Abstract
Background
ALOG (Arabidopsis LSH1 and Oryza G1) family with a conserved domain widely exists in plants. A handful of ALOG members have been functionally characterized, suggesting their roles as key developmental regulators. However, the evolutionary scenario of this gene family during the diversification of plant species remains largely unclear.
Methods
Here, we isolated seven ALOG genes from Torenia fournieri and phylogenetically analyzed them with different ALOG members from representative plants in major taxonomic clades. We further examined their gene expression patterns by RT-PCR, and regarding the protein subcellular localization, we co-expressed the candidates with a nuclear marker. Finally, we explored the functional diversification of two ALOG members, TfALOG1 in euALOG1 and TfALOG2 in euALOG4 sub-clades by obtaining the transgenic T. fournieri plants.
Results
The ALOG gene family can be divided into different lineages, indicating that extensive duplication events occurred within eudicots, grasses and bryophytes, respectively. In T. fournieri, seven TfALOG genes from four sub-clades exhibit distinct expression patterns. TfALOG1–6 YFP-fused proteins were accumulated in the nuclear region, while TfALOG7-YFP was localized both in nuclear and cytoplasm, suggesting potentially functional diversification. In the 35S:TfALOG1 transgenic lines, normal development of petal epidermal cells was disrupted, accompanied with changes in the expression of MIXTA-like genes. In 35S:TfALOG2 transgenic lines, the leaf mesophyll cells development was abnormal, favoring functional differences between the two homologous proteins. Unfortunately, we failed to observe any phenotypical changes in the TfALOG1 knock-out mutants, which might be due to functional redundancy as the case in Arabidopsis.
Conclusion
Our results unraveled the evolutionary history of ALOG gene family, supporting the idea that changes occurred in the cis regulatory and/or nonconserved coding regions of ALOG genes may result in new functions during the establishment of plant architecture.
Similar content being viewed by others
Background
Surveying the evolutionary history of plants, gene duplication plays a major role in promoting speciation [1,2,3]. The occurrence of duplicated genes is often related to the achievement of new functions, including those involved in plant architectures, which cause the extraordinary abundance of plant morphologies [2, 4,5,6].
Gene duplication generates a large proportion of genes and further forms different sizes of gene families with diverse functions, evident in TCP (TEOSINTE BRANCHED 1, CYCLOIDEA and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR), KNOX (KNOTTED HOMEOBOX) and MADS (MINI CHROMOSOME MAINTENANCE 1, AGAMOUS, DEFICIENS, and SERUM RESPONSE FACTOR) families [6]. TCP family genes encoding bHLH (basic helix-loop-helix) transcription factors are divided into three sub-clades, involved in plant developmental processes such as branching, cell size and flower shape [7,8,9,10]. KNOX family members consist in the class I KNOX proteins, which mainly regulate shoot apical meristems and leaf complexity, and the class II KNOX proteins, involved in cell wall biosynthesis [11,12,13,14,15,16]. The MADS gene family is another large family well-known for its functions in regulating different floral organs identities [17,18,19,20]. Although the evolutionary history and functional diversification have been well studied among some classical gene families, there are numerous families yet to be studied.
ALOG (Arabidopsis LSH1 and Oryza G1) proteins are key developmental regulators among land plants, which share a highly conserved ALOG domain (also known as DOMAIN OF UNKNOWN FUNCTION 640 / DUF640) [21]. However, functional studies were only carried out in several limited species, including rice (Oryza sativa), Arabidopsis thaliana, as well as tomato (Solanum lycopersicum). TAWAWA1 (TAW1) and LONG STERILE LEMMA1 (G1) are two ALOG genes in rice: TAW1 affects inflorescence architecture while G1 represses the growth of sterile lemma in spikelet [22, 23]. In A. thaliana, ORGAN BOUNDARY 1 (OBO1) and OBO4, encoding ALOG family proteins, are involved in boundary formation, which function redundantly with other genes [24, 25]. An ALOG member, TERMINATING FLOWER (TMF) in tomato, acts as a putative transcriptional regulator, affecting flower and leaf architecture [26,27,28]. These studies suggest functional divergence within ALOG gene family; however, there is still a lack of evolutionary analysis of this gene family linked to the diversified functions.
In this study, we aimed to investigate the evolutionary history of ALOG gene family, and for that, we used T. fournieri as material due to its importance in horticulture. Based on the phylogenetic analysis, the extensive gene duplication events were observed among three major taxonomic plant lineages: eudicots, grasses and bryophytes. In T. fournieri, seven TfALOG genes from four subclades showed distinct expression patterns and we further ectopically expressed two ALOG members, TfALOG1 and TfALOG2 from distinct sub-clades. Our results showed different functions of two ALOG proteins, favoring that changes in cis-regulatory and nonconserved coding regions would be important for the neo-functionalization of ALOG gene family.
Methods
Molecular cloning
Plant DNA was extracted by 2% CTAB from juvenile leaves and total RNA was extracted from different tissues, using Plant RNA Kit (Omega Bio-Tek, Guangzhou, China) according to the manufacturer’s instructions. The first strand cDNA were synthesized using PrimeScript RT reagent Kit with gDNA Eraser (Takara, Beijing, China). To isolate ALOG and MIXTA-like genes in T. fournieri, protein sequences of LSHs (ALOG members from A. thaliana) and AmMIXTAs (MIXTA-like factors from Antirrhinum majus) were used to blast in the in-house transcriptome of T. fournieri. The corresponding cDNA sequences were obtained and their relative ORFs (Open Reading Frames) were predicted through an online program ORF-Finder (https://www.ncbi.nlm.nih.gov/orffinder/). After further blasting in Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi), the candidate genes were maintained. To verify the sequences from the transcriptome, gene-specific primers were designed for PCR, and genomic DNA and cDNA were used as templates. The PCR products were cloned into pMD19-T vector (Takara, Beijing, China) prior to sequencing. Sequences were then analyzed in MEGA7 to identify the gene structures [29]. Primers used in this study were listed in Additional file 1: Table S1.
Phylogeny analysis and motif-based sequence analysis
ALOG genes from T. fournieri were cloned in this study. To figure out the evolutionary history of ALOG genes in plants, we collected homologous gene sequences from Phytozome version 12 and other databases (https://phytozome.jgi.doe.gov). The reference database versions from Phytozome were as follows: Physcomitrella patens: version 3.3; Sphagnum fallax: version 0.5; Oryza sativa: version 7.0; Zea mays: version PH207 v1.1; Sorghum bicolor: version 3.1.1; Brachypodium distachyom: version Bd21–3 v1.1; Aquilegia coerulea: version 3.0; Arabidopsis thaliana: version TAIR10; Populus trichocarpa: version 3.0; Daucus carota: version 2.0; Mimulus guttatus: version 2.0; Solanum lycopersicum: version iTAG2.4.). Other databases of two Lamiales species including Sesamum indicum and Utricularia gibba were also referred [30, 31].
Alignments of nucleotide and amino acid sequences were performed in MEGA7 [29]. Using MRBAYES v.3.2.1 with the GTR + I + G nucleotide substitution model, the aligned sequences were analyzed with 300,000,000 generations and a 1000 generation sample frequency [32]. ALOG gene sequences of the representative plant species were submitted to motif-based sequence analysis website (MEME; http://meme-suite.org/tools/meme) for motif mining under the parameters: -nmotifs 50, −minw 6, −maxw 50.
RT-PCR and quantitative RT-PCR (qRT-PCR)
The templates used in the RT-PCR were synthesized by the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Shanghai, China). Seven cloned ALOG genes were examined. β-actin (TfACT3) was the internal control of the RT-PCR as previously reported [33]. For qRT-PCR, we obtained the cDNA under the protocol of PrimeScript RT reagent Kit with gDNA Eraser (Takara, Beijing, China). Using LightCycler 480 Real-Time PCR System (Roche, Shanghai, China), qRT-PCR was performed based on the manual. β-actin (TfACT3) was used as an internal control of the qRT-PCR. Three biological replicates were performed to calculate the SD in qRT-PCR assays. Graphs were produced with GraphPad Prism 5. Primers used in this study were listed in Additional file 1: Table S1.
Protein signal prediction and subcellular localization
The nuclear localization signals were predicted in http://mleg.cse.sc.edu/seqNLS/. The open reading frame (ORF) of each T. fournieri ALOG gene was fused with a yellow fluorescent protein (YFP) in the C-terminal and was inserted into pA7 plasmid (provided by Prof. Hongwei Xue from Institute of Plant Physiology and Ecology, Chinese Academy of Science). The leaf mesophyll protoplasts of Arabidopsis thaliana were extracted from two-week plants and followed by PEG-induced transformation, as previously described [34]. All the ALOG-YFP plasmids were co-transformed with a nuclear marker, ARF19IV-mCherry, as previously used [35]. The YFP and mCherry signals were observed using a confocal laser scanning microscopy Zeiss7 DUO NLO.
Plant growth and transformation of T. fournieri
Plants were kept under 16-h-light and 8-h-dark greenhouse with 70–80% relative humidity at 22–24 °C. Sterile seedlings were obtained as previously reported [33]. The binary vector, pCAMBIA1302, was used for transformation. The ORFs of two target genes TfALOG1 and TfALOG2 were driven by 35S promoter. The construction of TfALOG1-Cas9 vector followed the protocol provided by Prof. Yaoguang Liu from South China Agricultural University [36]. Agrobacterium tumefaciens strain EHA105 was used for plant transformation as previously described [37]. Transgenic plants were screened on the 1/2 MS media containing hygromycin and were identified by PCR.
Scanning electron microscopy (SEM)
Mature petals of T. fournieri were dissected and taped on slide glasses. Multiple layers of impression materials were used to generate the epoxy replicas of petals as previously described in [38]. The epoxy replicas were sputtered with gold particles before observing under a scanning electron microscope Jeol JSM 6360LV (Jeol, Tokyo, Japan). The images were adjusted by Adobe PHOTOSHOP CS6 (Adobe, San Jose, CA, USA).
Results
Isolation of ALOG genes from T. fournieri
We used T. fournieri as a representative model of Asterids lineage eudicot, from which we isolated seven ALOG members that we named as TfALOG1–7 (Additional file 1: Figure S1A). Generally, the ALOG genes from T. fournieri encode small proteins with about 200 amino acids. Only two members out of seven, namely TfALOG3 and TfALOG7, have an intron with 374 bp and 255 bp, respectively (Additional file 1: Figure S1A). Multiple alignment indicated these ALOG proteins contain one conserved ALOG domain, with highly variable N-/C- terminals (Additional file 1: Figure S1B).
ALOG gene family has undergone multiple times of independent duplication and/or loss events during the diversification of land plants
To reconstruct the evolutionary history of ALOG gene family, we in-depth mined the ALOG members using BLAST (Basic Local Alignment Search Tool) in different plants genomes. ALOG genes are present in the basal land plants like Physcomitrella patens and Sphagnum fallax (Fig. 1). There are 4 and 5 ALOG members characterized from P. patens and S. fallax genome, respectively (Fig. 1). Members from one species clustered together, indicating that these duplication events happened independently after speciation (Fig. 1). Due to the differences in their sequences, we designated the PpALOGs and SfALOGs as out-groups in later phylogeny studies.
Bayesian phylogram (left) and motif analysis (right) of ALOG genes in three plants lineages. Physcomitrella patens and Sphagnum fallax were selected as outer groups. The Bayesian posterior probability is located in each node and the accession number can be found in each sequence. Seven ALOG genes cloned from Torenia fournieri were presented in red. The motif diagrams were generated in MEME and different colors represent different motifs
In angiosperms, ALOG genes from monocots and eudicots clustered independently (Fig. 1). The grass ALOG genes from Zea mays, Oryza sativa, Sorghum bicolor and Brachypodium distachyon, were divided into three clades named GrassALOG1/2/3 (Fig. 1). The GrassALOG3 clade was further divided into three sub-clades because of its motif differences (Fig. 1). The phylogenetic tree of grass ALOGs also provided us two indications: first, ALOG genes have been duplicated in their common ancestors; second, independent duplication and/or loss events occurred together with the divergence of Poaceae (Fig. 1). There has been a whole genome duplication event in Z. mays after its divergence from S. bicolor [39]. However, only GrassALOG2 and GrassALOG3 clades members supported the duplication event, as there were two ZmALOG paralogues corresponding to one SbALOG gene in these two clades (Fig. 1).
We further focused on eudicots, and selected five representative species for phylogeny analysis: Aquilegia coerulea, a basal eudicots; A. thaliana and Populus trichocarpa, two Rosids lineage species; S. lycopersicum and T. fournieri, two Asterids lineage species. The phylogenetic tree of the ALOG genes from eudicots separated into four clades, designated as euALOG1, euALOG2, euALOG3 and euALOG4 (Fig. 2). These four clades of euALOGs were also evident when more Asterids ALOG genes (from Daucus carota, Mimulus guttatus, S. lycopersicum, Sesamum indicum, Utricularia gibba and T. fournieri) were added to the phylogenetic tree (Additional file 1: Figure S2). Apart from the ancestral duplication events before the divergence of eudicots, frequent duplication and/or loss of euALOG members also occurred along with speciation (Fig. 2). Since different clades of euALOGs have been duplicated independently, we inferred that the ALOG gene family might play various roles in different biological processes.
Bayesian phylogram of ALOG genes in eudicots. Physcomitrella patens and Sphagnum fallax were chosen as outer groups. The Bayesian posterior probability is located in each node and the accession number can be found in each sequence. Seven ALOG genes cloned from Torenia fournieri were presented in red
Expressional analysis of ALOG genes in T. fournieri
Since the expression pattern of one gene is always closed related to its function, we examined the expression of 7 ALOG genes from T. fournieri by performing RT-PCR in different plant tissues. The phylogenetic tree indicated that TfALOG1 and TfALOG3 clustered closely, together with TfALOG5, belonging to the euALOG1 clade; TfALOG6 and TfALOG7 clustered in the euALOG3 clade; while TfALOG2 and TfALOG4 were found in euALOG2 and euALOG4 clades, respectively (Fig. 2). The expression patterns of ALOG genes were highly correlated with their phylogeny, except for TfALOG5, which showed a different expression pattern compared with its paralogues (Fig. 3). TfALOG1 and TfALOG3 were highly expressed in different flower buds, while TfALOG5 was expressed in roots, leaves and vegetative and reproductive apexes; TfALOG6 and TfALOG7 were highly expressed in roots and vegetative and reproductive apexes; TfALOG2 was highly accumulated in leaf and TfALOG4 was found almost in all tissues we examined (Fig. 3). The distinct expression patterns of T. fournieri ALOG genes implied that this family might have generated various functions in the establishment of plant architecture.
Reverse transcription polymerase chain reaction analysis (RT-PCR) of TfALOG genes. Gene names are shown on the left sides; PCR cycles are shown on the right sides in each gene. β-actin (TfACT3) was used as an internal control. R, root; S, stem; L, leaf; VA, vegetative apexes; RA, reproductive apexes; F9, stage 9 flower bud; F10, stage 10 flower bud; F11, stage 11 flower bud
Subcellular localization of TfALOG-YFP fused proteins
The subcellular localization of a protein is linked to its function, thus we analyzed the potential nuclear localization signals within the seven TfALOG proteins using online tools. The results showed that all the TfALOG proteins had nuclear localization signals in their N-terminals (Additional file 1: Figure S3). To verify the predicted results, we fused the open reading frame of each TfALOG gene with a yellow florescence protein (YFP) tag and co-transformed into Arabidopsis mesophyll protoplasts with a nuclear marker ARF19IV-mCherry. The empty plasmid with an YFP was used as a control, which existed in both nuclear and cytoplasm regions (Fig. 4). TfALOG1–6 YFP-fused proteins were all co-localized with the nuclear marker, indicating their accumulation in the nuclear region (Fig. 4). One exception was from TfALOG7 YFP-fused protein, which was localized both in nuclear and cytoplasm, suggesting potentially functional diversification (Fig. 4). These results supported that TfALOGs might basically work as transcriptional factors / cofactors, as previously reported.
Phenotypic analysis of 35S:TfALOG1 and 35S:TfALOG2 transgenic plants
To explore the functions of TfALOG genes, we ectopically expressed TfALOG1 from euALOG1 clade and TfALOG2 from euALOG4 clade in T. fournieri. In the 35S:TfALOG1 transgenic plants (14 lines showed stable phenotypes out of 30 transgenic lines), the pigmentation and shape of flowers became abnormal compared with the wild type (Fig. 5 a). The light-violet petal background color was less intensely than WT (Fig. 5 a). We further observed the epidermal cells of petals by scanning electronic microscopy, and found that the conical cells in the lobe regions became flat together with an increase in cell size (Fig. 5 a). Furthermore, we observed other phenotypes in 35S:TfALOG1 transgenic lines, which included changes in petal shape, elongated yellow region in petal tube, as well as changed leaf color (Additional file 1: Figure S4A). These results demonstrated TfALOG1 as an important player during plant organ development. However, we failed to observe any phenotypical change in the TfALOG1 knock-out mutants (among 10 genetically edited lines), which may be due to functional redundancy among this family, consistent with previous studies from Arabidopsis (Additional file 1: Figure S6) [24, 25, 40].
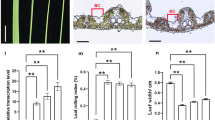
Phenotypic analysis of wild type (WT), 35S:TfALOG1 and 35S:TfALOG2 transgenic plants. a Three rows represent the over-all plants, flowers (bars: 10 mm) and epidermal cells in petal lobes (bars: 20 μm), two independent lines 35S:TfALOG1 16# and 35S:TfALOG1 26# were used for analysis. b Two rows represent the over-all plants and leaves (bars: 10 mm), two independent lines, 35S:TfALOG2 2# and 35S:TfALOG2 3# were used for analysis; white arrows indicate developmental defects on leaves
When we ectopically expressed TfALOG2 (10 lines showed stable phenotypes out of 19 transgenic lines), normal leaf development was affected, which was consistent with the high expression of TfALOG2 in leaves (Fig. 5b). The mesophyll cells of marginal leaf regions failed to form in the 35S:TfALOG2 transgenic plants and this was always linked with changes in leaf shape, suggesting its function in leaf development (Fig. 5b). We did not observe any difference in the flowers of 35S:TfALOG2 transgenic plants (Additional file 1: Figure S4B).
It has been reported that MIXTA-like MYB transcription factors were responsible for the differentiation of conical epidermal cells [41,42,43], thus we cloned four MIXTA-like factors from T. fournieri (Additional file 1: Figure S1C). All four MIXTA-like genes were highly expressed in the petal lobe regions, consistent with the existence of conical epidermal cells (Additional file 1: Fig. S5). We also found that the three flower-expressed TfALOG1/3/4 genes were preferentially expressed in the petal tube regions, which differed from the expression of MIXTA-like genes (Additional file 1: Figure S5). Hence, we inferred antagonistic roles between TfALOGs and TfMIXTAs. To test this hypothesis, we checked the expression of TfMIXTAs in the 35S:TfALOG1 transgenic lines. Three out of four MIXTA-like genes, TfMIXTA1, TfMIXTA3 and TfMIXTA4, were significantly down-regulated in the 35S:TfALOG1 transgenic flowers, while TfMIXTA2 was up-regulated in the transgenic plants (Fig. 6). These indicated functional differences among the MIXTA-like genes [41, 44,45,46]. These data threw lights on how TfALOG1 was involved in the regulation of flower development.
Relative expression of TfMIXTA genes in wild type (WT) and 35S:TfALOG1 transgenic plants. Relative expression of TfMIXTA genes in different plants was determined by qRT-PCR in the stage 10 flower buds. Error bars represent ±1 SD from three biological replicates; statistically significant differences were marked by asterisks with *: P < 0.05. ***: P < 0.001
Discussion
Evolutionary scenario of ALOG gene family
The ALOG domain was originated from the XerC/D-clade of tyrosine recombinases containing a novel class of DIRS-1 transposon, which belongs to the tyrosine recombinase/phage integrase N-terminal DNA-binding domain superfamily [21]. We found that the ALOG genes evolved from the basal land plants and independently duplicated multiple times during the diversification of plant species, which was consistent with a previously study [40].
The phylogenetic analysis showed that, ALOG family members from different large plant lineages could be easily distinguished (Figs. 1 and 2). For example, the grass ALOG members were distinct from ALOGs in either eudicots or bryophytes. We did not detect any ALOG member from published genomes of the green algae lineage (chlorophytes) indicating that ancient ALOG gene emerged after the generation of land plants. Although multiple ALOG members are found from the basal land plant genomes, they have never been functionally studied. Phylogenetic analysis suggested that these multiple bryophyta ALOG paralogues share a common ancestor with distinct motifs in the N-terminal. In grasses, there has been a significant increase in the number of ALOG members, companying with the increased complexity in plant architecture. We were surprised by the phylogenetic tree in which all the grass ALOG members were separated from the eudicots, suggesting a possible functional divergence (Fig. 1). In eudicots, the ALOG family members were further divided into four clades, although independent loss of genes occurred in different eudicots species (Fig. 2). The general four euALOG clades were also supported by the Asterids ALOG phylogenetic tree, which indicated functional conservation within different clades of euALOG genes (Additional file 1: Figure S2).
Functions of ALOG family members
The predicted secondary structure of an ALOG domain shares four conserved helices with a zinc ribbon inserted between the helices two and three [21]. This secondary structure is likely to confer a DNA binding activity to the ALOG domain, possibly making the protein function as a transcriptional factor [21].
Until now, there are only three rice ALOG members that have been functionally characterized: G1 from grassALOG1 clade, TAW1/G1 L5 from grassALOG3 clade and G1 L6/TH1/BLS1/BSG1/AFD1 from grassALOG3 clade [22, 23, 47,48,49,50,51]. The expression patterns and functions of different grass ALOG gene clades are divergent: The mRNA of G1 is found in sterile lemma as well as in the basal region of palea, and is involved in the repression of lemma identity during the sterile lemma development [22]; TAW1 is expressed in various meristems including shoot apical, axillary, inflorescence and branch meristems; gain-of-function taw1-D mutants result in an increase of inflorescence branches [23]; TH1 transcripts are ubiquitous in multiple tissues, and the th1 mutant plants show defects both in vegetative and reproductive organ development [47, 50]. Transcriptional activity assays suggest that G1 and TAW1 likely act as transcriptional activators, while TH1 works as a transcriptional repressor [22, 23, 51]. These results indicate that grass ALOG genes functions evolved after gene duplication events.
In the eudicots, to our knowledge, only five ALOG members have been functionally addressed: LSH1, LSH3 and LSH4 from Arabidopsis, TMF from tomato and LjALOG1 from Lotus japonicus [24,25,26,27, 40, 52]. Over-expression of Arabidopsis ALOG genes leads to changes in seedling development and boundary formation; however, no obvious phenotype could be detected in either LSH1 antisense transgenic lines or lsh3 lsh4 double mutants, indicating functional redundancy in Arabidopsis [24, 25, 40]. From our phylogenetic tree, there were four euALOG1 clade members from Arabidopsis, which might function redundantly (Fig. 1). Although multiple closed paralogues exist in tomato, the single mutant of TMF shows strong phenotypes with early flowering time and conversion from an inflorescence to single abnormal flower [26]. The TMF protein can physically interact with multiple transcription factors such as BOP-like proteins, supporting the hypothesis of TMF working as a transcriptional cofactor that controls inflorescence architecture [26, 27]. In Lotus japonicus, an ALOG gene family member LjALOG1, highly expressed in nodule, has a novel role in controlling nodulation of legume species [52].
Functional divergence in T. fournieri ALOG family
In this study, we isolated seven ALOG genes from T. fournieri and noticed that the N/C-terminals of these ALOG proteins were rather divergent from each other even among those from the same clade (Fig. 1). In order to detect their functional divergence, we ectopically expressed two TfALOG members from different clades. Ectopic expression of TfALOG2 led to abnormal leaf development, while in 35S:TfALOG1 transgenic lines, normal flower development was affected (Fig. 5). These results indicated that different functions have been evolved from changes in the variable regions among these ALOG proteins.
TfALOG1 may be involved in petal differentiation
MIXTA-like MYB genes play various roles in plant epidermis including trichome development in Gossypium hirsutum, cuticle formation in Arabidopsis and T. fournieri, and epidermal cell differentiation in Thalictrum [42, 53, 54]. As an important pollination syndrome, different studies conclude that conical cells on petal epidermis are also determined by MIXTA-like MYB genes [41,42,43,44,45,46].
In the 35S:TfALOG1 transgenic lines, conical epidermal cells in the lobe regions became flat with large amount of granules adhered onto their surface (Fig. 5 a). We also found multiple MIXTA-like genes down-regulated in the 35S:TfALOG1 transgenic lines, supporting that TfALOG1 may regulate petal epidermis differentiation by directly or indirectly inhibiting the expression of MIXTA-like genes in the petal tube region (Fig. 6).
Conclusions
Our study unveiled the evolutionary scenario of ALOG family in land plants. We observed that multiple gene duplication events independently occurred in different plant linages. We also systematically studied seven ALOG genes from an Asterids species, T. fournieri, and found functional differences of TfALOG proteins in the establishment of plant architecture. In the future, it is of interest to explore how TfALOG1 is involved in petal epidermal cell differentiation and how TfALOG2 is recruited to leaf development. Our work offers new insights of functional divergence among plant ALOG gene family.
Abbreviations
- ALOG:
-
Arabidopsis LSH1 and Oryza G1
- DUF640:
-
DOMAIN OF UNKNOWN FUNCTION 640
- G1:
-
LONG STERILE LEMMA1
- KNOX:
-
KNOTTED HOMEOBOX
- MADS:
-
MINI CHROMOSOME MAINTENANCE 1, AGAMOUS, DEFICIENS, and SERUM RESPONSE FACTOR
- OBO:
-
Organ Boundary
- ORF:
-
Open reading frame
- SEM:
-
Scanning electron microscopy
- TAW1:
-
TAWAWA1
- TCP:
-
TEOSINTE BRANCHED 1, CYCLOIDEA and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR
- TMF:
-
TERMINATING FLOWER
- YFP:
-
Yellow fluorescent protein
References
Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5.
Conant GC, Wolfe KH. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 2008;9:938–50.
Rensing SA. Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol. 2014;17:43–8.
Baum DA. The evolution of plant development. Curr Opin Plant Biol. 1998;1:79–86.
Jiang X, Assis R. Natural selection drives rapid functional evolution of young Drosophila duplicate genes. Mol Biol Evol. 2017;34:3089–98.
Zhang J. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–8.
Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature. 1996;383:794–9.
Doebley JAS, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–8.
Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice PROLIFERATING CELL NUCLEAR ANTIGEN gene. Plant Cell. 1997;9:1607–19.
Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–22.
Vollbrecht E, Veit B, Sinha N, Hake S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature. 1991;350:241–3.
Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–76.
Hareven D, Gutfinger T, Parnis A, Eshed Y, Lifschitz E. The making of a compound leaf: genetic manipulation of leaf architecture in tomato. Cell. 1996;84:735–44.
Waites R, Selvadurai HR, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell. 1998;93:779–89.
Nishimura A, Tamaoki M, Sato Y, Matsuoka M. The expression of tobacco Knotted1-type class 1 homeobox genes correspond to regions predicted by the cytohistological zonation model. Plant J. 1999;18:337–47.
Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296:1858–60.
Schwarz-Sommer Z, Huijser P, Nacken W, Saedler H, Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus. Science. 1990;250:931–6.
West AG, Shore P, Sharrocks AD. DNA binding by MADS-box transcription factors: a molecular mechanism for differential DNA bending. Mol Cell Biol. 1997;17:2876–87.
Becker A. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol. 2003;29:464–89.
Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010;11:214.
Iyer LM, Aravind L. ALOG domains: provenance of plant homeotic and developmental regulators from the DNA-binding domain of a novel class of DIRS1-type retroposons. Biol Direct. 2012;7:39.
Yoshida A, Suzaki T, Tanaka W, Hirano HY. The homeotic gene long sterile lemma (G1) specifies sterile lemma identity in the rice spikelet. Proc Natl Acad Sci U S A. 2009;106:20103–8.
Yoshida A, Sasao M, Yasuno N, Takagi K, Daimon Y, Chen R, Yamazaki R, Tokunaga H, Kitaguchi Y, Sato Y, et al. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proc Natl Acad Sci U S A. 2013;110:767–72.
Cho E, Zambryski PC. ORGAN BOUNDARY1 defines a gene expressed at the junction between the shoot apical meristem and lateral organs. Proc Natl Acad Sci U S A. 2011;108:2154–9.
Takeda S, Hanano K, Kariya A, Shimizu S, Zhao L, Matsui M, Tasaka M, Aida M. CUP-SHAPED COTYLEDON1 transcription factor activates the expression of LSH4 and LSH3, two members of the ALOG gene family, in shoot organ boundary cells. Plant J. 2011;66:1066–77.
MacAlister CA, Park SJ, Jiang K, Marcel F, Bendahmane A, Izkovich Y, Eshed Y, Lippman ZB. Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat Genet. 2012;44:1393–8.
Xu C, Park SJ, Van Eck J, Lippman ZB. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016;30:2048–61.
Ichihashi Y, Aguilar-Martinez JA, Farhi M, Chitwood DH, Kumar R, Millon LV, Peng J, Maloof JN, Sinha NR. Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci U S A. 2014;111:E2616–21.
Kumar SGS, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Wang L, Yu S, Tong C, Zhao Y, Liu Y, Song C, Zhang Y, Zhang X, Wang Y, Hua W, et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014;15:R39.
Carretero-Paulet L, Chang T, Librado P, Ibarra-Laclette E, Herrera-Estrella L, Rozas J, Albert V. Genome-wide analysis of adaptive molecular evolution in the carnivorous plant Utricularia gibba. Genome Biol Evol. 2015;7:444–56.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Su S, Xiao W, Guo W, Yao X, Xiao J, Ye Z, Wang N, Jiao K, Lei M, Peng Q, et al. The CYCLOIDEA–RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 2017;215:1582–93.
Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–72.
Zhai C, Zhang Y, Yao N, Lin F, Liu Z, Dong Z, Wang L, Pan Q. Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. PLoS One. 2014;9:e98067.
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–84.
Li H, Kang P, Li M, Li M. Genetic transformation of Torenia fournieri using the PMI/mannose selection system. Plant Cell Tissue Org Cult. 2007;90:103–9.
Su S, Shao X, Zhu C, Xu J, Lu H, Tang Y, Jiao K, Guo W, Xiao W, Liu Z, et al. Transcriptome-wide analysis reveals the origin of peloria in Chinese Cymbidium (Cymbidium sinense). Plant Cell Physiol. 2018. https://doi.org/10.1093/pcp/pcy130.
Blanc G. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 2004;16:1667–78.
Zhao L, Nakazawa M, Takase T, Manabe K, Kobayashi M, Seki M, Shinozaki K, Matsui M. Overexpression of LSH1, a member of an uncharacterised gene family, causes enhanced light regulation of seedling development. Plant J. 2004;37:694–706.
Noda K, Glover BJ, Linstead P, Martin C. Flower colour intensity depends on specialized cell shape controlled by a Myb-related transcription factor. Nature. 1994;369:661–4.
Di SVS, Martin C, Schulfer AF, Connelly CF. An ortholog of MIXTA-like2 controls epidermal cell shape in flowers of Thalictrum. New Phytol. 2009;183:718–28.
Brockington SF, Alvarez-Fernandez R, Landis JB, Alcorn K, Walker RH, Thomas MM, Hileman LC, Glover BJ. Evolutionary analysis of the MIXTA gene family highlights potential targets for the study of cellular differentiation. Mol Biol Evol. 2013;30:526–40.
Perez-Rodriguez M, Jaffe FW, Butelli E, Glover BJ, Martin C. Development of three different cell types is associated with the activity of a specific MYB transcription factor in the ventral petal of Antirrhinum majus flowers. Development. 2005;132:359–70.
Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134:1691–701.
Jaffe FWAT, Glover BJ. A truncated MYB transcription factor from Antirrhinum majus regulates epidermal cell outgrowth. J Exp Bot. 2007;58:1515–24.
Li X, Sun L, Tan L, Liu F, Zhu Z, Fu Y, Sun X, Sun X, Xie D, Sun C. TH1, a DUF640 domain-like gene controls lemma and Palea development in rice. Plant Mol Biol. 2012;78:351–9.
Yan D, Zhou Y, Ye S, Zeng L, Zhang X, He Z. Beak-shaped grain 1/TRIANGULAR HULL 1, a DUF640 gene, is associated with grain shape, size and weight in rice. Sci China Life Sci. 2013;56:275–83.
Sato DS, Ohmori Y, Nagashima H, Toriba T, Hirano HY. A role for TRIANGULAR HULL1 in fine-tuning spikelet morphogenesis in rice. Genes Genet Syst. 2014;89:61–9.
Ren D, Rao Y, Wu L, Xu Q, Li Z, Yu H, Zhang Y, Leng Y, Hu J, Zhu L, et al. The pleiotropic ABNORMAL FLOWER AND DWARF1 affects plant height, floral development and grain yield in rice. J Integr Plant Biol. 2016;58:529–39.
Peng P, Liu L, Fang J, Zhao J, Yuan S, Li X. The rice TRIANGULAR HULL1 protein acts as a transcriptional repressor in regulating lateral development of spikelet. Sci Rep. 2017;7:13712.
Lei Y, Su S, He L, Hu X, Luo D. A member of the ALOG gene family has a novel role in regulating nodulation in Lotus japonicus. J Integr Plant Biol. 2018. https://doi.org/10.1111/jipb.12711.
Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M. MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell. 2013;25:1609–24.
Tan J, Walford S, Dennis ES, Llewellyn D. Trichomes control flower bud shape by linking together young petals. Nature Plants. 2016;2:16093.
Acknowledgments
We thank Ms. Xiaohe Hu (Sun Yat-sen University) for her help on plant care; Prof. Xia Huang (Sun Yat-sen University) for her instructions on T. fournieri transformation. Miss Maria João Ferreira (Faculdade de Ciências da Universidade do Porto) for her kindly help on manuscript revision. We also thank two anonymous reviewers for their valuable comments on the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of the People’s Republic of China (grant no. 2013BAD01B0702) and the Ministry of Agriculture of the People’s Republic of China for Transgenic Research (grant no. 2014ZX0800943B).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article. Sequences cloned in this study have been uploaded to the GenBank with the accession numbers from MG797503 to MG797517.
Author information
Authors and Affiliations
Contributions
WX, ZY and SS wrote the manuscript. WX, ZY, XY and SS performed the experiments. WX, YL, LH, and SS carried out bioinformatics analyses. WX, DL and SS designed the study. XY, YL, LH and DL provided helpful discussion. All authors approved the submission and revision of this manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Figure S1. Structures of TfALOGs and TfMIXTAs. (A) Gene structures of TfALOGs. (B) Multiple alignment of TfALOG proteins. The red line indicates the conserved DUF 640 domain. (C) Gene structures of TfMIXTAs. Box represents exon region and line represents intron region. Figure S2. Bayesian phylogram of ALOG genes in Asterids. Physcomitrella patens and Sphagnum fallax were chosen as outgroups. The Bayesian posterior probability is located in each node and the accession number can be found in each sequence. Seven ALOG genes cloned from Torenia fournieri were presented in red. Figure S3. The predicted NLSs (Nuclear Localization Signal) of TfALOG proteins. The NLSs were highlighted in red. Figure S4. Other phenotypes of transgenic plants. (A) Petal and leaf phenotypes analysis of wild type (WT) and 35S:TfALOG1 transgenic plants; two independent lines 35S:TfALOG1 16# and 35S:TfALOG1 26# were used for analysis. Two dorsal, two laterals and one ventral petal were dissected from the flower. (B) Flower phenotypes of 35S:TfALOG2 transgenic plants; two independent lines 35S:TfALOG2 2# and 35S:TfALOG2 3# were used for analysis. Figure S5. Relative expression of TfMIXTA and TfALOG genes. Relative expression of TfMIXTA and TfALOG genes in different tissues of wild type flowers were determined using qRT-PCR. WT10-L, petal lobes from stage 10 flowers; WT10-T, petal tubes from stage 10 flowers; WT11-L, petal lobes from stage 11 flowers; WT11-T, petal tubes from stage 11 flowers; WT12-L, petal lobes from stage 12 flowers; WT12-T, petal tubes from stage 12 flowers. Error bars represent ±1 SD from three biological replicates. Figure S6. Details of transgenic plants. (A) Relative expression of TfALOG1 in WT and two independent lines 35S:TfALOG1 16# and 35S:TfALOG1 26#. Error bars represent ±1 SD from three biological replicates. (B) Relative expression of TfALOG2 in WT and two independent lines 35S:TfALOG2 2# and 35S:TfALOG1 3#. Error bars represent ±1 SD from three biological replicates. (C) Designed CRISPR/Cas9 targets. Targets were highlighted in yellow, start and end codons were marked in red. (D) Sequence alignments of TfALOG1 in two TfALOG1 knock-out lines, designed targets were marked by red boxes. Table S1. Primers used in this study. (DOCX 5469 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Xiao, W., Ye, Z., Yao, X. et al. Evolution of ALOG gene family suggests various roles in establishing plant architecture of Torenia fournieri. BMC Plant Biol 18, 204 (2018). https://doi.org/10.1186/s12870-018-1431-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-018-1431-1