Abstract
Background
Abscisic acid (ABA) signaling plays a crucial role in developmental and environmental adaptation processes of plants. However, the PYL-PP2C-SnRK2 families that function as the core components of ABA signaling are not well understood in banana.
Results
In the present study, 24 PYL, 87 PP2C, and 11 SnRK2 genes were identified from banana, which was further supported by evolutionary relationships, conserved motif and gene structure analyses. The comprehensive transcriptomic analyses showed that banana PYL-PP2C-SnRK2 genes are involved in tissue development, fruit development and ripening, and response to abiotic stress in two cultivated varieties. Moreover, comparative expression analyses of PYL-PP2C-SnRK2 genes between BaXi Jiao (BX) and Fen Jiao (FJ) revealed that PYL-PP2C-SnRK2-mediated ABA signaling might positively regulate banana fruit ripening and tolerance to cold, salt, and osmotic stresses. Finally, interaction networks and co-expression assays demonstrated that the core components of ABA signaling were more active in FJ than in BX in response to abiotic stress, further supporting the crucial role of the genes in tolerance to abiotic stress in banana.
Conclusions
This study provides new insights into the complicated transcriptional control of PYL-PP2C-SnRK2 genes, improves the understanding of PYL-PP2C-SnRK2-mediated ABA signaling in the regulation of fruit development, ripening, and response to abiotic stress, and identifies some candidate genes for genetic improvement of banana.
Similar content being viewed by others
Background
In plants, phytohormone abscisic acid (ABA) regulates numerous developmental processes, such as seedling development, seed dormancy, and fruit ripening [1,2,3,4,5]. In addition, ABA plays a central role in the adaptation of plants to environmental stresses, such as drought, salinity, and cold [6, 7]. Due to the biological and agricultural importance of ABA, many studies have focused on plant responses to ABA at the level of cytology and molecular biology. Since 2009, the ABA signaling pathway began to be better understood [6]. PYR/PYL/RCARs (ABA receptors), Group A PP2Cs (negative regulators), and SnRK2s (positive regulators) were confirmed as crucial components of ABA signaling in Arabidopsis. Finally, a double negative regulatory model is constituted by these components. SnRK2s activities are repressed by direct dephosphorylation by Group A PP2Cs in the absence of ABA. When responding to developmental or environmental clues, the ABA signal induces PYR/PYL/RCAR interaction with Group A PP2Cs, including ABI1, ABI2, AHG3, and HAB1, leading to inhibition of Group A PP2Cs and activation of SnRK2s [6, 8,9,10]. This results in phosphorylation or activation of downstream targets, such as ABF/AREB/ABI5, SLAC1, and other ABA-responsive gene products [6, 11]. The ABA-mediated interaction model between PYLs and PP2Cs was validated by in vitro reconstitution in Arabidopsis protoplasts [12].
Additionally, the function of PYL-PP2C-SnRK2 genes in developmental processes and in response to ABA and abiotic stress were characterized in plants. PYL9, PYL5 or PYL8 overexpression improved drought tolerance or ABA responses in Arabidopsis [9, 13, 14]. In contrast, an ABA insensitive phenotype was observed in the quadruple mutant of pyr1 pyl1 pyl2 pyl4 [10]. Double and triple mutation of several crucial members of Group A PP2Cs (ABI1, ABI2, HAB1, HAB2, AHG1, and PP2CA) resulted in enhanced ABA sensitivity, indicating the negative roles of Group A PP2Cs in ABA signaling [15,16,17,18,19]. Interference of AtPP2CA increased tolerance to freezing stress and ABA sensitivity in Arabidopsis [20]. Mutation of abi2-1 resulted in enhanced tolerance to salt stress and ABA insensitivity in Arabidopsis [21]. Overexpression of SnRK2.8 improved tolerance to drought stress in Arabidopsis [22]. Conversely, mutation of snrk2.2, snrk2.3, and snrk2.6 decreased drought stress tolerance and ABA responses, such as seed germination, plant growth, stomatal behavior [6]. Besides, the similar roles of PYL and SnRK2 genes were also observed in rice. Overexpression of OsPYL3 or OsPYL9 positively regulated the ABA response during seed germination and improved drought and cold stress tolerances in rice [23]. OsPYL/RCAR5 overexpressing rice plants showed hypersensitivity to ABA during seed germination [24]. Overexpression of SAPK4 in rice resulted in improved germination, growth and development under salt stress both in seedlings and mature plants [25]. OsSAPK9 was reported to improve drought tolerance and grain yield through regulating cellular osmotic potential, stomatal closure and stress-responsive gene expression in rice [26]. Interestingly, Arabidopsis plants overexpressing OsPP108 (a Group A PP2C gene in rice) showed highly insensitivity to ABA and tolerance to salt and osmotic stresses during seed germination, root growth and overall seedling growth. This indicated that OsPP108 negatively regulates ABA signaling and positively regulates abiotic stress tolerance [27]. Together, this evidence suggests that Group A PP2Cs negatively regulate ABA signaling and negatively/positively regulate ABA-mediated biological processes; and PYLs and SnRK2s could positively regulate the response of plants to these processes.
To date, genes that encode the crucial components of ABA signaling have been identified in several species based on genome sequencing. There are 14 PYLs in Arabidopsis, 13 in rice, 10 in Selaginella moellendorffi, and 4 in Physcomitrella patens; 9 Group A PP2Cs in Arabidopsis, 10 in rice, 5 in Selaginella moellendorffi, and 2 in Physcomitrella patens; and 10 SnRK2s in Arabidopsis, 11 in rice, 6 in Selaginella moellendorffi, and 4 in Physcomitrella patens [6]. In spite of the economic and social importance of banana and the critical role of PYL-PP2C-SnRK2s in the plant development and stress responses, no information is known about the PYL-PP2C-SnRK2 gene family in banana. Banana is the largest fruit crop and vital for food security for millions of people around the world [28, 29]. Because it is mainly cultivated as a staple food in many impoverished continents, such as Africa, banana studies have proceeded slowly [30]. Investigation of genes in the signal transduction pathways on the basis of complete genome sequences is of benefit for revealing the cellular biological processes [31]. The banana genome sequencing was finished in 2012 [32], which supplies full genome data for us to perform systematic analyses of PYL-PP2C-SnRK2 gene families.
In this study, we identified 24 PYLs, 87 PP2Cs, and 11 SnRK2s from the banana genome and investigated their phylogenetic relationships, protein motifs, gene structure, and expression patterns in different tissues, in diverse stages of fruit development and ripening, and under abiotic stress. Further, we studied the interaction networks and co-expression profiles of Group A PP2Cs in response to cold, salt, and osmotic stresses. This systematic study increases the understanding of the core components of ABA signaling associated with developmental processes and abiotic stress responses and builds a solid foundation for genetic improvement of banana.
Results
Identification and phylogenetic analyses of banana PYL-PP2C-SnRK2s
To identify all PYL-PP2C-SnRK2 family members in banana, both Hidden Markov Model and BLAST searches were carried out to search the banana genome database with PYL-PP2C-SnRK2 sequences from Arabidopsis and rice as queries. After confirming their conserved domain using the PFAM and CDD databases, a total of 24 PYL, 87 PP2C, and 11 SnRK2 proteins were identified from the banana genome. The predicted features of the PYL, PP2C and SnRK2 proteins are summarized in Additional file 1: Table S1.
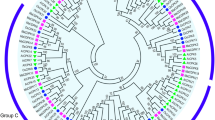
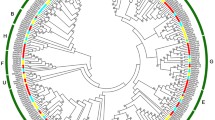
To understand the phylogenetic relationship of PYL-PP2C-SnRK2 proteins, neighbor-joining (NJ) trees were reconstructed with the complete PYL-PP2C-SnRK2 protein sequences from banana, Arabidopsis and rice (Figs. 1, 2, and 3). According to the phylogenetic analyses, the PYL, PP2C, and SnRK2 families were divided into 4 (group 1-4), 13 (group A-L), and 3 (group 1-3) subgroups, respectively. Some orthologous PYL-PP2C-SnRK2s between banana and rice were identified, which implied that some ancestral PYL-PP2C-SnRK2s existed prior to the divergence of banana and rice. Generally, banana PYL-PP2C-SnRK2s showed closer relationships with PYL-PP2C-SnRK2s in rice than those in Arabidopsis, which is accordance with the current understanding of plant evolutionary history.
Conserved motifs and gene structure analyses of banana PYL-PP2C-SnRK2
To get insight into the structural features of the banana PYL-PP2C-SnRK2 proteins, conserved motifs were analyzed based on the phylogenetic relationship. Ten conserved motifs were acquired for each gene family with MEME and InterPro databases (Fig. 4). For the banana PYL family, motifs 1-3 were annotated as the START-like domain. All the identified MaPYLs contained motifs 1 and 2. The subgroup 1-3 also showed the conserved motif 3 (Fig. 4b). For the banana PP2C family, motifs 1-5 were annotated as the PPM-type phosphatase domain. Almost all of the PP2Cs contain the motifs 1, 2, 4, and 5, except for subgroup K showing motifs 1, 2, and 4. Interestingly, subgroup C specially showed motif 3, and subgroup D uniquely had motif 3, 7, 8, and 10 (Fig. 4a). For the banana SnRK2 family, motifs 1-5 were annotated as the Protein kinase domain. All the MaSnRK2s have motifs 1-5. Motif 10 was especially pronounced in subgroup 1 and motifs 8 and 9 were only found in subgroup 3 (Fig. 4c). This indicates that all the identified PYL-PP2C-SnRK2s have typical family features and the proteins classified into the same subgroup share similar amino acid sequences.
To better understand the gene structure of banana PYL-PP2C-SnRK2s, exon-intron organizations of these genes were tested (Fig. 5). For the banana PYL family, subgroups 1, 3, and 4 have 2, 0, and 1 introns, respectively; and subgroup 2 showed 0-2 introns (Fig. 5b). For the banana PP2C family, subgroups A, B, D, F1, G, and K contain 2-5 introns; subgroups C, E, F2, and H have 3-9 introns; and subgroup L shows 1-15 introns (Fig. 5a). For the banana SnRK2 family, subgroups 1, 2, and 3 show 8-9, 8-13, and 8 introns, respectively (Fig. 5c). These results indicate that PYL-PP2C-SnRK2 genes in the same subgroup show similar exon-intron organization.
Expression analyses of PYL-PP2C-SnRK2 genes in different banana tissues
To examine the expression profiles of PYL-PP2C-SnRK2 genes in different tissues of banana, roots, leaves, and fruits from BaXi Jiao (Musa acuminate L. AAA group cv. Cavendish, BX) and Fen Jiao (Musa ABB PisangAwak, FJ) were collected to perform trancriptomic assays (Fig. 6; Additional file 1: Tables S2; S3; S4; S5). Generally, most of the PYL-PP2C-SnRK2 genes showed similar tissue expression patterns between BX and FJ. For example, several genes (MaPYL-14, MaPP2C-14, −34, −37, −38, −45, −47, and MaSnRK2-6) displayed high transcript abundance (FPKM value > 20) in both BX and FJ. In contrast, some genes (MaPYL-5, −16, −17, −18, −21, and MaPP2C-16, −20, −22, −23, −29, −46, −59, −63, −64, −80, −81, −84) had low transcript abundance (FPKM value < 3) in both BX and FJ.
In addition, we also found different expression patterns of PYL-PP2C-SnRK2 genes between BX and FJ. For the PYL family, the number of genes with high expression levels (FPKM value > 10) in roots and leaves was greater in BX (10/22 and 8/21, respectively) than in FJ (7/22 and 5/20, respectively). For the PP2C family, the number of genes with high expression levels (FPKM value > 10) in roots and fruits was less in BX (48/86 and 19/82, respectively) than in FJ (51/86 and 33/84, respectively). This phenomenon was also observed in the tissue expression patterns of the SnRK2 family. Taken together, the tissue expression patterns of PYL-PP2C-SnRK2 genes in two cultivated varieties could lay a foundation for further investigation of tissue development and function.
Expression analyses of PYL-PP2C-SnRK2 genes in different stages of fruit development and ripening
To get some clues on the function of the PYL-PP2C-SnRK2 genes in fruit development and ripening of banana, total RNA was extracted during different stages of fruit development and ripening for transcriptomic analyses (Fig. 7; Additional file 1: Tables S6; S7; S8; S9).
Expression profiles of banana PP2Cs, PYLs, and SnRK2s in different stages of fruit development and ripening in BX and FJ varieties. The heat map was constructed according to the FPKM value of banana PP2Cs, PYLs, and SnRK2s from two independent experiments. FPKM value is shown in color as the scale. Group A PP2Cs are marked with purple dot
According to the transcriptomic data, most of PYL-PP2C-SnRK2 genes showed similar expression patterns at different stages of fruit development and ripening in both BX and FJ. Some genes showed high expression levels (FPKM value > 10) at different stages of fruit development and ripening. For the PYL family, 7/22, 7/22, 5/16, 4/19, and 4/17 PYL genes showed high expression levels (FPKM value > 10) at 0 days after flower (DAF), 20 DAF, 80 DAF, 8 days post-harvest (DPH), and 14 DPH in BX, respectively; and 7/21, 9/22, 5/17, 5/17, and 3/19 PYL genes showed high expression levels (FPKM value > 10) at the corresponding stages in FJ, respectively. For the PP2C family, 47/85, 45/87, 19/82, 30/83, and 27/79 PP2C genes showed high expression levels (FPKM value > 10) at 0 DAF, 20 DAF, 80 DAF, 8 DPH, and 14 DPH in BX, respectively; and 51/85, 52/85, 33/84, 35/84, and 28/82 PP2C genes showed high expression levels (FPKM value > 10) at the corresponding stages in FJ, respectively. For the SnRK2 family, 6/11, 6/11, 6/11, 7/11, and 5/10 SnRK2 genes showed high expression levels (FPKM value > 10) at 0 DAF, 20 DAF, 80 DAF, 8 DPH, and 14 DPH in BX, respectively; and 6/11, 7/11, 7/11, 6/11, and 4/11 SnRK2 genes showed high expression levels (FPKM value > 10) at the corresponding stages in FJ, respectively. These results indicated the possible involvement of PYL-PP2C-SnRK2 genes in banana development and ripening.
The number of PP2C genes in BX with high expression levels (FPKM value > 10) was more at 0 (47/85) and 20 (45/87) DAF than at subsequent stages, including 80 DAF (19/82), 8 DPH (30/83), and 14 DPH (27/79). Also, similar expression patterns for PP2C genes were observed in FJ. These results indicate that PP2C genes play an important role during early fruit development.
Notably, FJ showed more PYL genes with high expression levels (FPKM value > 10) than BX at 20 DAF and 3 DPH. PP2C genes with high expression levels (FPKM value > 10) were more in FJ than in BX during all the tested stages, except for 6 DPH. More SnRK2 genes with high expression levels (FPKM value > 10) was also observed in FJ relative to BX at 20 and 80 DAF. These results imply that PYL-PP2C-SnRK2 genes may be more active in FJ than in BX during fruit development and ripening stages.
A total of 17 PYL-PP2C-SnRK2 genes, including MaPYL-9, −10, −12, MaPP2C-7, −14, −32, −37, −45, −47, −49, −55, −67, −69, −72, and SnRK2-4, −5, −6, showed high expression levels (FPKM value > 10) during all the tested stages in both BX and FJ, indicating the extensive and vital role of these genes during fruit developmental and ripening processes.
Most of the Group A PP2Cs, including PP2C-24, −40, −43, −45, and −47, showed high expression levels (FPKM value > 10) in the majority of the development and ripening stages of BX and FJ, whereas PP2C-16, −20, −22, −23, −46, −59, −60, −62, −63, −82, and −83 had extremely low expression (FPKM value < 3) during all the stages of fruit developmental and ripening in both BX and FJ. In addition, 8, 4, 7, 7, 8 Group A PP2C genes showed higher expression levels (FPKM value > 10) in FJ than in BX at each stages, respectively.
Expression analyses of PYL-PP2C-SnRK2 genes in response to cold, salt, and osmotic stresses
To gain insight into the role of PYL-PP2C-SnRK2 genes in banana in response to abiotic stress, the leaves of banana after cold, salt, and osmotic treatments were collected for transcriptomic analyses (Fig. 8; Additional file 1: Tables S10; S11; S12; S13).
Under the cold treatment, 4/21 PYLs, 17/84 PP2Cs, and 0/11 SnRK2s showed significant upregulation (Log2 based fold change >1; P-value < 0.05) in BX, whereas 5/21 PYLs, 19/84 PP2Cs, and 2/11 SnRK2s were significantly upregulated in FJ. Under the salt treatment, 1/21 PYLs, 10/84 PP2Cs, and 0/11 SnRK2s showed significant induction in BX, while 1/21 PYLs, 6/84 PP2Cs, and 0/11 SnRK2s were significantly upregulated in FJ. Under the osmotic treatment, 1/21 PYLs, 10/84 PP2Cs, and 1/11 SnRK2s were significantly induced in BX, whereas 1/21 PYLs, 21/84 PP2Cs and 2/11 SnRK2s were significantly upregulated in FJ. These results suggest that the number of PYL-PP2C-SnRK2 genes upregulated by cold and osmotic stresses was more in FJ than in BX, implying that these genes may be more active in FJ than in BX in response to cold and osmotic stresses.
Notably, 2 PYL genes (MaPYL8 and MaPYL15) and 12 PP2C genes (MaPP2C-3, −4, −21, −52, −53, −61, −62, −74, −75, −85, −86, and −87) were strongly induced (Log2 based fold change >2; P-value < 0.05) after cold treatment in FJ. Six PP2C genes (MaPP2C-2, −8, −25, −52, −83, and −87) and 1 SnRK2 genes (MaSnRK2-11) were strongly upregulated (Log2 based fold change >2; P-value < 0.05) by osmotic treatments in FJ. These genes may be crucial candidates for further use to improve abiotic stress tolerance of banana.
In addition, 10 genes (MaPYL-8, −24, MaPP2C-20, −39, −47, −52, −53, −57, and MaSnRK2-9, −10), 5 genes (MaPYL24 and MaPP2C-67, −77, −80, −83), and 14 genes (MaPP2C-87, −83, −67, −52, −57, −61, −85, −25, −9, −8, −51, −13, −76 and SnRK2-9) were significantly induced (Log2 based fold change >1; P-value < 0.05) by cold, salt, and osmotic treatments, respectively in FJ, but were not significantly induced in BX. These results indicate that these genes may uniquely function on the tolerance of FJ to abiotic stress.
Several Group A PP2Cs showed different expression patterns between BX and FJ in response to abiotic stress. MaPP2C-25, −43, −44, −45, −46, and −63 were upregulated in FJ after cold treatment, whereas in BX, were downregulated or did not show any change. MaPP2C-44 and −63 showed upregulation in FJ after salt treatment, whereas downregulation or no change in BX. MaPP2C43 showed induction in FJ after osmotic treatment, but showed repression in BX.
PYL-PP2C-SnRK2 interaction networks and their co-expression after abiotic stress treatment
To better understand the biological function of PYL-PP2C-SnRK2s in banana, the possible interaction networks and co-expression of Group A banana PP2Cs were investigated based on experimentally validated interactions of Group A PP2Cs in Arabidopsis and transcriptomic data in banana (Figs. 9, 10 and 11; Additional file 1: Table S14). Firstly, an Arabidopsis Group A PP2C-mediated interaction network was created and 29 interactive proteins (with high confidence; score > 0.9), including 9 PP2Cs and 20 other interactive proteins, were identified with STRING. Secondly, homologs of these interacting proteins in banana were identified with reciprocal BLASTP analyses. Lastly, the expression profiles of the banana genes in BX and FJ under abiotic stress were extracted from RNA-seq data sets.
Under the cold and salt treatments in BX, no gene pair was found to be co-expressed (Figs. 9a and 10a). Under the osmotic treatment in BX, gene pairs HAB1:Ma6270-PYL2:Ma940/PYL6:Ma790/RCAR1:Ma460 showed uniform downregulation (Fig. 11a). Under the cold treatment in FJ, gene pairs HAB1:Ma6270-PYL4:Ma0270/PYL6:Ma790/PYL11:Ma320/PYL1:Ma780 had upregulated co-expression, whereas HAI1:Ma130-PYR1:Ma9170/RCAR1:Ma460/RCAR3:Ma490 showed co-expression of uniform downregulation (Fig. 9b). Under the salt treatment in FJ, gene pairs HAI2:Ma600- CIPK23:Ma540/PYL10:Ma9170, HAI3:Ma9000- PYL10:Ma9170, and PYL1:Ma780-PP2CA:Ma050 showed uniform upregulation (Fig. 10b). Under the osmotic treatment in FJ, HAI2:Ma600- CIPK23:Ma540 had upregulated co-expression (Fig. 11b). Collectively, these results suggest that more gene pairs were uniformly upregulated in FJ than in BX under cold, salt, and osmotic treatments, indicating the crucial roles of Group A PP2C-mediated network in stress signaling.
Discussion
ABA signaling plays a crucial role in regulating developmental processes and in adaptation to environmental stresses in plants [6, 7]. Investigation of the core regulatory network in the ABA pathway would advance the understanding of the roles of ABA signaling and the function of ABA-associated genes. Currently, no information is known regarding the PYL-PP2C-SnRK2 gene family in banana. Herein, a total of 24 PYLs, 87 PP2Cs, and 11 SnRK2s were identified from the banana genome, which was classified into 4, 13, and 3 subgroups respectively according to phylogenetic relationship (Figs. 1, 2, and 3). This classification is in accordance with previous phylogenetic analyses of PYL, PP2C, or SnRK2s in Arabidopsis, rice, Brassica napus, and Brachypodium distachyon [6, 33,34,35]. Moreover, the phylogenetic classification of PYL-PP2C-SnRK2 was also supported by conserved motif anslysis (Fig. 4). Conserved motif analyses showed that all the PYLs, PP2Cs, and SnRK2s had START-like, PPM-type phosphatase, and protein kinase domains, respectively, and each subfamily shared similar motifs. These typical characteristics of PYL-PP2C-SnRK2s were also observed in other plant species, such as Arabidopsis, apple, and Brachypodium distachyon [6, 7, 35, 36].
As one of the most popular fruits, fruit development and ripening process are crucial for banana fruit quality. ABA signaling has been demonstrated to participate in the fruit development process and ripening of many plant species, including sweet cherries, strawberry, and tomato [2,3,4,5]; however, whether PYL-PP2C-SnRK2s participate in fruit development and post-harvest ripening of banana is unclear. In the present study, we found that more than 4/19 MaPYLs, 19/82 MaPP2Cs, and 5/10 MaSnRK2s showed high expression levels (FPKM value >10) in BX at any one stage of fruit development and ripening. Also, in FJ, more than 3/19 MaPYLs, 28/82 PP2Cs, and 4/11 MaSnRK2s showed high expression levels (FPKM value >10) at any one stage of fruit development and ripening (Fig. 7; Additional file 1: Tables S6; S7; S8; S9). Moreover, a total of 17 PYL-PP2C-SnRK2 genes, including MaPYL-9, −10, −12, MaPP2C-7, −14, −32, −37, −45, −47, −49, −55, −67, −69, −72, and SnRK2-4, −5, −6, showed high expression levels (FPKM value > 10) during all the tested stages in both BX and FJ. Considering the negative role of PP2C in ABA signaling, we also found 11 MaPP2C genes (PP2C-16, −20, −22, −23, −46, −59, −60, −62, −63, −82, and −83) showing extremely low expression (FPKM value < 3) during all the stages of fruit developmental and ripening in both BX and FJ. These results imply that PYL-PP2C-SnRK2 genes may be involved in the fruit development and ripening processes of banana.
The number of PP2C genes with high expression levels (FPKM value > 10) was more at 0 and 20 DAF than at subsequent stages in both BX and FJ, implying their regulatory role during early fruit development (Fig. 7; Additional file 1: Tables S6; S7; S8; S9). This is consistent with the expression of CsPP2C1 that reached the first peak value at early stages during cucumber fruit development [37].
Accumulated evidences suggests that exogenous application of ABA could accelerate fruit ripening of banana [38]; however, the role of the core components of ABA signaling, PYL-PP2C-SnRK2, in banana development and ripening is unknown. By comparing the PYL-PP2C-SnRK2 expression profiles at different stages of fruit development and ripening between BX and FJ, an interesting phenomenon was observed. The number of PYL-PP2C-SnRK2 genes with high expression levels (FPKM value > 10) was more in FJ than in BX at several stages, which implied that PYL-PP2C-SnRK2 genes may be more active in FJ than in BX during fruit development and ripening stages (Fig. 7; Additional file 1: Tables S6; S7; S8; S9). Previously, we observed that FJ ripened faster than BX during postharvest ripening. It took 8 and 14 DPH to reach more green than yellow and full yellow degrees of ripening for BX, respectively, whereas it only took 3 and 6 DPH for FJ, respectively [28, 29]. In tomato, RNA interference-mediated repression of ABA biosynthesis resulted in delay of fruit senescence and extension of shelf life [39]. In strawberry, inhibition of FaNCED1 led to a significant decrease of ABA levels and delay of fruit ripening by gene silencing and RNA interference [40]. In grape, fruit development and quality were improved by exogenous application of ABA [41]. This evidence demonstrates that ABA signaling plays a positive role in fruit development and ripening. Additionally, down-regulation of the FaPYR1 gene significantly delayed fruit ripening and repressed the expression of ABI1 and SnRK2 genes in strawberry, which implied that PYL-PP2C-SnRK2 genes may positively regulate fruit development and ripening [42]. Therefore, these findings suggest that PYL-PP2C-SnRK2-mediated ABA signaling may contribute to fruit development and ripening in banana.
Because banana has shallow roots, permanent green canopy, and rapid growth rate, it is usually subjected to water stress caused by abiotic stress such as cold, drought, or salt [43]. Investigation of the mechanism underlying banana response to abiotic stress is of great importance for banana breeding. Although ABA plays a predominant role in regulating plants’ tolerance to abiotic stress, the role of the core components of ABA signaling, PYL-PP2C-SnRK2, in banana responding to abiotic stress is unknown. In the present study, we found that many PYL-PP2C-SnRK2 genes showed transcriptional changes after cold, salt, or osmotic treatment in both BX and FJ, indicating that these genes may function on the regulation of banana tolerance to abiotic stress (Fig. 8; Additional file 1: Tables S10; S11; S12; S13).
By comparing the expression patterns of PYL-PP2C-SnRK2 genes under abiotic stress between BX and FJ, it was clear that more genes were significantly upregulated (Log2 based fold change >1) in FJ than in BX under the cold and osmotic treatments (Fig. 8; Additional file 1: Tables S10; S11; S12; S13). Furthermore, from the interaction network and co-expression analyses, more gene pairs were uniformly upregulated in FJ than in BX in response to the osmotic, cold, and salt stresses (Figs. 9, 10, and 11; Additional file 1: Table S14). The B-genome has been considered to be related to tolerance to abiotic stresses. The banana species M. balbisiana with the B-genome is demonstrated to have strong resistance to drought or water stress [44, 45]. Moreover, the “ABB” banana genotypes are more tolerant to drought and other abiotic stresses than other genotypes [46]. Thus, the banana varieties based on the “ABB” genotype can be used as a crucial genetic resource for crop improvement for abiotic stress. FJ (ABB genotype), containing the B-genome, has been reported to have strong tolerance to abiotic stress [28, 29]. Much evidence confirms that PYL- and SnRK2-mediated ABA signaling play a positive role in plants response to abiotic stress [6, 9, 13, 14, 22]. Together, these findings suggest that more PYL-PP2C-SnRK2 genes and gene pairs upregulated by abiotic stress in FJ could contribute to the tolerance of banana to abiotic stress.
Previously, Group A PP2Cs were demonstrated to be negative factors of ABA signaling [6, 27], whereas the function of Group A PP2Cs in ABA-mediated biological processes seem to be different in different species [20, 21, 27]. For example, mutation of abi2-1 resulted in enhanced tolerance to salt stress in Arabidopsis [21], while Arabidopsis plants overexpressing OsPP108 showed increased tolerance to salt and osmotic stresses [27]. Most of the Group A PP2Cs displayed high expression levels during fruit development and ripening in tomato [18]. Moreover, most of the Group A PP2C members were induced at transcriptional levels under osmotic, cold, salt, and drought treatments in Arabidopsis [47]. Based on our transcriptomic data, most of the Group A PP2Cs showed high expression levels (FPKM value > 10) in the majority of the development and ripening stages of BX and FJ, and Group A PP2C genes were found to be more active in FJ than in BX at transcriptional levels after cold, salt, and osmotic treatments. The function and mechanism of PP2Cs in ABA signaling transduction and ABA-mediated biological processes need to be further clarified in future studies.
Conclusions
In this study, we identified 24 PYL, 87 PP2C, and 11 SnRK2 genes from banana and studied their classification and evolutionary relationships by evolutionary, conserved protein motif, and gene structure analyses. The expression analyses reveal the involvement of PYL-PP2C-SnRK2 genes in banana fruit development, ripening, and responses to abiotic stress. Additionally, comparison of the differential expression profiles of PYL-PP2C-SnRK2 genes between BX and FJ suggested that PYL-PP2C-SnRK2-mediated ABA signaling might positively regulate banana fruit ripening and responses to abiotic stress. Furthermore, interaction networks and co-expression assays demonstrated the strong transcriptional response of core components of ABA signaling in FJ responding to abiotic stress, further supporting the crucial role of the genes for banana tolerance to abiotic stress. These data will supply abundant information for functional characterization of PYL-PP2C-SnRK2 genes, advance the understanding of PYL-PP2C-SnRK2-mediated ABA signaling in the regulation of fruit development, ripening, and response to abiotic stress, and lay a solid foundation for further research on banana breeding.
Methods
Plant materials and treatments
Two banana cultivars of BaXi Jiao (Musa acuminate L. AAA group cv. Cavendish, BX) and Fen Jiao (Musa ABB PisangAwak, FJ) were used in this study. BX is widely planted in China due to its virtues of long storage and high production. FJ is widely cultivated in the Hainan province of China. FJ has stronger tolerance to abiotic stress, including drought, salt, and cold, and ripened faster than BX during postharvest ripening (unpublished data). BX and FJ seedlings at the five-leaf stage were acquired from the banana tissue culture center (Institute of Bananas and Plantains, Chinese Academy of Tropical Agricultural Sciences, Danzhou). Seedlings with consistent growth state were cultured in soil under the conditions of 70% relative humidity and 200 μmol m−2 s−1 light intensity in 16 h light/8 h dark cycle, 28 °C. Roots and leaves from the five-leaf stage plants, and fruits of 80 DAF were sampled for expression analysis in different organs. Fruits from 0 DAF (budding), 20 DAF (cutting flower) and 80 DAF (harvest stage) were collected to study the expression profiles of genes during fruit development process. Fruits from 8 DPH and 14 DPH in BX and 3 DPH and 6 DPH in FJ were sampled to investigate gene expression patterns during post-harvest ripening stages because FJ reach full yellow degree faster than BX after harvesting [28, 29]. Banana seedlings at the five-leaf stage were irrigated with 200 mM mannitol or 300 mM NaCl for 7 days to study gene expression in response to osmotic and salt stresses, respectively. Banana seedlings were incubated in 4 °C for 22 h to detect gene expression upon cold stress.
Identification and phylogenetic analyses
The whole protein sequences of banana were downloaded from the banana genome database [32]. The PYL, PP2C, and SnRK2 protein sequences from rice and Arabidopsis were obtained from RGAP and UniProt databases, respectively [48, 49]. The HMM profiles built from the known PYL-PP2C-SnRK2s were used as queries to search the banana dataset with HMMER software [50, 51]. BLAST was also employed to identify the predicted banana PYL-PP2C-SnRK2s with all PYL-PP2C-SnRK2s from rice and Arabidopsis as queries. Then, the conserved domains of predicted banana PYL-PP2C-SnRK2s were further confirmed with PFAM and CDD databases [52, 53]. The accession numbers of identified banana PYLs, PP2Cs, and SnRK2s are displayed in Table S1. The phylogenetic tree was reconstructed with the PYL-PP2C-SnRK2 proteins from Arabidopsis, rice, and banana using MEGA 5.0 and Clustal X2.0 softwares (bootstrap values for 1000 replicates) [54, 55].
Protein properties and sequence analyses
Using the ExPASy database, the isoelectric points and molecular weights of the banana PYL-PP2C-SnRK2s were predicted [56]. MEME software was used to identify motifs of banana PYL-PP2C-SnRK2 proteins, and then the motifs were annotated with InterProScan [57, 58]. The optimum width of motifs ranged from 6 to 50, the maximum number of motifs was 10, and the other parameter settings used were default values. The PYL-PP2C-SnRK2 gene structure was analyzed by GSDS [59]. With the help of STRING software, the Group A PP2Cs-mediated protein interactions in Arabidopsis were explored with the confidence score > 0.9 and no more than 20 interactors.
Transcriptomic analysis
Total RNA of each sample was extracted with plant RNA extraction kit (TIANGEN, China) and used for cDNA library construction. The sequencing was performed with an Illumina GAII following manufacturer’s instructions. Using FASTX-toolkit, adapter sequences in the raw sequence reads were removed. After examining the sequence quality and removing low quality sequences by FastQC, clean reads were generated. Using Tophat v.2.0.10, clean reads were maped to the DH-Pahang genome (Musa acuminate, A-genome, 2n = 22) [32]. The transcriptome assemblies were performed by Cufflinks [60]. The RNA-seq reads status was listed in Additional file 1: Tables S2; S6; S10. Genes were scored as not expressed if the corresponding RNA-seq reads could not align to the genome. Calculation the ratio of PYL-PP2C-SnRK2 genes with high expression levels or showing significantly changes after abiotic stress treatments was performed according to the genes that is expressed. Gene expression levels were calculated as Reads Per Kilobase of exon model per Million mapped reads (FPKM). DEGseq was used to identify differentially expressed genes (Log2 based fold change >1 or Log2 based fold change <−1; P-value < 0.05) in response to cold, salt, and osmotic stresses [61]. There are two biological replicates, which showed good consistency (Additional file 2: Figures S1; S2; S3; Additional file 1: Table S3; S7; S11).
Abbreviations
- ABA:
-
Abscisic acid
- BX:
-
BaXi Jiao
- DAF:
-
Days after flower
- DPH:
-
Days post-harvest
- FJ:
-
Fen Jiao
- FPKM:
-
Reads Per Kilobase of exon model per Million mapped reads
- NJ:
-
Neighbor-joining
References
Feng CZ, Chen Y, Wang C, Kong YH, Wu WH, Chen YF. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014;80(4):654–68.
Tijero V, Teribia N, Munoz P, Munne-Bosch S. Implication of Abscisic acid on ripening and quality in sweet cherries: differential effects during pre- and post-harvest. Front Plant Sci. 2016;7:602.
Li D, Mou W, Luo Z, Li L, Limwachiranon J, Mao L, Ying T. Developmental and stress regulation on expression of a novel miRNA, fan-miR73, and its target ABI5 in strawberry. Sci Rep. 2016;6:28385.
Mou W, Li D, Bu J, Jiang Y, Khan ZU, Luo Z, Mao L, Ying T. Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PLoS One. 2016;11(4):e0154072.
Jia H, Jiu S, Zhang C, Wang C, Tariq P, Liu Z, Wang B, Cui L, Fang J. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol J. 2016;14(10):2045–65.
Umezawa T, Nakashima K, Miyakawa T, Kuromori T, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Molecular basis of the Core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51(11):1821–39.
Ben-Ari G. The ABA signal transduction mechanism in commercial crops: learning from Arabidopsis. Plant Cell Rep. 2012;31(8):1357–69.
Hu W, Yan Y, Hou XW, He YZ, Wei YX, Yang GX, He GY, Peng M. TaPP2C1, a group F2 protein Phosphatase 2C gene, confers resistance to salt stress in transgenic tobacco. PLoS One. 2015;10(6):e0129589.
Ma Y, Szostkiewicz I, Korte A, Moes D, Yang Y, Christmann A, Grill E. Regulators of PP2C Phosphatase activity function as Abscisic acid sensors. Science. 2009;324(5930):1064–8.
Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TFF, et al. Abscisic acid inhibits type 2C protein Phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–71.
Bhaskara GB, Nguyen TT, Verslues PE. Unique drought resistance functions of the highly ABA-induced Clade a protein Phosphatase 2Cs. Plant Physiol. 2012;160(1):379–95.
Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462(7273):660–4.
Santiago J, Rodrigues A, Saez A, Rubio S, Antoni R, Dupeux F, Park SY, Marquez JA, Cutler SR, Rodriguez PL. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade a PP2Cs. Plant J. 2009;60(4):575–88.
Saavedra X, Modrego A, Rodriguez D, Gonzalez-Garcia MP, Sanz L, Nicolas G, Lorenzo O. The nuclear Interactor PYL8/RCAR3 of Fagus Sylvatica FsPP2C1 is a positive regulator of Abscisic acid signaling in seeds and stress. Plant Physiol. 2010;152(1):133–50.
Allen GJ, Kuchitsu K, Chu SP, Murata Y, Schroeder JI. Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell. 1999;11(9):1785–98.
Saez A, Apostolova N, Gonzalez-Guzman M, Gonzalez-Garcia MP, Nicolas C, Lorenzo O, Rodriguez PL. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37(3):354–69.
Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140(1):127–39.
Sun HL, Wang XJ, Ding WH, Zhu SY, Zhao R, Zhang YX, Xin Q, Wang XF, Zhang DP. Identification of an important site for function of the type 2C protein phosphatase ABI2 in abscisic acid signalling in Arabidopsis. J Exp Bot. 2011;62(15):5713–25.
Rubio S, Rodrigues A, Saez A, Dizon MB, Galle A, Kim TH, Santiago J, Flexas J, Schroeder JI, Rodriguez PL. Triple loss of function of protein Phosphatases type 2C leads to partial constitutive response to endogenous Abscisic acid. Plant Physiol. 2009;150(3):1345–55.
Tahtiharju S, Palva T. Antisense inhibition of protein phosphatase 2C accelerates cold acclimation in Arabidopsis Thaliana. Plant J. 2001;26(4):461–70.
Ohta M, Guo Y, Halfter U, Zhu JK. A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci U S A. 2003;100(20):11771–6.
Umezawa T, Yoshida R, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K. SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis Thaliana. Proc Natl Acad Sci U S A. 2004;101(49):17306–11.
Tian X, Wang Z, Li X, Lv T, Liu H, Wang L, Niu H, Bu Q. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice. 2015;8(1):1–13.
Kim H, Hwang H, Hong JW, Lee YN, Ahn IP, Yoon IS, Yoo SD, Lee S, Lee SC, Kim BG. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J Exp Bot. 2012;63(2):1013–24.
Diédhiou CJ, Popova OV, Dietz KJ, Golldack D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008;8(1):49.
Dey A, Samanta MK, Gayen S, Maiti MK. The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 2016;16(1):1–20.
Singh A, Jha Bagri J, Pandey GK. ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS One. 2015;10(4):156–62.
Hu W, Zuo J, Hou XW, Yan Y, Wei YX, Liu JH, Li MY, Xu BY, Jin ZQ. The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci. 2015;6:742.
Hu W, Hou XW, Huang C, Yan Y, Tie WW, Ding ZH, Wei YX, Liu JH, Miao HX, Lu ZW, et al. Genome-wide identification and expression analyses of Aquaporin gene family during development and Abiotic stress in banana. Int J Mol Sci. 2015;16(8):19728–51.
Sreedharan S, Shekhawat UKS, Ganapathi TR. Transgenic banana plants overexpressing a native plasma membrane aquaporin MusaPIP1;2 display high tolerance levels to different abiotic stresses. Plant Biotechnol J. 2013;11(8):942–52.
Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61(6):1041–52.
D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. The banana (Musa Acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–7.
Xue T, Wang D, Zhang S, Ehlting J, Ni F, Jakab S, Zheng C, Zhong Y. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9:550.
Yoo MJ, Ma T, Zhu N, Liu L, Harmon AC, Wang Q, Chen S. Genome-wide identification and homeolog-specific expression analysis of the SnRK2 genes in Brassica Napus guard cells. Plant Mol Biol. 2016;91(1-2):211–27.
Cao JM, Jiang M, Li P, Chu ZQ. Genome-wide identification and evolutionary analyses of the PP2C gene family with their expression profiling in response to multiple stresses in Brachypodium Distachyon. BMC Genomics. 2016;17:175.
Shao Y, Qin Y, Zou YJ, Ma FW. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus Prunifolia. Gene. 2014;552(1):87–97.
Wang YP, Wu Y, Duan CR, Chen P, Li Q, Dai SJ, Sun L, Ji K, Sun YF, Xu W, et al. The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J Plant Physiol. 2012;169(18):1874–82.
Jiang Y, Joyce DC, Macnish AJ. Effect of Abscisic acid on banana fruit ripening in relation to the role of ethylene. J Plant Growth Regul. 2000;19(1):106–11.
Sun L, Sun YF, Zhang M, Wang L, Ren J, Cui MM, Wang YP, Ji K, Li P, Li Q, et al. Suppression of 9-cis-Epoxycarotenoid Dioxygenase, which encodes a key enzyme in Abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 2012;158(1):283–98.
Jia HF, Chai YM, Li CL, Lu D, Luo JJ, Qin L, Shen YY. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011;157(1):188–99.
Cantín CM, Fidelibus MW, Crisosto CH. Application of abscisic acid (ABA) at veraison advanced red color development and maintained postharvest quality of ‘crimson seedless’ grapes. Postharvest Biol Technol. 2007;46(3):237–41.
Chai YM, Jia HF, Li CL, Dong QH, Shen YY. FaPYR1 is involved in strawberry fruit ripening. J Exp Bot. 2011;62(14):5079–89.
van Asten PJA, Fermont AM, Taulya G. Drought is a major yield loss factor for rainfed east African highland banana. Agric Water Manag. 2011;98(4):541–52.
Davey MW, Gudimella R, Harikrishna JA, Sin LW, Khalid N, Keulemans J. A draft Musa Balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genomics. 2013;14:683.
Vanhove AC, Vermaelen W, Panis B, Swennen R, Carpentier SC. Screening the banana biodiversity for drought tolerance: can an in vitro growth model and proteomics be used as a tool to discover tolerant varieties and understand homeostasis. Front Plant Sci. 2012;3:176.
Ravi I, Uma S, Vaganan MM, Mustaffa MM. Phenotyping bananas for drought resistance. Front Physiol. 2013;4:9.
Chan ZL. Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics. 2012;100(2):110–5.
Bateman A, Martin MJ, O'Donovan C, Magrane M, Apweiler R, Alpi E, Antunes R, Ar-Ganiska J, Bely B, Bingley M, et al. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(D1):D204–12.
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu JZ, Zhou SG, et al. Improvement of the Oryza Sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4.
Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7(10):e1002195.
Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–37.
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu SN, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI's conserved domain database. Nucleic Acids Res. 2015;43(D1):D222–6.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8.
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9.
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31(13):3784–8.
Brown P, Baxter L, Hickman R, Beynon J, Moore JD, Ott S. MEME-LaB: motif analysis in clusters. Bioinformatics. 2013;29(13):1696–7.
Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70.
Hu B, Jin JP, Guo AY, Zhang H, Luo JC, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–7.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7(3):562–78.
Wang LK, Feng ZX, Wang X, Wang XW, Zhang XG. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Nonprofit Institute Research Grant of CATAS (1630052012012), the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1,630,052,016,005, 1,630,052,016,006), the Central Public-interest Scientific Institution Basal Research Fund for Innovative Research Team Program of CATAS (17CXTD-28), and the earmarked fund for Modern Agro-industry Technology Research System (CARS-32).
Availability of data and materials
All sequence information regarding banana is available at the banana genome database (http://banana-genome-hub.southgreen.fr/) and the accession numbers are listed in Table S1. The PYL, PP2C, and SnRK2 protein sequences from rice and Arabidopsis had been deposited in RGAP (http://rice.plantbiology.msu.edu/) and UniProt (http://www.uniprot.org/), respectively, and the accession numbers are listed in Table S15. The phylogenetic tree, sequence data and alignments was deposited in TreeBASE database (accession number: 20,795). The transcriptomic data was deposited in NCBI-SRA database (https://www.ncbi.nlm.nih.gov/sra/) (accession number: PRJNA394594). All datasets supporting the conclusions of this article are included as additional files. Banana seedlings at the five-leaf stage are available from the banana tissue culture center, Institute of Bananas and Plantains, Chinese Academy of Tropical Agricultural Sciences, Danzhou, Hainan, China.
Author information
Authors and Affiliations
Contributions
BX, ZJ, and HS conceived the study. WH, YY, JL, HM, WT, ZD, XD, CW, YL, and JW performed the experiments and carried out the analysis. WH, YY, and JL designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1: Table S1.
Characteristics of banana PYL, PP2C, and SnRK2 gene families. Table S2. Properties of transcriptome for RNA-seq analysis in different tissues. Table S3. FPKM value from each biological replicates of PYL-PP2C-SnRK2 genes in different tissues. Table S4. Expression data of the banana PYL-PP2C-SnRK2 genes in different tissues of BX and FJ varieties. Table S5. Statistical analyses of the expression data related to banana PYL-PP2C-SnRK2 genes in different tissues of BX and FJ varieties. Table S6. Properties of transcriptome for RNA-seq analysis in different stages of fruit development and ripening. Table S7. FPKM value from each biological replicates of PYL-PP2C-SnRK2 genes in different stages of fruit development and ripening. Table S8. Expression data of the banana PYL-PP2C-SnRK2 genes in different stages of fruit development and ripening in BX and FJ varieties. Table S9. Statistical analyses of the expression data related to banana PYL-PP2C-SnRK2 genes in different stages of fruit development and ripenings of BX and FJ varieties. Table S10. Properties of transcriptome for RNA-seq analysis under cold, salt, and osmotic treatments. Table S11. FPKM value from each biological replicates of PYL-PP2C-SnRK2 genes in response to cold, salt, and osmotic stresses. Table S12. Expression data (log2-based value) of the banana PYL-PP2C-SnRK2 genes after various abiotic stress treatment in BX and FJ. Table S13. Statistical analyses of the expression data related to banana PYL-PP2C-SnRK2 genes in response to abiotic stress. Table S14. Expression data of the genes involved in Group A PP2C-mediate interaction networks under abiotic stress in BX and FJ varieties. Table S15. The accession numbers and gene name of PYL-PP2C-SnRK2 gene families in Arabidopsis and rice. (XLS 216 kb)
Additional file 2: Figure S1.
Expression profiles of banana PP2Cs, PYLs, and SnRK2s in roots, leaves, and fruits of BX and FJ. The heat map was constructed according to the FPKM value of banana PP2Cs, PYLs, and SnRK2s from each replicates of two independent experiments. Figure S2. Expression profiles of banana PP2Cs, PYLs, and SnRK2s in different stages of fruit development and ripening in BX and FJ varieties. The heat map was constructed according to the FPKM value of banana PP2Cs, PYLs, and SnRK2s from each replicates of two independent experiments. Figure S3. Expression profiles of banana PP2Cs, PYLs, and SnRK2s in response to cold, salt, and osmotic treatments in BX and FJ varieties. The heat map was constructed according to the FPKM value of banana PP2Cs, PYLs, and SnRK2s from each replicates of two independent experiments. (PDF 1249 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hu, W., Yan, Y., Shi, H. et al. The core regulatory network of the abscisic acid pathway in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC Plant Biol 17, 145 (2017). https://doi.org/10.1186/s12870-017-1093-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-017-1093-4