the BLUP values were used for QTL mapping. QTL IciMapping [34] was used for QTL analysis. A LOD threshold of three was used to identify potential QTL and the mapping method was inclusive composite interval mapping with additive effect (ICIM-ADD) in the BIP (biparental populations) functionality. The software MapChart [35] was used to display the QTL results.
Abbreviations
- Add:
-
Estimated additive effect of the marker
- BLUP:
-
Best linear unbiased predictions
- GBS:
-
Genotyping by sequencing
- GWAS:
-
Genome-wide association studies
- ICIM-ADD:
-
Composite interval mapping with additive effect
- LG:
-
Linkage group
- PVE:
-
Phenotypic variation explained by QTL
- QTL:
-
Quantitative trait loci
- RAD-seq:
-
Restriction associated DNA sequencing
- RFLP:
-
Restriction fragment length polymorphism
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeats
- UNEAK:
-
Universal network enable analysis kit
References
Purcell S, Cherny SS, Sham PC. Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149–50.
Stanford EH. Tetrasomic inheritance in alfalfa 1. Agron J. 1951;43(5):222–5.
McCord P, Gordon V, Saha G, Hellinga J, Vandemark G, Larsen R, Smith M, Miller D. Detection of QTL for forage yield, lodging resistance and spring vigor traits in alfalfa (Medicago sativa L.). Euphytica. 2014;200(2):269–79.
Li X, Brummer EC. Applied genetics and genomics in alfalfa breeding. Agronomy. 2012;2(1):40–61.
Robins JG, Luth D, Campbell TA, Bauchan GR, He C, Viands DR, Hansen JL, Brummer EC. Genetic mapping of biomass production in tetraploid alfalfa. Crop Sci. 2007;47(1):1–10.
Kumar P, Gupta V, Misra A, Modi D, Pandey B. Potential of molecular markers in plant biotechnology. Plant Omics. 2009;2(4):141.
Li X, Wei Y, Acharya A, Jiang Q, Kang J, Brummer EC. A saturated genetic linkage map of autotetraploid alfalfa (Medicago sativa L.) developed using genotyping-by-sequencing is highly syntenous with the M. truncatula genome. G3. 2014. https://doi.org/10.1534/g3.114.012245.
Yu L-X. Identification of single-nucleotide polymorphic loci associated with biomass yield under water deficit in alfalfa (Medicago sativa L.) using genome-wide sequencing and association mapping. Front Plant Sci. 2017;8:1152.
Zhang T, Yu L-X, Zheng P, Li Y, Rivera M, Main D, Greene SL. Identification of loci associated with drought resistance traits in heterozygous autotetraploid alfalfa (Medicago sativa L.) using genome-wide association studies with genotyping by sequencing. PLoS One. 2015;10(9):e0138931.
Liu X-P, Yu L-X. Genome-wide association mapping of loci associated with plant growth and forage production under salt stress in alfalfa (Medicago sativa L.). Front Plant Sci. 2017;8:853.
Yu LX, Zheng P, Zhang T, Rodringuez J, Main D. Genotyping-by-sequencing-based genome-wide association studies on Verticillium wilt resistance in autotetraploid alfalfa (Medicago sativa L.). Mol Plant Pathol. 2017;18(2):187–94.
Wu J, Li L-T, Li M, Khan MA, Li X-G, Chen H, Yin H, Zhang S-L. High-density genetic linkage map construction and identification of fruit-related QTLs in pear using SNP and SSR markers. J Exp Bot. 2014;65(20):5771–81.
Yagi M, Shirasawa K, Waki T, Kume T, Isobe S, Tanase K, Yamaguchi H. Construction of an SSR and RAD marker-based genetic linkage map for carnation (Dianthus caryophyllus L.). Plant Mol Biol Report. 2017;35(1):110–7.
Barchi L, Lanteri S, Portis E, Valè G, Volante A, Pulcini L, Ciriaci T, Acciarri N, Barbierato V, Toppino L. A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS One. 2012;7(8):e43740.
Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S, Lorieux M, Ahmadi N, McCouch S. Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet. 2013;126(11):2699–716.
Tornqvist C-E, Taylor M, Jiang Y, Evans J, Buell CR, Kaeppler SM, Casler MD. Quantitative trait locus mapping for flowering time in a lowland× upland switchgrass pseudo-F 2 population. Plant Genome. 2018;11(2):1-9. https://doi.org/10.3835/plantgenome2017.10.0093.
Li X, Alarcón-Zúñiga B, Kang J, Tahir N, Hammad M, Jiang Q, Wei Y, Reyno R, Robins JG, Brummer EC. Mapping fall dormancy and winter injury in tetraploid alfalfa. Crop Sci. 2015;55(5):1995–2011.
Robins JG, Brummer EC. QTL underlying self-fertility in tetraploid alfalfa. Crop Sci. 2010;50(1):143–9.
Adhikari L, Lindstrom O, Markham J, Missaoui AMM. Dissecting key adaptation traits in the polyploid perennial Medicago sativa using GBS-SNP mapping. Front Plant Sci. 2018;9:934.
Ray IM, Han Y, Meenach CD, Santantonio N, Sledge MK, Pierce CA, Sterling TM, Kersey RK, Bhandari HS, Monteros MJ. Identification of quantitative trait loci for alfalfa forage biomass productivity during drought stress. Crop Sci. 2015;55(5):2012–33.
Chutimanitsakun Y, Nipper RW, Cuesta-Marcos A, Cistué L, Corey A, Filichkina T, Johnson EA, Hayes PM. Construction and application for QTL analysis of a restriction site associated DNA (RAD) linkage map in barley. BMC Genomics. 2011;12(1):4.
Wang N, Fang L, Xin H, Wang L, Li S. Construction of a high-density genetic map for grape using next generation restriction-site associated DNA sequencing. BMC Plant Biol. 2012;12(1):148.
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12(7):499.
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS One. 2014;9(2):e90346.
Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C. Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol. 2003;3(1):9.
Qiang H, Chen Z, Zhang Z, Wang X, Gao H, Wang Z. Molecular diversity and population structure of a worldwide collection of cultivated tetraploid alfalfa (Medicago sativa subsp. sativa L.) germplasm as revealed by microsatellite markers. PLoS One. 2015;10(4):e0124592.
Robins JG, Bauchan GR, Brummer EC. Genetic mapping forage yield, plant height, and regrowth at multiple harvests in tetraploid alfalfa (Medicago sativa L.). Crop Sci. 2007;47(1):11–8.
Van Ooijen J, Voorrips R. JoinMap 4.0. Software for the calculation of genetic linkage maps in experimental populations; 2006.
Brummer EC, Shah MM, Luth D. Reexamining the relationship between fall dormancy and winter hardiness in alfalfa. Crop Sci. 2000;40(4):971–7.
Wolfinger R, Federer WT, Cordero-Brana O. Recovering information in augmented designs, using SAS PROC GLM and PROC MIXED. Agron J. 1997;89(6):856–9.
Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD, Buckler ES, Costich DE. Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet. 2013;9(1):e1003215.
Diwan N, Bhagwat AA, Bauchan GB, Cregan PB. Simple sequence repeat DNA markers in alfalfa and perennial and annual Medicago species. Genome. 1997;40(6):887–95.
Broman KW, Wu H, Sen Ś, Churchill GA. R/QTL: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–90.
Meng L, Li H, Zhang L, Wang J. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3(3):269–83.
Voorrips R. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–8.
Acknowledgements
We thank Wenshan Guo and Fengqi Liu for their technical help in the field work and data collection.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31772656), the China Forage and Grass Research System (CARS-35-04), and the Agricultural Science and Technology Innovation Program (ASTIP-IAS14). The funding body played no role in the design of the study, the collection, analysis, and interpretation of data and the writing of the manuscript.
Availability of data and materials
All RAD raw sequence data were upload to NCBI with the BioProject ID of PRJNA503672. (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA503672). The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
QCY and TJZ conceived and designed the experiments. FZ, JMK, RCL, ZW, and ZXZ performed the experiments. JMK and RCL collected the data in Tongzhou in 2014 and 2015. FZ, ZW and ZXZ collected the data in Langfang in 2014, 2015 and 2016. FZ, TJZ, and LXY analyzed the data. FZ, JMK, and LXY wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhang, F., Kang, J., Long, R. et al. High-density linkage map construction and mapping QTL for yield and yield components in autotetraploid alfalfa using RAD-seq. BMC Plant Biol 19, 165 (2019). https://doi.org/10.1186/s12870-019-1770-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1770-6