Abstract
Background
Most studies of brain structure and function, and their relationships to cognitive ability, have relied on inter-individual variability in magnetic resonance (MR) images. Intra-individual variability is often ignored or implicitly assumed to be equivalent to the former. Testing this assumption empirically by collecting enough data on single individuals is cumbersome and costly. We collected a dataset of multiple MR sequences and behavioural covariates to quantify and characterize intra-individual variability in MR images for multiple individuals.
Methods and design
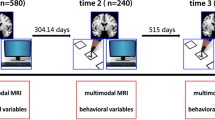
Eight participants volunteered to undergo brain scanning 40–50 times over the course of 6 months. Six participants completed the full set of sessions. T1-weighted, T2*-weighted during rest, T2-weighted high-resolution hippocampus, diffusion-tensor imaging (DTI), and proton magnetic resonance spectroscopy sequences were collected, along with a rich set of stable and time-varying physical, behavioural and physiological variables. Participants did not change their lifestyle or participated in any training programs during the period of data collection.
Conclusion
This imaging dataset provides a large number of MRI scans in different modalities for six participants. It enables the analysis of the time course and correlates of intra-individual variability in structural, chemical, and functional aspects of the human brain.
Similar content being viewed by others
Background
Magnetic resonance (MR) imaging can non-invasively quantify different aspects of brain structure with high spatial resolution. Different structural MR sequences are optimized to distinguish between different tissue types. For example, T1 and T2-weighted images distinguish between white and grey matter and cerebrospinal fluid (CSF). T2*-weighted images distinguish between different oxygen levels in brain tissue and are therefore an indirect measure of brain function [1]. Diffusion-tensor images (DTI) quantify water diffusivity and can therefore map major white matter tracts [2]; and proton magnetic resonance spectroscopy can be optimally set to measure the concentration of chemical groups, typically contained in neurotransmitters [3].
Within the last decades, neuroscientists have investigated different aspects of brain organization. First, associations between behaviour—e.g. cognitive ability, personality characteristics—and measures of grey matter thickness or density, white matter integrity, and functional connectivity between different brain areas have been identified in primarily cross-sectional studies [4]. The inter-individual variability in brain structure and function present in the normal population can explain some portion of variability in different cognitive processes, from low-level visual processing [5] to high-level cognition [6]. Further, clinical studies have identified large differences in these neurophysiological measures between patients and healthy control individuals [7]. Taken together, these results tend to paint a predominantly static, or trait-like, picture of brain organization and its association with behavior.
At the same time, other research, mostly based on the same measures, has revealed that the human brain is plastic [8,9,10]. For instance neuroimaging studies in clinical populations revealed that the course of disease, as in schizophrenia, is related to the trajectory of individual neural change [11]. Intervention studies have shown changes in brain structure and function in healthy individuals following diverse forms of explicit and often intense training [8, 12]. More strikingly, measures of brain structure and function have also been shown to depend on daily physiological variations that occur even in non-experimental settings like exercise [13], water [14, 15] and caffeine intake [16,17,18] or the menstrual cycle [19], revealing potentially important sources of intra-individual variability.
Cattell [20] usefully framed inter- and intra-individual variability with the concept of the Data Box, in which each data point represents the person under scrutiny, the specific measure taken, and the occasion (time point) at which the measure was taken [21, 22] (see Fig. 1). Importantly, in so-called ergodic data sets, that is, data sets that are drawn from a homogeneous population and are stationary, slicing this data box along different dimensions will lead to equal results [23]. But the variation of brain measures across individuals and time suggests that MR data might violate both assumptions. When data are non-ergodic, the structure of within-person variations over time cannot be derived from the structure of between-person differences at any given point in time. Hence, imaging results expressing between-person differences cannot be generalized to apply to within-person time series [23, 24].
Main differences between the Day2day dataset and other existing datasets. Most available MRI datasets that we are aware of investigate inter-individual variability only. By collecting MR data from the same participants across multiple occasions, we provide a dataset to investigate intra-individual variability (see also [23])
To summarize, cross-sectional data sets focus on between-person differences. Similarly, longitudinal data sets with few occasions, such as pretest–posttest intervention studies, focus on between-person differences in within-person change. Both research designs assume that individuals are drawn from a homogeneous population, and that the data structure does not change over time. Whether these assumptions are justified can only be known when between-person differences are compared to within-person changes. Hence, intra-individual variability must be studied explicitly, and appropriate methods for this purpose must be developed. Here, we report a dataset in which the same participants underwent MRI scanning on multiple (40–50) occasions. To the best of our knowledge, only one previous study collected imaging data of the same (single) individual with a large number of repetitions (MyConnectome project [18, 25]). The dataset presented here consists of a large number of scans for multiple individuals and is made available for public scientific use. It includes additional sequences and further behavioural covariates that make it comparable and complementary to the MyConnectome project and others of this nature.
Construction and content
Participants
Eight participants (2 male, mean age 29 years, SD = 2.58, range 24–32) volunteered to contribute to the dataset, for which they were scanned 40–50 times over the course of 6–8 months. Two of the participants (one male, one female) did not find the time to complete data acquisition. All participants were free of psychiatric disorder and had never previously suffered from a mental disease (see Table 1 for further details).
Scanning procedure
Images were collected on a 3T Magnetom Trio MRI scanner system (Siemens Medical Systems, Erlangen, Germany) using a 12-channel radiofrequency head coil. Participants kept their eyes closed during all image acquisitions.
T1 sequence
Structural images were collected using a three-dimensional T1-weighted magnetization prepared gradient-echo sequence (MPRAGE) with the following parameters: TR = 2500 ms, TE = 4.77 ms, TI = 1100 ms, FOV = 256 × 256 × 192 mm3, flip angle = 7°, bandwidth = 140 Hz/pixel, 1 × 1 × 1 mm3 voxel size, 9:20 min duration.
Resting state sequence
Functional images were collected using a T2*-weighted echo planar imaging (EPI) sequence sensitive to blood oxygen level dependent (BOLD) contrast with the following parameters: TR = 2000 ms, TE = 30 ms, FOV = 216 × 216 × 129 mm3, flip angle = 80°, bandwidth = 2042 Hz/pixel, voxel size 3 × 3 × 3 mm3, distance factor = 20%, 36 axial slices using GRAPPA acceleration factor 2, 5:08 min duration.
Hippocampus sequence
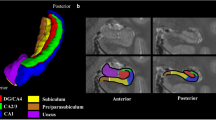
A high resolution T2-weighted turbo spin echo sequence was used to acquire high-resolution images of the hippocampus, to estimate subfield volumes with the following parameters: TR = 8150 ms, TE = 50 ms, 0.4 × 0.4 mm in plane resolution, 2 mm slice thickness, FOV = 150 × 150 × 62 mm3, flip angle = 120°, bandwidth = 99 Hz/pixel, turbo factor = 15, 31 slices covering the anterior three quarters and in some cases the whole hippocampus [26], 7:13 min duration.
Diffusion tensor imaging sequence
Diffusion-weighted images were obtained with a single-shot diffusion-weighted spin-echo-refocused echo-planar imaging sequence with the following parameters: TR = 8000 ms, TE = 93 ms, FOV 224 × 224 × 124 mm3, voxel size 2 × 2 × 2 mm3, 62 slices using GRAPPA acceleration factor 2, b-value 1000 s/mm2, 60 directions, 9:22 min duration.
Magnetic resonance spectroscopy sequence
To measure Proton (1H) magnetic resonance spectroscopy we used a point resolved spectroscopy (PRESS) sequence with the following parameters: TR = 3000 ms, TE = 80 ms, 128 averages, 90° flip angle, automatic shimming (advanced), vector size = 2048, 128/8 spectra with and without water-suppression, respectively, 6:36 min duration. All MRS voxels were individually positioned by anatomically trained MR operators on the bilateral ACC using the high-resolution three-dimensional T1-weighted MPRAGE (see above), which was collected in the same session.
Scanning schedule
Data collection took place between July 2013 and February 2014. While we aimed at collecting MR images from each participant two to three times a week to capture short-term variability, each participant was free to arrange a scanning regime that would optimally fit with his or her schedule. Additionally, scanning depended on availability of the MR scanner. As a result, MR data collection was not always done at regular intervals. Figure 2 provides an overview of the timing of each scanning session for each participant.
Covariates collected
We collected a number of covariates (see Table 2) related to scanner status, behaviour, and affect of the participants during the scan and in the 24 h prior to scanning. We aimed at collecting information for those covariates that had been shown or were expected to affect measures of brain structure and function. Participants did not modify their habits or engaged in any intensive training programs during the period of data collection, so the range of daily variation in covariates can be expected to roughly match that of the normal population.
We measured MR Room temperature and humidity with a digital indoor weather station (WS9410, TechnoTrade, Wildau, Germany). We collected weather data retrospectively from the German Weather Service (Deutsche Wetterdienst, [27]). We used standard devices to measure blood pressure and body weight. We recorded responses to the PANAS scales [28] to measure positive and negative affect at the moment of scanning. We tracked physical activity and hours of sleep with a Fitbit One activity tracker (Fitbit, San Francisco, USA).
We collected saliva samples using SaliCaps collection devices (IBL-International, Hamburg, Germany), which are validated for sampling of steroid hormones. Immediately after collection, saliva samples were frozen and stored at −25 °C. Oestrogen and testosterone concentrations were determined with the Saliva ELISA kit (IBL-International, using IBL Saliva Immunoassay -17ß-Estradiol) and the IBL Saliva Testosterone Luminescence Immunoassay.
Quality assurance
The technical image quality of the scanner was monitored by a quality assurance (QA) protocol defined after Friedman and Glover [29] including the Weisskoff noise coherence parameters [30]. Our QA protocol mainly focuses on scanner stability, signal-to-noise, drift, ghosting and other performance issues related to MR scanners. It was measured once per week, throughout the whole study period between July 2013 and February 2014. A BIRN agar phantom with 17-cm diameter was used.
Parameters that can be estimated in the Glover stability QA protocol include a signal image, temporal fluctuation noise image, signal-to-fluctuation-noise ratio (SFNR) image and a summary SFR value, a static spatial noise image, a signal to noise ration summary value, percent fluctuation and drift, a Fourier analysis of the residuals, and a Weisskoff analysis. Regular measurements of SNR, SFNR, percent fluctuations, and drift can provide critical feedback regarding scanner performance. The Fourier analysis can reveal periodic noise in time-series. Drifts in the RF amplifier gain settings and resonant frequency can also provide valuable feedback about the state of the scanner.
There were no major hardware or software changes made during the study period. There were no software upgrades as the scanner at our site is used solely for research purposes with numerous longitudinal studies, enabling a focus on continuity and therefore a neglect of software upgrades.
Utility and discussion
This dataset will allow researchers to quantify intra-individual variability in different measures of brain structure and function, characterize its time course, and identify its potential sources. It constitutes a unique effort to sample MR data multiple times from the same individuals, covering the variability over time, individuals and measurement variables that Cattell identified [21, 22]. It includes brain data over 40–50 measurement occasions for more than one person, and an array of time-varying covariates.
While several software tools are already available for the pre-processing and statistical analysis of MR images, new tools are also being developed. This dataset can also be of use to test the stability and sensitivity of measures of brain structure and function obtained through newly developed tools.
We have carried on analyses of these data and that we plan to publish in four different manuscripts. One article focuses on the reliability of measures of functional connectivity in resting-state [31] and a second article presents an analysis method to study functional brain dynamics (also on resting state data) on an individual level [32]. Two additional articles (currently in preparation) will quantify and describe the within-subject variability in structural measures.
Perhaps the main limitation of the dataset is that, due to the challenges of data collection, it includes a rather small and homogenous group of participants, compared to other MR studies that are focused on inter-individual differences. However, analyses of this relatively homogenous dataset might provide compelling evidence and arguments to extend this approach to larger, more heterogeneous populations. We recommend collecting a similar dataset in other age groups to examine the tenability of the ergodicity assumptions data across the lifespan.
Conclusion
This dataset, unique in the field of MRI, will allow researchers to address the important dimension of intra-individual variability in MR images. Because behavioural and physiological measures are included, multi-variate analyses can be performed to identify potential sources of variability. This dataset can therefore be used to inform MRI studies, in general, about potential confounds that should be taken into account when collecting and analysing MR data.
References
Hillman EMC. Coupling mechanism and significance of the BOLD signal: a status report. Annu Rev Neurosci. 2014;37:161–81.
Assaf Y, Pasternak O. Diffusion tensor imaging (DTI)-based white matter mapping in brain research: a review. J Mol Neurosci. 2008;34:51–61.
Soares DP, Law M. Magnetic resonance spectroscopy of the brain: review of metabolites and clinical applications. Clin Radiol. 2009;64:12–21.
Kanai R, Rees G. The structural basis of inter-individual differences in human behaviour and cognition. Nat Rev Neurosci. 2011;12:231–42.
Song C, Schwarzkopf DS, Rees G. Variability in visual cortex size reflects tradeoff between local orientation sensitivity and global orientation modulation. Nat Commun. 2013;4:2201.
Fleming SM, Weil RS, Nagy Z, Dolan RJ, Rees G. Relating introspective accuracy to individual differences in brain structure. Science. 2010;329:1541–3.
Freitag CM, Luders E, Hulst HE, Narr KL, Thompson PM, Toga AW, Krick C, Konrad C. Total brain volume and corpus callosum size in medication-naïve adolescents and young adults with autism spectrum disorder. Biol Psychiatry. 2009;66:316–9.
Lövdén M, Wenger E, Mårtensson J, Lindenberger U, Bäckman L. Structural brain plasticity in adult learning and development. Neurosci Biobehav Rev. 2013;37:2296–310.
Lövdén M, Bodammer NC, Kühn S, Kaufmann J, Schütze H, Tempelmann C, Heinze HJ, Düzel E, Schmiedek F, Lindenberger U. Experience-dependent plasticity of white-matter microstructure extends into old age. Neuropsychologia. 2010;48:3878–83.
Kühn S, Lindenberger U. Research on human plasticity in adulthood: A lifespan agenda. In: Schaie KW, Willis SL, editors. Handbook of the psychology of aging. 8th ed. Amsterdam: Academic Press;2016. p.105–123
Andreasen NC, Nopoulos P, Magnotta V, Pierson R, Ziebell S, Ho B-C. Progressive brain change in schizophrenia: a prospective longitudinal study of first-episode schizophrenia. Biol Psychiatry. 2011;70:672–9.
Zatorre RJ, Fields RD, Johansen-Berg H. Plasticity in gray and white. Nat Neurosci. 2012;15:528–36.
Thomas AG, Dennis A, Bandettini PA, Johansen-Berg H. The effects of aerobic activity on brain structure. Front Psychol. 2012;3:86.
Duning T, Kloska S, Steinsträter O, Kugel H, Heindel W, Knecht S. Dehydration confounds the assessment of brain atrophy. Neurology. 2005;64:548–50.
Kempton MJ, Ettinger U, Schmechtig A, Winter EM, Smith L, McMorris T, Wilkinson ID, Williams SCR, Smith MS. Effects of acute dehydration on brain morphology in healthy humans. Hum Brain Mapp. 2009;30:291–8.
Koppelstaetter F, Poeppel TD, Siedentopf CM, Ischebeck A, Kolbitsch C, Mottaghy FM, Felber SR, Jaschke WR, Krause BJ. Caffeine and cognition in functional magnetic resonance imaging. J Alzheimers Dis JAD. 2010;20(Suppl 1):S71–84.
Field AS, Laurienti PJ, Yen Y-F, Burdette JH, Moody DM. Dietary caffeine consumption and withdrawal: confounding variables in quantitative cerebral perfusion studies? Radiology. 2003;227:129–35.
Poldrack RA, Laumann TO, Koyejo O, Gregory B, Hover A, Chen M-Y, Gorgolewski KJ, Luci J, Joo SJ, Boyd RL, Hunicke-Smith S, Simpson ZB, Caven T, Sochat V, Shine JM, Gordon E, Snyder AZ, Adeyemo B, Petersen SE, Glahn DC, Reese Mckay D, Curran JE, Göring HHH, Carless MA, Blangero J, Dougherty R, Leemans A, Handwerker DA, Frick L, Marcotte EM, et al. Long-term neural and physiological phenotyping of a single human. Nat Commun. 2015;6:8885.
Lisofsky N, Mårtensson J, Eckert A, Lindenberger U, Gallinat J, Kühn S. Hippocampal volume and functional connectivity changes during the female menstrual cycle. NeuroImage. 2015;118:154–62.
Cattell RB. Abilities: their structure, growth, and action, vol. xxii. Oxford: Houghton Mifflin; 1971.
Cattell RB. The three basic factor-analytic research designs—their interrelations and derivatives. Psychol Bull. 1952;49:499–520.
Cattell RB. Factor analysis: an introduction and manual for the psychologist and social scientist. New York, NY: Harper & Row; 1952.
Molenaar PCM, Campbell CG. The new person-specific paradigm in psychology. Curr Dir Psychol Sci. 2009;18:112–7.
Raz N, Lindenberger U, Rodrigue KM, Kennedy KM, Head D, Williamson A, Dahle C, Gerstorf D, Acker JD. Regional brain changes in aging healthy adults: general trends, individual differences and modifiers. Cereb Cortex. 2005;15:1676–89.
Mueller SG, Chao LL, Berman B, Weiner MW. Evidence for functional specialization of hippocampal subfields detected by MR subfield volumetry on high resolution images at 4 T. NeuroImage. 2011;56:851–7.
Watson D, Clark LA, Tellegen A. Development and validation of brief measures of positive and negative affect: the PANAS scales. J Pers Soc Psychol. 1063;1988:54.
Friedman L, Glover GH. Report on a Multicenter fMRI quality assurance protocol. J Magn Reson Imaging. 2006;23:827–39.
Weisskoff RM. Simple measurement of scanner stability for functional NMR imaging of activation in the brain. Magn Reson Med. 1996;36:643–5.
Pannunzi M, Hindriks R, Bettinardi RG, Wenger E, Lisofsky N, Martensson J, et al. Resting-state fMRI correlations: from link-wise unreliability to whole brain stability. Neuroimage. 2017;157:250–62.
Donnelly-Kehoe PA, Saenger VM, Lisofsky N, Kühn S, Kringelbach ML, Schwarzbach J, Deco G. Consistent local dynamics in the brain across sessions are revealed by whole brain modeling of resting state activity. BioRxiv (Submitted). doi:10.1101/104232.
Authors’ contributions
All authors designed and planned the study. EF, NL, MB, OB, ML, JM, EW and SK collected the data and were study participants. EF and NL prepared the data for publication. EF, NL, SK, and UL wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
We are grateful for the assistance of the MRI team at the MPI Berlin, consisting of Sonali Beckmann, Nils Bodammer, Thomas Feg, Sebastian Schröder and Nadine Taube.
Competing interests
The authors have no competing interests to report.
Availability of data and materials
The day2day dataset is freely available for usage in scientific research. To prevent its circulation unrelated to research usage, we ask that scientists interested in obtaining the dataset email the corresponding author (SK) directly.
Consent to publish
All participants provided written consent to make the of their pseudonymized data open for research purposes. Finally, all participants involved agreed to publish each of the variables included in the dataset. Two variables containing sensitive information were excluded from the dataset.
Ethics approval and consent to participate
The Ethics Committee of Charité University Clinic, Berlin, approved of the study. Participants were members of the research group, and expressed interest in the scientific motivation of the study. In light of the unique nature of the data collection, ethical considerations were addressed with special care. Participants provided written consent for participation and publication after being informed of the details and purpose of the study, and were informed that they can end study participation at any time. Two participants terminated the study early due to lack of time. Some members of the group did not express interest in participating in the study, and accordingly did not participate. Importantly, no member of the research group was coerced to take part in the study.
Funding
The Max Planck Society funded this research. The design of the study, the collection, analysis, and interpretation of the data, and the writing of the manuscript was the sole responsibility of the authors.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Filevich, E., Lisofsky, N., Becker, M. et al. Day2day: investigating daily variability of magnetic resonance imaging measures over half a year. BMC Neurosci 18, 65 (2017). https://doi.org/10.1186/s12868-017-0383-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12868-017-0383-y