Abstract
Background
Dof (DNA binding with one finger) proteins, a class of plant-specific transcription factors which contain a conserved C2-C2-type zinc finger domain, are involved in many fundamental processes. In the Arabidopsis photoperiod response pathway, CDF (CYCLING DOF FACTOR) proteins have a primary role as acting via transcriptional repression of the direct FLOWERING LOCUS T (FT) activator CONSTANS (CO). Our previous study indicated that one of CDF homologs, OsDOf12, was involved in photoperiodic flowering. However, the functional characterization of other rice CDF like genes is still in progress. Here, we characterized the function of OsDof4 in rice.
Results
Phylogenic analysis indicated that OsDof4 is closely clustered into the same subgroup with CDFs and OsDof12. The subcellular localization experiment and transcriptional activity assay suggested that OsDof4 may function as a transcription factor. The diurnal expression pattern indicated that OsDof4 was regulated by endogenous circadian clock. Overexpression of OsDof4 led to earlier flowering under natural long-day field conditions (NLDs) and late flowering under natural short-day field conditions (NSDs), respectively. We compared the expression level of key floral genes in vector line and OsDof4-ox lines grown under long-day conditions (LDs) and short-day conditions (SDs). Real-time q-PCR results demonstrated that under LDs, Hd3a, RFT1 and Ehd1 were up-regulated whereas under SDs they were down-regulated. Hd1 was down-regulated at dusk period independent of photoperiods.
Conclusions
Taken these results together, we may speculate that the abnormal flowering responses in OsDof4-ox plants under LDs and SDs might be mediated by Ehd1 and Hd1.
Similar content being viewed by others
Background
Plants experience floral transition at the most favorable time to maximize their reproductive success [1,2,3]. Environmental cues are aligned with endogenous signals to determine the timing of flowering, of which photoperiod has largely been proven to be a principal determinant [4, 5]. Extensive studies have revealed Arabidopsis thaliana, a facultative long-day plant (LDP), and rice (Oryza sativa), a facultative short-day plant (SDP), share a conserved photoperiodic flowering signaling cascade: GIGANTEA (GI)-CONSTANS (CO)-FLOWERING LOCUS T (FT) [6, 7]. In Arabidopsis, under inductive long-day conditions (LDs) GI and FLAVIN-BINDING, KELCH REPEAT, F-Box 1 (FKF1) form more stable complexes to increase the CO expression level, a direct activator of FT, by post-transcriptionally degrading CYCLING DOF FACTOR1 (CDF1) which is a CO transcriptional repressor, thus promoting flowering [8, 9]. In rice, under inductive short-day conditions (SDs) OsGI activates Heading date 1 (Hd1), the counterpart of CO, to induce the homolog of FT-Heading date 3a (Hd3a), causing early flowering [10,11,12]. Nevertheless, there indeed exist some distinct floral molecular mechanisms between these two species [6, 7, 13]. For instance, when switched from inductive SDs to non-inductive LDs, Hd1 converts into a repressor of Hd3a by the modification of red-light photoreceptor Phytochrome B (PhyB) [14].
Moreover, compared with Arabidopsis, rice has evolved another unique Grain number, plant height and heading date 7 (Ghd7)-Early heading date 1(Ehd1)-Hd3a/Rice FT-like 1 (RFT1) flowering procedure because Ghd7 and Ehd1 are rice specific genes [6, 7]. Ehd1 acts as a positive regulator of Hd3a and RFT1 under both SDs and LDs [15]. Ghd7 is induced by red light and acts as a repressor of Ehd1 [16, 17]. Therefore, under LDs, Ghd7 accumulated more and subsequently down-regulates Ehd1 [17]. In comparison, under SDs, less-induced Ghd7 attenuates the repression on Ehd1, thus activating Hd3a and RFT1 [17]. Furthermore, Early heading date 2 (Ehd2)/Rice Indeterminate 1 (RID1)/Oryza sativa Indeterminate 1 (OsId1), Early heading date 3 (Ehd3), and Early heading date 4 (Ehd4) function as a positive regulator upstream of Ehd1 under both SDs and LDs [18,19,20,21,22]. On the other hand, OsMADS51 promotes Ehd1 under SDs, whereas OsMADS50 and OsMADS56 act antagonistically via OsLFL1-Ehd1 to control flowering under LDs [23, 24]. Days to heading 8 (DTH8) and Early flowering 1 (EL1)/Heading date 16 (Hd16) are inhibitors of Ehd1 and Hd3a under LDs [25,26,27]. Hence, those regulators upstream of Ehd1 and Ghd7 constitute novel flowering network in rice.
Dof (DNA binding with one finger) proteins, a class of plant-specific transcription factors which contain a conserved C2C2-type zinc finger domain, are involved in many fundamental processes, such as phytohormone response, seed germination, and photoperiodic flowering [28, 29]. In Arabidopsis, systematic and genetic study revealed 4 Dofs (including CDF1, CDF2, CDF3, and CDF5) acted redundantly to delay flowering by depressing the transcription of CO [30]. In rice, 2 Dofs, including rice Dof daily fluctuations 1 (RDD1) and OsDof12, were previously reported to participate in rice flowering time regulation [31, 32]. RDD1-RNAi plants flowered later than wild type [31]. Overexpression of OsDof12 leads to earlier flowering under LDs by up-regulating Hd3a [32].
In present study, we characterized the function of another Dof member-OsDof4 in rice by ectopic expression approaches. Phylogenic analysis showed that protein OsDof4 belonged to the same clade with proteins OsDof12 and CDF1–5. The subcellular localization and transcriptional activity analysis indicated OsDof4 is a plausible transcription factor. Tissue specific profiling showed OsDof4 is enriched in developing leaves and young panicles, and the diurnal expression rhythm indicated OsDof4 might be regulated by endogenous circadian clock. Constitutive expression of OsDof4 gave rise to earlier flowering but delayed flowering, under LDs and SDs, respectively. Further analysis on flowering genes expression levels suggested overexpression of OsDof4 might affect the flowering time by a regulating Hd3a and RFT1 via Ehd1 and Hd1.
Methods
Plant materials and growth conditions
Rice (Oryza sativa L.) subspecies japonica cultivar Nipponbare was used in this study. The seeds of wild type Nipponbare and the derived transgenic rice lines are maintained in our lab. Rice seeds were sterilized by 3% NaClO solutions, and immersed in water for 2 days, then the germinated seeds were moved onto the soil seed bed in greenhouse with 14 h light supplied. After growth for 28 days, the young seedlings were transplanted to the paddy field. For evaluation of the performance under natural-day conditions, plants were grown in the Experimental Stations (Beijing and Hainan Island) of the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, China (Beijing: 40°10′N, 116°42′E; Hainan Island: 18°32′N, 110°02′E). The average day lengths in Beijing were approximately 14.3 h at germination, 15 h at 30 days after germination (DAG), 14.8 h at 60 DAG, 13.7 h at 90 DAG. The average day lengths in Hainan Island were approximately 11 h at germination, 11.1 h at 30 DAG, 11.3 h at 60 DAG, 11.7 h at 90 DAG.
Various WT tissues were collected at heading stage to determine the OsDof4 mRNA levels. For flowering genes expression analysis, the germinated seeds were grown in the growth chamber under SD conditions (10 h light, 28 °C/14 h dark, 24 °C) and LD conditions (14 h light, 28 °C/10 h dark, 24 °C), with the humidity set at approximately 70%. The sampling happened for SD and LD respectively at 28 DAG and 35 DAG. Leaf blades were collected every 3 h under both conditions and stored in liquid nitrogen before total RNA extraction.
Protein sequence alignment and phylogenic analysis
All the rice Dof protein sequences were downloaded from Rice Genome Annotation Project (RGAP) (http://rice.plantbiology.msu.edu/); the protein sequences of 5 AtDofs were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/). ClustalW program with default parameters was used for protein sequence alignment [33]. The phylogenic tree was constructed using MEGA v5 software by the neighbor-joining method with a bootstrap value of 1000 [34].
Subcellular localization and rice transformation
The coding sequence without a termination code of OsDof4 was amplified by PCR using full length-cDNA(FL-cDNA) clone (AK066984) as the template. The FL-cDNA clone was kindly provided by the National Institute of Agrobiological Sciences, Japan (NIAS DNA BANK; http://www.dna.affrc.go.jp/). The PCR products was digested by EcoR I and BamH I, and inserted into the binary vector pEZR(K)-LN to create the overexpression vector of OsDof4 driven by a CaMV 35S promoter with GFP tagged at the C-terminus of OsDof4. The primers used were list in Additional file 1: Table S1. To construct OsDof4 overexpression lines, the overexpression vector (p35S::OsDof4:GFP) and control vector (p35S::GFP) were transformed to Agrobacterium tumefaciens strain LBA4404 (Shanghai Weidi Biotechnology, China) by electroporation, and introduced into wild type cultivator Nipponbare following the protocol as described previously [35].
For subcellular localization of OsDof4, the empty vector p35S::GFP or p35S::OsDof4:GFP with nuclear marker construct pSAT6-mCherry-VirD2NLS [36] were transiently co-expressed in rice protoplasts by means of PEG–mediated transformation [37]. The fluorescent signals were observed by a fluorescence confocal microscope (Zeiss Axio Imager.Z2, http://www.zeiss.com).
Transcriptional activation assay
The transcriptional activity of OsDof4 was evaluated by Dual-Luciferase Reporter Assay System (Promega, http://www.promega.com). The plasmids used in this assay were kindly provided by Prof. Shouyi Chen (Institute of Genetics and Biology, Chinese Academy of Science). pGAL4-LUC in which a minimal 35S plus five copies of GAL4 binding elements driving the firefly luciferase coding sequence was used as reporter. pTRL with AtUbiquitin3 promoter driving the Renilla reniformis luciferase gene was used as internal control [38]. The full-length coding sequence of OsDof4 was amplified with the primers (Additional file 1: Table S1) by PCR, and recombinated using NEBuilder® HiFi DNA Assembly Master Mix Kit (NEB, http://www.neb.com) into the sites Bam HI and Sal I at the 3′ end of GAL4 DNA binding domain coding sequence in pGAL4DBD to generate the effector plasmid pGAL4DBD-OsDof4. pGAL4DBD and pGAL4DBD-VP16 were used respectively as the negative and positive effector [38].
The plasmids with the content ratio of effector: reporter: internal control plasmids at 6:6:1(6μg:6μg:1μg) were co-transfected into rice leaf protoplasts via PEG mediated treatment [37]. After incubation in dark at 28 °C for 16 h, the relative luciferase activity(represented by the ratio of Firefly LUC activity to Renilla LUC activity) was measured by the GloMax 20–20 luminometer based on a Dual-Luciferase Reporter Assay System [38].
RNA extraction and real-time PCR analysis
The Total RNA was isolated from different rice tissues using Trizol reagent (Invitrogen, California, USA) according to the manufacturer’s instruction. DNA digestion was done by DnaseI (Takara, Japan), and first-strand cDNA was obtained by GoScript Reverse Transcription System Kit (Promega, http://www.promega.com). Real-time PCR was performed using EvaGreen qPCR Master Mix (Abm, Canada) on a real-time PCR System (Bio-Rad CFX96) with the specific primers. The real-time PCR program consists of 95 °C for 3 min and 42 cycles of 95 °C for 5 s, 60 °C for 15 s. Three replicates were performed for each gene and for each analysis two biological repeats were done. All the specific primers used were listed in Additional file 1: Table S1.
Statistical analysis
The data of transcriptional activity assay in Fig. 3b and the agronomic traits in Fig. 5c-f was subjected to Student’s t-test analysis using Microsoft Excel 2015.
Results
OsDof4 encodes a Dof transcription factor
We are quite interested in rice Dof genes because of their importance on various biological processes. In Arabidopsis, CDFs play key roles in photoperiodic flowering time [30, 39]. To explore the relationship between CDFs and rice Dof homologs, we conducted phylogenetic analysis with a neighbor-joining method. The phylogenic tree indicated that OsDof2, OsDof4, OsDof5, OsDof6, OsDof12, OsDof23 and OsDof26 are closely clustered with CDFs (Fig. 1a), thus we named them OsCDFs (Oryza sativa CDFs). In our previous study, we characterized the function of OsDof12 and found that, overexpression of OsDof12 led to earlier flowering [32]. To further investigate the biological roles of CDFs, we obtained OsDof4 overexpression lines. We screened and found the transgenic lines harboring OsDof4 (Locus Number: LOC_Os01g17000) coding sequence (CDS) showed abnormal flowering responses. According to the Rice Genome Annotation Project database (http://rice.plantbiology.msu.edu/), the reading frame of LOC_01g17000 totals 1473 base pairs (bp) with an intron of 2140 bp, encoding a protein of 490 amino acids. By searching the Pfam database(http://pfam.xfam.org), we found that the deduced protein is characteristic of DNA binding with one finger (Dof) domain (PF02701) located at N-terminus from 112 to 168 amino acids. Besides, the protein sequences aligned with CDF1, CDF2 and CDF3 indicated that OsDof4 shares highly conserved Dof domain and two motifs at the C-terminus with CDFs (Fig. 1b).
Phylogenic analysis and protein structure of OsDof4 homologs. a Phylogenic relationship between OsDof4 homologs from rice and Arabidopsis. An unrooted neighbor-joining tree was reconstructed based on the full–length amino acid sequences of rice Dofs and CDFs from Arabidopsis. Numbers stands for the bootsrap values along the branches. The protein sequences of rice Dofs were downloaded from RGAP, and the protein sequences of 5 Arabidopsis Dofs were downloaded from TAIR. b Protein sequence comparasion between OsDof4 and CDF1–3. The identical residues are highlighted in black,and the similar ones are in gray. The conserved Dof domain and two putative motifs were underlined
OsDof4 is localized in nucleus
In higher plants, several cases have shown Dof proteins by binding to promotor region regulate the expression of downstream targets in nucleus [40]. To gain more insight into the function of OsDof4, the full coding region of OsDof4 was fused with GFP, and introduced into rice leaf protoplasts, where the genes were transiently expressed under the control of CaMV 35S promotor. The fluorescent signal of OsDof4-GFP was specifically detected in nucleus where exclusively co-localized with mCherry-VirD2NLS (a nuclear marker) (Fig. 2a). In comparison, the free GFP signal was distributed in both nucleus and cytoplasm (Fig. 2a). Meanwhile, we analyzed the in vivo localization of OsDof4-GFP in transgenic plant roots. Similar to the results obtained in rice leaf protoplasts, OsDof4-GFP was predominantly distributed in nucleus whereas free GFP equally appeared in nucleus and cytoplasm (Fig. 2b), further confirming OsDof4 is nuclear-localized. Consequently, we proposed that OsDof4 might function as a nuclear transcription factor.
Subcellular localization of OsDof4. a Subcellular localization of OsDof4 in rice protoplasts. The vector 35S::OsDof4-GFP or 35S::GFP with the nuclear marker plasmids pSAT6-mCherry-VirD2NLS were co-transformed into and transiently expressed in rice protoplasts. Left to right: GFP fluorescence signal, RFP fluorescence signal, Bright-field signal, and Merged signal. Top panel: localization of OsDof4-GFP fusion proteins. Bottom panel: localization of free GFP proteins. Scale bar = 5um. The excitation wavelengths for GFP and RFP detection were respectively 488 nm and 587 nm. b Microscopic observation of the fresh root tips in 40-day-old transgenic plants. The top panel indicated the images acquired from 35S::GFP plants; The bottom panel indicated the images acquired from 35S::OsDof4-GFP plants. Left: GFP fluorescence signal; Right: Merged signal. Bar = 20um
OsDof4 has the capability of transcriptional activation
To confirm whether OsDof4 is a transcription factor, we tested the transcriptional activation ability by using a dual-luciferase reporter (DLR) assay system in rice protoplasts. The full coding sequence of OsDof4 was fused in-frame with pGAL4DBD to generate effector construct pGAL4DBD-OsDof4. With pGAL4DBD and pGAL4DBD-VP16 setting respectively as negative and positive control (Fig. 3a), we conducted a transcriptional activity assay. We found that pGAL4DBD-OsDof4 exerted obviously enhanced transcriptional activation ability compared with pGAL4DBD (Fig. 3b), implying that OsDof4 is capable of activating transcription, which further confirmed that OsDof4 encodes a rice transcription factor.
Transcriptional activity assay of OsDof4. a Schematic representation of effectors, reporter, and internal control plasmids structure. b The transcriptional activity of each effector. GAL4DBD and GAL4DBD-VP16 represented respectively negative and positive control effector. The plasmids of effector, reporter, and internal control were co-transformed into rice protoplasts, after overnight incubation in dark at 28 °C, the relative LUC activity of each effector was measured. With the relative LUC activity of GAL4DBD normalized to 1, obviously GAL4DBD-VP16 and GAL4DBD-OsDof4 showed higher transcriptional activity. Values are means ± SE (n = 5). Error bar indicates SE. The double asterisks indicate significant difference (p < 0.01) compared to GAL4DBD value according to student’s t test
Expression of OsDof4 is ubiquitous and diurnal
To determine the tissue specific expression profiles of OsDof4, we determined the OsDof4 mRNA abundance in various rice tissues by real-time PCR. OsDof4 showed constitutive expression pattern, and was most abundantly expressed in developing leaves (leaf 1) and young panicles, followed by root, stem, sheath and developed leaves (leaf 2, leaf 3) (Fig. 4a). OsDof4 exhibited diurnal expression pattern under both SDs and LDs, the mRNA abundance was much higher at day time than that at night time (Fig. 4b). The diurnal rhythm of OsDof4 mRNA level indicated that OsDof4 might be regulated by an endogenous circadian clock. To investigate whether OsDof4 is driven by circadian clock, we examined the OsDof4 transcript level in constant light conditions. Four-week-old wild type (WT,cv. Nipponbare) seedlings were firstly entrained in LDs for two weeks and then moved to constant light conditions (LLs). The leaf blades were harvested every three hours for 24 h in LDs and for 48 h in LLs. Then the collected samples were subjected to RNA extraction and real-time PCR analysis. The result showed that, though the amplitude in LLs was attenuated than that in LDs, the phases in both conditions were very alike (Fig. 4c). Thus, we postulated that OsDof4 is regulated by circadian clock.
Tissue-specific and diurnal expression profiles of OsDof4 in WT. a OsDof4 mRNA expression in various rice tissues. All the rice tissues were harvested from WT at heading stage. b Diurnal expression pattern of OsDof4. Leaf blades from 28- and 35-day-old WT grown under SDs and LDs, respectively, were collected for expression analysis. The solid line standed for the expression values under SDs, and the dotted line standed for the ones under LDs. c Free running OsDof4 expression curve in continuous light conditions. 4-week-old WT were firstly entrained in LDs for 2 weeks and then moved to constant light conditions (LLs). The leaf blades were harvested every three hours for 24 h in LDs and for 48 h in LLs. Values are means ± SE (n = 3). Error bar indicates SE
Overexpression of OsDof4 promotes flowering under NLDs but delays flowering under NSDs
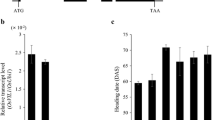
To elucidate the biological function of OsDof4, we constructed transgenic lines in which OsDof4 was driven under the control of CaMV 35S promotor (35S::OsDof4), and the empty vector lines were set as control. In total, we obtained eighteen and four positive lines harboring 35S::OsDof4 and empty vector, respectively. We randomly chose eight 35S::OsDof4 (including line OsDof4-ox-1/2/4/6/9/10/13/18) lines and one empty vector line to analyze the expression level of OsDof4 by using real-time PCR, and found all OsDof4-ox lines showed obviously much higher OsDof4 mRNA level than that in WT or empty vector plants (Additional file 2: Figure S1).
To investigate the biological effects that OsDof4 may bring, we grew the OsDof4-ox lines and vector lines in Beijing and Hainan experimental fields, respectively. We observed and statistically analyzed the agronomic traits of two OsDof4-ox lines (OsDof4-ox-13, OsDof4-ox-18), WT, and one vector line. Comparison of morphologic phenotypes between WT, vector plants and OsDof4-ox-13, OsDof4-ox-18 plants revealed that the plant height of WT or vector plants reached about 84 cm, while OsDof4-ox-13, OsDof4-ox-18 plants only were 78 cm(cm) in height which were significantly dwarf than WT or vector plants (Fig. 5a-c). Meanwhile, we found the tiller numbers in each lines were comparable (Fig. 5d).
Agronomic traits comparison between OsDof4-ox and control plants. a-b Morphologic comparision between WT, Vector and OsDof4-ox plants at heading stage. NLD and NSD represent for the pictures acquired in Beijing and Hainan Island, respectively. c-d Statistical data of plant height (c) and tiller number (d). The plant height and tiller number were measured after mature stage. e-f Statistical data of flowering time under NLD (e) and NSD (f). DTF represents for the days from germination to flowering. Values are means ± SE (n = 20). Error bar indicates SE. The double asterisks indicate significant difference (p < 0.01) compared to control according to student’s t test
Besides, we found the flowering time in OsDof4-ox-13, OsDof4-ox-18 plants were significantly different from that in WT or vector plants. When grew in Beijing (40°10′N, 116°42′E), which was more like natural long-day field conditions (NLDs), OsDof4-ox-13, OsDof4-ox-18 plants promoted flowering by 18 days than WT or vector plants on average (Fig. 5a, e). Interestingly, when grew in Hainan Island (18°32′N, 110°02′E), which was more like natural short-day field conditions (NSDs), OsDof4-ox-13, OsDof4-ox-18 plants delayed flowering by about 10 days than WT or vector plants (Fig. 5b, f). These results implied constitutive expression of OsDof4 affected the photoperiodic flowering response under both conditions.
Flowering genes expression are affected in OsDof4-ox plants
To gain more insight into the abnormal flowering responses and elucidate the corresponding molecular mechanisms in OsDof4-ox plants, we examined the key flowering regulators expression levels by conducting real-time PCR. OsDof4-ox plants and vector plants were grew under controlled conditions. Leaf blades from 28-day-old and 35-day-old seedlings cultivated respectively under SDs and LDs were harvested for expression analysis.
Under long-day conditions (LDs), two florigen genes, Hd3a and RFT1, were significantly up-regulated in OsDof4-ox-13, OsDof4-ox-18 plants compared with vector plants (Fig. 6a). Furthermore, we analyzed the expression levels of flowering genes upstream of Hd3a and RFT1, including Ghd7, Ehd1, and Hd1. The transcription level of Ghd7 was unimpaired in OsDof4-ox-13, OsDof4-ox-18 plants (Fig. 6a). Hd1 in OsDof4-ox-13, OsDof4-ox-18 plants was down-regulated in the dusk period but was not obviously affected in the light period than that in vector plants (Fig. 6a). Strikingly, the expression level of Ehd1 in OsDof4-ox-13, OsDof4-ox-18 plants was obviously much higher than that in vector plants (Fig. 6a).
Expression analysis of rice flowering genes in vector and OsDof4-ox plants. a-b mRNA levels of flowering genes under LDs (a) and SDs (b) were detected by real-time PCR. Leaf blades from 28- and 35-day-old WT grown under SDs and LDs, respectively, were collected for expression analysis. Blue lines represent for the expression curves of flowering genes in vector plants; Red and green lines respectively represent for the ones in OsDof4-ox-13/−18 plants. Values are means ± SE (n = 3). Error bar indicates SE
Under SD conditions (SDs), the transcription levels of Hd3a and RFT1 mRNAs were severely lower in OsDof4-ox-13, OsDof4-ox-18 plants (Fig. 6b). Similar to the observation under LDs, Ghd7 mRNA levels were comparable in OsDof4-ox plants and vector plants (Fig. 6b), and Hd1 in OsDof4-ox-13, OsDof4-ox-18 plants was down-regulated in the dusk period but was not obviously affected in the light period than that in vector plants (Fig. 6b). However, the expression level of Ehd1 in OsDof4-ox-13, OsDof4-ox-18 plants was strongly depressed than that in vector plants (Fig. 6b).
Taken together, these results indicated overexpression of OsDof4 may up-regulate but down-regulate Hd3a and RFT1 under LDs and SDs, respectively, by modulating the expression levels of Ehd1 and Hd1.
Discussion
In plants, Dof genes have been well documented participating in various important developmental events, including flowering [28, 29]. There has much progress in understanding Dof genes involved floral mechanisms in model dicot plant-Arabidopsis, however, the counterpart in rice remain rather limited. Our previous study showed overexpression of OsDof12 promoting flowering by up-regulating Hd3a under LDs in rice [32]. In the present study, we demonstrated that constitutive expression of another Dof member, OsDof4 in rice, could lead to distinct flowering phenotypes under LDs and SDs, respectively. Further evidences implied affected expression of Ehd1 might be responsible for the abnormal flowering time in OsDof4 overexpression plants. Taken together, our study clearly reveals that OsDof4 plays a pivotal role in rice flowering pathway.
Tissue specific expression profile displayed that OsDof4 was ubiquitously expressed in root, stem, leaf and young panicle (Fig. 4a), suggesting OsDof4 might be a versatile regulator. Indeed, OsDof4-ox lines showed significantly reduced plant height than control lines (Fig. 5a-c). Interestingly, our recent study indicated that another Dof member in rice-OsDof12 may be involved in plant height regulation [41]. Considering proteins OsDof4 and OsDof12 are clustered into one clade, we surmised that OsDof4 may play an analogic role as OsDof12 does in plant height regulation. However, its underlying molecular mechanisms remain to be further investigated. Endogenous circadian clock is involved in many biological processes by regulating various downstream transcriptional regulators [42, 43]. The five Arabidopsis Dof members CDF1, CDF2, CDF3, CDF4 and CDF5 all showed circadian clock-regulated rhythms [30, 39], meanwhile, the daily oscillations in Rdd1 expression also indicated it was circadian-regulated in rice [31]. By observing the expression rhythm of OsDof4 in LLs, we speculated that OsDof4 might be also controlled by endogenous circadian clock. Notably, mRNA level of Rdd1 peaked at subjective light period and, OsDof12 transcriptional level decreased under dark condition. By contrast, OsDof4 mRNA transcripts accumulated and peaked at light period but, declined at dark period (Fig. 4b-c). These observations imply that though OsDof4 shares similar protein sequence with Rdd1 and OsDof12, still, there might exist distinguished biological function between them because of the differences in diurnal expression profiles.
In Arabidopsis, Fornara et al. (2009) systematically misexpressed 26 Dof members in companion cells by using the SUCROSE TRANSPORTER 2 (SUC2) promoter, and found that 5 Dof members overexpressors transgenic lines displayed delayed flowering time under LDs [30]. Phylogenic analysis suggested those 5 Dof members, including CDF2, CDF3, CDF4, CDF5 and COG1, clustered in the same clade with CDF1 which had been reported to delay flowering when overexpressed by CaMV 35S promoter [30, 39]. Coincidently, our result indicated OsDof4 also belongs to the same subgroup, suggesting OsDof4 might share similar conserved role in flowering pathway with those Arabidopsis Dof members. We conducted reverse-genetics approaches to characterize the function of OsDof4 because there were no corresponding mutants in the rice mutant database. Observation of the OsDof4-ox lines flowering time come to a conclusion that overexpression of OsDof4 caused opposite effects on flowering under LDs and SDs, respectively. It is of interest to note that OsDof12-ox lines also flowered earlier under LD but no clear difference occurred under SDs [32]. Moreover, CDFs overexpressors flowered later under LDs rather than under SDs in Arabidopsis [30, 39]. Therefore, as a homolog of OsDof12 and CDFs, OsDof4 displayed novel roles in flowering regulation especially under SDs.
To decipher the mechanisms underlying the affected photoperiod sensitivity in OsDof4-ox plants, we tested the expression of key floral regulators. Up to now, the largely accumulated evidences have suggested that Hd3a and RFT1 are indispensable for rice flowering [44, 45]. We found that Hd3a and RFT1 transcript-level expression in OsDof4-ox were higher but lower than that in control plants under LDs and SDs (Fig. 6a-b), respectively, which were closely corresponding with the flowering time. Considering Hd1 and Ehd1 are the main transcriptional regulators upstream of Hd3a and RFT1 [11, 15, 17], we further carried out real-time PCR to detect their expression. The results showed that Ehd1 was positively regulated in OsDof4-ox plants under LDs (Fig. 6a). However, under SDs the scenario was opposed where Ehd1 was strongly repressed in OsDof4-ox plants (Fig. 6b). Meanwhile, in OsDof4-ox plants, Hd1 was not obviously affected in the light period but, repressed at dust period independent of photoperiod (Fig. 6a-b). Previous studies have revealed that, Ghd7, OsGI, OsMADS50, OsMADS51, OsMADS56, Ehd2 and DTH8 are involved in regulation of Hd1 or Ehd1 [12, 16,17,18,19,20, 23,24,25, 46]. Therefore, we also tried to figure out whether the expression variation in Ehd1 or Hd1 in OsDof4-ox plants were mediated by these regulators under LDs but, found that all these regulators displayed robust expression as control plants did (Additional file 3: Figure S2). Previous reports have revealed that Ehd1 functions as a floral promotor under both LDs and SDs and, Hd1 acts as a florigen activator under SDs but converts into a repressor under LDs [10, 11, 14, 15, 45]. Consequently, in combination with our experimental data, we deduced that the impaired transcript-level expression of Hd3a and RFT1 or flowering time in OsDof4-ox plants was probably mediated by Ehd1, Hd1 and some other factors.
In rice, a near-isogenic line carrying a weak photoperiod-sensitive allele of EL1/Hd16 (Koshihikari) was reported to promote flowering under LDs and inhibit flowering under SDs [26]. Meanwhile, Hd16-RNAi plants showed earlier flowering under LDs and showed later flowering under SDs than WT plants [26]. The flowering phenotypes in Hd16-RNAi lines under SDs and LDs are very similar to that in OsDof4-ox lines. Further analysis showed that Hd16 controls the flowering time via modulating the expression of Ehd1 and its downstream Hd3a and RFT1, which indicates Hd16 and OsDof4 might share the same pathway. However, from our transcriptome date (not published) we found Hd16 was not affected under SDs and LDs in OsDof4-ox lines, thus we speculated that OsDof4 may not regulate Hd16 at transcriptional level. Anyway, the exact genetic relationship between OsDof4, Hd16 and Ehd1 should be investigated in future work.
Subcellular localization (Fig. 2a-b) and transcriptional activity assay (Fig. 3b) together strongly indicated OsDof4 functions as a transcription factor, but that whether OsDof4 directly regulates Ehd1 and Hd1 or not should be further revealed. Besides, more recently, Nemoto et al. [47] suggested that, under LDs Hd1 and Ghd7 form a complex binding to the cis-regulatory elements in Ehd1 and repress it. Therefore, to which extent the expression variation in Hd1 accounts for the impaired Ehd1 expression, at least under LDs, remains to be investigated.
Overexpression of OsDof4 leads to earlier but late flowering under LDs and SDs, respectively. On the contrary, whether the plants harboring down-expressed or knock-outed OsDof4 induce opposed effects should be further validated. Actually, to fully understand the role of OsDof4, we successfully constructed OsDof4 knockout mutant-osdof4 in wild type Nipponbare background, by using gene editing tool-CRISPR/Cas9 [48]. Comparison of the OsDof4 allele sequences between WT and osdof4-KO lines revealed that there was a 4-bp deletion in line SG1546–3 and multi-site deletion in another line SG1546–6, resulting in a premature stop codon in the open reading frame (data not shown). Thus, we concluded that OsDof4 was specifically knocked out in osdof4-KO lines. Therefore, these results suggested that loss-of-function in OsDof4 do not affect the flowering time which may result from the functional redundancy of other Dof family genes. Multi-Dof-mutation approaches in future study will provide more cues to determine which Dof genes compensate the loss-of-function effect of OsDof4 on flowering time.
Conclusions
Collectively, we demonstrate that OsDof4 plays important roles in regulating rice flowering time. Overexpression of OsDof4 led to earlier flowering under LDs and late flowering under SDs, respectively. Our results imply that the abnormal flowering responses in OsDof4-ox plants under LDs and SDs might be mediated by Ehd1 and Hd1. This finding could help us better understand the photoperiodic flowering in rice.
Abbreviations
- CDF:
-
CYCLING DOF FACTOR
- CO:
-
CONSTANS
- Dof:
-
DNA binding with one finger
- DTH8:
-
Days to heading 8
- Ehd1:
-
Early heading date 1
- Ehd2:
-
Early heading date 2
- Ehd3:
-
Early heading date 3
- Ehd4:
-
Early heading date 4
- EL1:
-
Early flowering 1
- FKF1:
-
FLAVIN-BINDING KELCH REPEAT, F-Box 1
- FT:
-
FLOWERING LOCUS T
- Ghd7:
-
Grain number, plant height and heading date 7
- GI:
-
GIGANTEA
- Hd1:
-
Heading date 1
- Hd16:
-
Heading date 16
- Hd3a:
-
Heading date 3a
- LDs:
-
long-day conditions
- NLDs:
-
natural long-day field conditions
- NSDs:
-
natural short-day field conditions
- OsId1:
-
Oryza sativa Indeterminate 1
- RDD1:
-
Dof daily fluctuations 1
- RFT1:
-
Rice FT-like 1
- RID1:
-
Rice Indeterminate 1
- SDs:
-
short-day conditions
References
Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125(4):655–64.
Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11(11):550–8.
Zhou M, Luo H. MicroRNA-mediated gene regulation: potential applications for plant genetic engineering. Plant Mol Biol. 2013;83(1–2):59–75.
Fornara F, de Montaigu A, Coupland G. SnapShot: control of flowering in Arabidopsis. Cell. 2010;141(3):550. 550 e551-552
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013;161(3):1375–91.
Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr Opin Plant Biol. 2011;14(1):45–52.
Tsuji H, Taoka K, Shimamoto K. Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr Opin Plant Biol. 2013;16(2):228–35.
Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318(5848):261–5.
Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336(6084):1045–9.
Yano M, Katayose Y, Ashikari M, Yamanouchi U, Monna L, Fuse T, Baba T, Yamamoto K, Umehara Y, Nagamura Y, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12(12):2473–83.
Kojima S, Takahashi Y, Kobayashi Y, Monna L, Sasaki T, Araki T, Yano M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002;43(10):1096–105.
Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422(6933):719–22.
Hayama R, Coupland G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 2004;135(2):677–84.
Ishikawa R, Aoki M, Kurotani K-i, Yokoi S, Shinomura T, Takano M, Shimamoto K. Phytochrome B regulates heading date 1 (Hd1)-mediated expression of rice florigen Hd3a and critical day length in rice. Mol Gen Genomics. 2011;285(6):461–70.
Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18(8):926–36.
Xue WY, Xing YZ, Weng XY, Zhao Y, Tang WJ, Wang L, Zhou HJ, SB Y, CG X, Li XH, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–7.
Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet. 2010;42(7):635–8.
Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M. Ehd2, a Rice Ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 2008;148(3):1425–35.
Park SJ, Kim SL, Lee S, Je BI, Piao HL, Park SH, Kim CM, Ryu CH, Park SH, Xuan YH, et al. Rice indeterminate 1 (OsId1) is necessary for the expression of Ehd1 (early heading date 1) regardless of photoperiod. Plant J. 2008;56(6):1018–29.
Wu C, You C, Li C, Long T, Chen G, Byrne ME, Zhang Q. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Natl Acad Sci. 2008;105(35):12915–20.
Matsubara K, Yamanouchi U, Nonoue Y, Sugimoto K, Wang ZX, Minobe Y, Yano M. Ehd3, encoding a plant homeodomain finger-containing protein, is a critical promoter of rice flowering. Plant J. 2011;66(4):603–12.
Gao H, Zheng XM, Fei G, Chen J, Jin M, Ren Y, Wu W, Zhou K, Sheng P, Zhou F, et al. Ehd4 encodes a novel and oryza-genus-specific regulator of photoperiodic flowering in rice. PLoS Genet. 2013;9(2):e1003281.
Kim SL, Lee SY, Kim HJ, Nam HG, An GH. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 2007;145(4):1484–94.
Ryu CH, Lee S, Cho LH, Kim SL, Lee YS, Choi SC, Jeong HJ, Yi J, Park SJ, Han CD, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant Cell Environ. 2009;32(10):1412–27.
Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153(4):1747–58.
Hori K, Ogiso-Tanaka E, Matsubara K, Yamanouchi U, Ebana K, Yano M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013;76(1):36–46.
Kwon CT, Yoo SC, Koo BH, Cho SH, Park JW, Zhang Z, Li J, Li Z, Paek NC. Natural variation in early flowering1 contributes to early flowering in japonica rice under long days. Plant Cell Environ. 2014;37(1):101–12.
Yanagisawa S. The Dof family of plant transcription factors. Trends Plant Sci. 2002;7(12):555–60.
Noguero M, Atif RM, Ochatt S, Thompson RD. The role of the DNA-binding one zinc finger (DOF) transcription factor family in plants. Plant Sci. 2013;209(0):32–45.
Fornara F, Panigrahi KCS, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17(1):75–86.
Iwamoto M, Higo K, Takano M. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant Cell Environ. 2009;32(5):592–603.
Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L. Functional characterization of rice OsDof12. Planta. 2009;229(6):1159–69.
Zhang B, Chen W, Foley RC, Buttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell. 1995;7(12):2241–52.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza Sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6(2):271–82.
Lee LY, Fang MJ, Kuang LY, Gelvin SB. Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods. 2008;4:24.
Liang Z, Zhang K, Chen K, Gao C. Targeted mutagenesis in Zea Mays using TALENs and the CRISPR/Cas system. J Genet Genom. 2014;41(2):63–8.
Wei W, Huang J, Hao Y-J, Zou H-F, Wang H-W, Zhao J-Y, Liu X-Y, Zhang W-K, Ma B, Zhang J-S, et al. Soybean GmPHD-type transcription regulators improve stress tolerance in transgenic Arabidopsis plants. PLoS One. 2009;4(9):e7209.
Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309(5732):293–7.
Whiteside ST, Goodbourn S. Signal transduction and nuclear targeting: regulation of transcription factor activity by subcellular localisation. J Cell Sci. 1993;104(Pt 4):949–55.
Wu Q, Li D, Li D, Liu X, Zhao X, Li X, Li S, Zhu L. Overexpression of OsDof12 affects plant architecture in rice (Oryza Sativa L.). Front Plant Sci. 2015;6:833.
Wijnen H, Young MW. Interplay of circadian clocks and metabolic rhythms. Annu Rev Genet. 2006;40:409–48.
Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–77.
Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–6.
Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135(4):767–74.
Lee S, Kim J, Han JJ, Han MJ, An G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004;38(5):754–64.
Nemoto Y, Nonoue Y, Yano M, Izawa T. Hd1,a CONSTANS Orthlog in Rice, functions as an Ehd1 repressor through interaction with monocot-specific CCT-domain protein Ghd7. Plant J. 2016;86:221–3.
Belhaj K, Chaparro-Garcia A, Kamoun S, Patron NJ, Nekrasov V. Editing plant genomes with CRISPR/Cas9. Curr Opin Biotechnol. 2015;32:76–84.
Acknowledgements
We would like to thank Prof. Shouyi Chen for providing assistance on transcriptional activity assay. We also thank Dr. Jun Fang (Northeast Institute of Geography and Agroecology, Chinese Academy of Sciences, China) and Dr. Man Zhou (Department of Genetics and Biochemistry, Clemson University, USA) for critical reading on the manuscript.
Funding
This research was supported by the National Key Research and Development Program of China (2016YFD0100904), the Major Project of New Varieties of Genetically Modified Organism of China (2016ZX08009–001), the State Key Laboratory of Plant Genomics, China (2017A0407–19), the National Natural Science Foundation of China (31,471,475; 31,571,634) and the State Key Laboratory of Hybrid Rice.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files. Sequence data used in this manuscript can be found in the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org) and the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu) under the following accession numbers: OsDof1(LOC_Os01g64590),OsDof2(LOC_Os01g15900), OsDof3(LOC_Os01g09720),OsDof4(LOC_Os01g17000),OsDof5(LOC_Os01g48290),OsDof6(LOC_Os01g55340),OsDof7(LOC_Os02g47810),OsDof8(LOC_Os02g49440),OsDof9(LOC_Os02g45200),OsDof10(LOC_Os02g15350),OsDof11(LOC_Os03g38870),OsdDof12(LOC_Os03g07360),OsDof13(LOC_Os03g42200),OsDof14(LOC_Os03g16850),OsDof15(LOC_Os03g55610),OsDof1 6(LOC_Os03g60630),OsDof17(LOC_Os04g58190),OsDof18(LOC_Os04g47990),OsDof19(LOC_Os05g02150),OsDof20(LOC_Os06g17410),OsDof21(LOC_Os07g13260),OsDof22(LOC_Os07g32510), OsDof23(LOC_Os07g48570),OsDof24(LOC_Os08g38220),OsDof25(LOC_Os09g29960),Dof26(LOC_Os10g26620),OsDof27(LOC_Os10g35300),OsDof28(LOC_Os12g38200),OsDof29(LOC_Os05g36900),OsDof30(LOC_Os12g39990),CDF1(At5g62430),CDF2(At5g39660),CDF3(At3g47500),CDF4(At2g34140),CDF5(At1g69570). The neighbor-joining tree in Fig. 1 and related sequences were deposited in TreeBASE (http://treebase.org) under the following URL: http://purl.org/phylo/treebase/phylows/study/TB2:S21270.
Author information
Authors and Affiliations
Contributions
LZ, SL and DL conceived and designed the experiments. QW and XuL performed most of the experiments. QW, LZ, SL and DL wrote the manuscript. QX constructed the rice transgenic lines. DY, HY, XiL, and XZ performed transcriptional activity assay, phenotypes observation and statistical analysis. All authors have read and approved the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
The primers sequences used in this study. (DOCX 19 kb)
Additional file 2: Figure S1.
OsDof4 expression levels in WT, vector line and OsDof4-ox lines. Leaf blades from plants before heading stage were collected for real-time PCR. Values are means ± SE (n = 3). Error bar indicates SE. (TIFF 1080 kb)
Additional file 3: Figure S2.
Expression analysis of key floral genes in vector and OsDof4-ox plants under LDs. mRNA levels of flowering genes under LDs were detected by real-time q-PCR. Leaf blades from 35-day-old WT grown under LDs were collected for expression analysis. Blue lines represent for the expression curves of flowering genes in vector plants; Red and green lines respectively represent for the ones in OsDof4-ox-13/−18 plants. Values are means ± SE (n = 3). Error bar indicates SE. (TIFF 2564 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, Q., Liu, X., Yin, D. et al. Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza sativa L.). BMC Plant Biol 17, 166 (2017). https://doi.org/10.1186/s12870-017-1109-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-017-1109-0