Abstract
Background
Thrips tabaci is a severe pest of onion and cotton. Due to lack of information on its genome or transcriptome, not much is known about this insect at the molecular level. To initiate molecular studies in this insect, RNA was sequenced; de novo transcriptome assembly and analysis were performed. The RNAseq data was used to identify reference and RNAi pathway genes in this insect. Additionally, feeding RNAi was demonstrated in T. tabaci for the first time.
Results
From the assembled transcriptome, 27,836 coding sequence (CDS) with an average size of 1236 bp per CDS were identified. About 85.4% of CDS identified showed positive Blast hits. The homologs of most of the core RNAi machinery genes were identified in this transcriptome. To select reference genes for reverse-transcriptase real-time quantitative PCR (RT-qPCR) experiments, 14 housekeeping genes were identified in the transcriptome and their expression was analyzed by (RT-qPCR). UbiCE in adult, 28s in nymphs and SOD under starvation stress were identified as the most stable reference genes for RT-qPCR. Feeding dsSNF7 and dsAQP caused 16.4- and 14.47-fold reduction in SNF7 and AQP mRNA levels respectively, when compared to their levels in dsGFP fed control insects. Feeding dsSNF7 or dsAQP also caused 62 and 72% mortality in T. tabaci. Interestingly, simultaneous feeding of dsRNAs targeting SNF7 or AQP and one of the RNAi pathway genes (Dicer-2/Aubergine/Staufen) resulted in a significant reduction in RNAi of target genes. These data suggest the existence of robust RNAi machinery in T. tabaci.
Conclusion
The current research is the first report of the assembled, analyzed and annotated RNAseq resource for T. tabaci, which may be used for future molecular studies in this insect. Reference genes validated across stages and starvation stress provides first-hand information on stable genes in T. tabaci. The information on RNAi machinery genes and significant knockdown of the target gene through dsRNA feeding in synthetic diet confirms the presence of efficient RNAi in this insect. These data provide a solid foundation for further research on developing RNAi as a method to manage this pest.
Similar content being viewed by others
Background
Thrips belong to order Thysanoptera comprising of about 7400 described species [1]. These are minute insects (about 1 to 4 mm in length) with unique characteristics such as a haplodiploid genetic system, parthenogenesis, female-biased sex ratio, high fecundity, postembryonic remetaboly, asymmetrical mouthparts, short generation time, high locomotor activity and preference for concealed spaces [2, 3]. Thrips are polyphagous pests that cause economic losses to the onion, tomato, cotton, peanut, potato, pepper, corn, wheat, several ornamental plants and vegetable crops either by direct feeding or by transmitting plant viruses. The damage is caused by extensive feeding which empties the cytoplasmic and other cellular contents of plant cells [4, 5]. Due to damage perpetrated by their feeding, oviposition and ability to transmit plant viruses of genus Orthotospovirus, they have attained the status of a major agricultural pest globally [6,7,8].
Several management practices including cultural, mechanical and biological tactics are being followed worldwide to manage thrips. However, the mainstay control relies on the chemical insecticides. The development of insecticide resistance and failure of insecticides to reach the eggs in plant tissues, as well as fourth instars and pupae in the soil, hampers the effective chemical control of thrips [9]. Insecticide applications also have well-documented effects on non-target organisms and the environment. Thus, development of novel and environmentally safe control strategies are the need of the hour. RNAi, also known as PTGS (post-transcriptional gene silencing) is being developed for control of insect pests [10]. Identification of target genes for RNAi requires knowledge about genes and their expression patterns. Unfortunately, annotated and assembled transcriptome or genome sequence data are not available for T. tabaci in open access public databases. Therefore, RNA isolated from T. tabaci was sequenced, and assembled as a de novo transcriptome. We were able to retrieve T. tabaci homologs for most of the genes associated with RNAi response in other insects. RNAi target genes, as well as 14 genes, have also been identified and used to demonstrate RNAi and identification of stable reference genes for RT-qPCR analysis, respectively.
Results
De novo transcriptome
The cDNA libraries from T. tabaci mRNA were sequenced using Illumina paired-end sequencing technology and assembled to obtain transcriptome (Table 1). Sequencing of RNA yielded 16,737,058 (2 × 150 bp) high-quality reads. These reads were assembled into 36,342 contigs containing 200 to 16,083 nucleotides (nt) with an average length of 1816 nt per contig and the N50 of 3543. Using TransDecoder, a total of 27,836 CDS with a size between 297 and 13,161 nt and the average length of 1236 nt per CDS were identified. About 31% of the transcripts were between 200 and 500 nt in length, and a total of 8% were of size more than 5000 nt. The coding sequences predicted from these transcripts comprised of more than 31% sequences between 500 and 1000 nt and 1.43% above 5000 nt in length.
For functional annotation, the assembled CDS were analyzed for gene ontology by BLASTx searches in NCBI non-redundant (nr) protein database using Blast2GO [11]. About 85.4% (23,775) of CDS showed positive blast hit matches with sequences derived from insects including Zootermopsis nevadensis Hagen, Athalia rosae Linnaeus, Tribolium castaneum Herbst, Halyomorpha halys Stål, Cimex lectularius Linnaeus, Pediculus humanus Linnaeus, Orussus abietinus Scopoli, Nasonia vitripennis Ashmead, Diaphorina citri Kuwayama, and Megachile rotundata Fabricius (Additional file 1). The predicted CDS from T. tabaci showed the highest similarity with Z. nevadensis sequences with over 7968 blast hits followed by hemipteran, A. rosae with over 6504 hits (Fig. 1).
The annotation of T. tabaci transcriptome revealed main gene ontology (GO) categories under ‘Biological Processes’, ‘Molecular Function’ and ‘Cellular Component’ (Fig. 2). The most enriched terms for Biological processes are organic substance metabolic process (8.8%), primary metabolic process (8.3%) and cellular metabolic process (8.2%). In Molecular function, the major contributions were towards organic cyclic compound binding (7.7%) and heterocyclic compound binding (7.7%). Under Cellular component, 14.06% sequences comprised of intracellular, followed by intracellular part (11.6%), intracellular organelle (8.4%) (Additional file 2).
Classification of Thrips tabaci transcripts based on predicted gene ontology terms via WEGO plot associated with the cellular component, molecular function, and biological process. GO terms were determined using Blast2Go with an E-value cutoff of 10−5. In total, 7484 CDS with BLAST matches to known proteins were assigned to gene ontology
Functional annotation of transcripts using Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis assigned KEGG Orthology (KO) IDs under four biological processes, i.e., Metabolism, Cellular, Genetic information processing and Environmental information (Fig. 3). The majority of transcripts grouped into Metabolism (37.68%) followed by Genetic information processing (23.08%). For pathways listed under this major processes, the majority of transcripts (16.02%) were associated with Signal transduction pathway, followed by Transport and catabolism (9.54%). The maximum transcripts (16.24%) under Metabolism were associated with Carbohydrate metabolism and under Genetic information processing in Folding, sorting and degradation (37.46%). Out of 27836 transcripts, only 26.88% were assigned the KO IDs using KEGG pathway analysis (Additional file 3). The completeness and quality of our assembled T. tabaci transcripts were evaluated by BUSCO analysis, which revealed 82.6% and 84.2% representation compared to insect and metazoan lineages, respectively [12, 13]. About 10.8% and 9.4% were “fragmented”, while 6.6% and 6.4% were missing compared to insect and metazoan lineages, respectively.
Identification and validation of reference genes
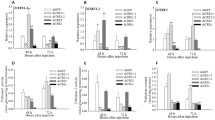
Based on published reports from other insects, fourteen candidate genes (TUB, Actin, EF, 18s, 28s, RPL17, TATA, E2F, Hist3, UbiCE, GAPDH, vATPase, GSTD2 and SOD) were selected for identification of stable reference genes in T. tabaci. The presence of selected genes in T. tabaci was reconfirmed by PCR amplification followed by agarose gel electrophoresis. The amplification efficiency of candidate reference genes was calculated by 3-point standard curve obtained by serial dilutions of known concentration of cDNA template. Amplification efficiency varied between 82.96 and 100.71% except for Actin (78.56%), GADPH (72.57%), Hist3 (144.68%) and vATPase (175.1%) (Additional file 4: Table S1). The correlation coefficient values (R2) ranged between 0.95 and 1.00 (Table 3). Melt-curve analysis confirmed the amplification specificity, as a single peak was detected for each amplified product. The agarose gel electrophoresis of these amplified products further confirmed primer specificity through a single crisp band of the respective gene. Relative expression levels of all selected genes analyzed using RT-qPCR ranged from 17.71 (Actin) to 31.12 (vATPase) Ct value. The expression of the vATPase gene was considerably low (29.73–31.12) in all developmental stages tested. The Ct value of 18s, 28s, SOD and GSTD2 ranged between 22 and 25 across all experimental samples. However, expression of TATA, 18s, and Hist3 showed variations among these samples (Fig. 4). The expression EF, RPL17, TATA, E2F, Hist3, UbiCE, GAPDH, vATPase and Actin was stable among insect stages and under starvation stress. However, the expression of E2F was substantially lower in starved animals when compared to that in normal ones.
BestKeeper analysis identifies the most stable gene based on its standard deviation (SD) calculated from raw Ct values across biological replicates. SD > 1 signifies an unstable expression of the candidate gene due to high variation in its expression within a sample of the same origin. The expression of all candidate genes was stable in adult samples (SD < 1). However, in nymphs, instability was observed in the expression of 18s, 28s, TUB, EF and RPL17 (Additional file 4: Table S2). Based on BestKeeper analysis, RPL17 (SD value = 0.02) in adults, UbiCE in nymphs (SD = 0.15) and TUB (SD = 0.20) under starvation stress were found to be highly stable. The candidate genes were ranked according to descending order of their stability and presented in Fig. 5. The geNorm analysis is based on two parameters, expression stability value (M) and pairwise variation (V). The highest M value indicates the least stable gene and vice versa. SOD and GSTD2 (M value 0.159) in nymphs, 18s and TUB in adults (M value 0.06) and under starvation stress (M value 0.24) were found to be the most stable genes (Fig. 6).
NormFinder designates the stable gene based on stability value as well as identifies the best combination of reference genes. GSTD2 (stability value ~ 0.231) was identified as the most stable gene across all experimental samples (Additional file 4: Table S3). The overall best combination of stable genes is 18s and GSTD2 (stability value ~ 0.191), however, individually under starvation stress, 18s and TUB were highly stable genes (stability value 0.007). Similarly, UbiCE (stability value—0.005) in nymphs, and SOD and GSTD2 (stability value—0.003) in adults showed the highest expression. The genes ranked based on their stability values are presented in Fig. 7. Based on interpretations of both NormFinder and geNorm analysis, 18s and TUB are the most stably expressed genes in starved animals. RefFinder assembles the output of BestKeeper, geNorm, and NormFinder along with Delta CT to assign an overall rank to genes based on its geometric mean (Additional file 4: Table S4 and Table 2). These analyses identified GSTD2 and 28s as the common stable genes.
Impact of reference gene normalization on expression analysis in RNAi studies
To study the impact of identified stable reference genes on mRNA expression in RNAi experiments, the relative expression of SNF7 gene was compared after normalization with two most and two least stable genes. The expression of SNF7 gene was calculated in adult thrips fed with dsSNF7 after normalization with individual or combination of most stable (GSTD2, SOD) and least stable (TUB, EF) genes. The dsSNF feeding in thrips caused 93.98% and 93.4% knockdown of SNF7 when normalized with GSTD2 and SOD, respectively (Fig. 8). The use of these two genes together for normalization showed almost similar knockdown (93.3%) of SNF7. However, normalization with the least stable genes, i.e. TUB and EF showed 61.1% and 70.9% knockdown of SNF7, respectively. The normalization with individual least stable gene resulted in 22–32% less knockdown compared to normalization with stable genes. Normalization with two least stable genes (TUB and EF) showed 85.5% reduction in SNF7 mRNA levels (Fig. 8), which was 7.8% less compared to combination of most stable genes.
Relative expression of SNF7 in Thrips tabaci after feeding dsRNA incorporated in the liquid diet through membrane feeding assay as compared control with respect to most stable and least stable reference genes. a Normalization studies with most stable genes (GSTD2—93.98%, SOD—93.4%, GSTD2 + SOD—93.3% Knockdown efficiency. b Normalization studies with least stable genes EF—70.0%, TUB—61.1%, TUB + EF—85.05% knockdown efficiency. *Indicates significant levels of mRNA transcripts of target genes compared to GFP (P ≤ 0.05, Student’s t-test)
Identification of homologs of RNAi pathway genes
Thrips tabaci transcriptome was also mined to identify RNAi pathway genes. Homologs of 21 genes [SID1, Dicer1, Dicer2, Drosha, Argonaute 1, Argonaute 2, Apoptosis linked genes (ALG), PIWI, Aubergine, abnormal spindle, Mut-7, HPS4, Gawky, DEAD-Box helicase, Tudor (TDR), RISC, dsRNAse2, Belle, Clathrin heavy chain, innexin, Staufen and Clp-1] which have been shown to function in RNAi pathway in other insects were identified in T. tabaci transcriptome (Additional file 4: Table S5). The homologs of genes involved in siRNA (Argonaute 2, Dicer2), miRNA (Argonaute1, Dicer1, Drosha,) and piRNA (Aubergine) pathways were identified in T. tabaci transcriptome. However, further analysis is required to validate their function in this insect. The core RNAi pathway genes from T. tabaci showed similarity with their homologs from other insect species in phylogenetic analysis (Fig. 9). The expression of 11 RNAi genes was also confirmed using RT-qPCR both in nymphs and adults of T. tabaci (Fig. 10). Slight variation in the expression level of these genes was observed between the life stages viz. the expression of TDR was high in adult compared to nymph. Within genes, the expression of Ago-1, Mut-7 and TDR was higher (Cq value: 24–27) compared to other genes tested. These data provide evidence for the existence of RNAi machinery in T. tabaci. However, some of the genes such as RNA-dependent RNA polymerase (RDRP), Loquacious, and Pasha associated with RNAi were missing in this transcriptome. This may be attributed to failure of transcriptome sequencing to include low abundant transcripts [14] or the presence of alternate proteins that can perform these functions in T. tabaci, which needs further investigation.
Demonstration of feeding RNAi in T. tabaci
To date, feeding RNAi has not been reported for any member of the insect order Thysanoptera. To confirm RNAi functioning in T. tabaci, 500 ng/μl of dsRNA targeting SNF or AQP gene was evaluated in membrane feeding assay. The dsRNA feeding caused 16.4- and 14.47-fold reduction in the target gene mRNA levels in dsSNF7 and dsAQP fed insects respectively, when compared to their levels in dsGFP fed control insects (Fig. 11). Feeding dsSNF7 or dsAQP also caused 62 and 72% mortality, respectively compared to 20% observed in dsGFP-fed control thrips (Fig. 14).
Relative expression of targeted genes in Thrips tabaci after feeding dsRNA incorporated in the liquid diet through membrane feeding assay. a SNF7 (multivesicular protein) knockdown. b AQP (Aquaporin) knockdown. The expression level has been normalized with GSTD2. Error bar represents standard deviation (n = 6) and * indicates significant levels of mRNA transcripts of target genes compared to GFP (P ≤ 0.05, Student’s t-test)
The impact of knockdown of RNAi pathway genes on RNAi efficiency of target genes (AQP/SNF7) was evaluated through simultaneous knockdown of Dicer-2, Aubergine and Staufen. Feeding dsDicer-2 or, dsAubergine and dsStaufen (500 ng/ul) resulted in 78.8% (4.7-fold) and 56.8% (2.3-fold) significant reduction in the mRNA levels of target genes compared to those in dsGFP fed control insects, respectively (Figs. 12, 13). Co-administration of 500 ng/ul each of dsDicer-2 and dsAubergine in combination with 500 ng/μl dsAQP in the diet reduced the knockdown of AQP by 42.4 -59.8%. Feeding of a mixture of dsStaufen and dsSNF reduced 88.2% RNAi efficiency compared to dsSNF fed insects (Fig. 13). Further, the expression of AQP in dsDicer-2 and dsAubergine + dsAQP fed insects was 7.8–14.9% lower compared to dsGFP control, while the corresponding figure for dsStaufen + dsSNF was 7.1% (Figs. 12, 13). In addition, the feeding of dsGFP + dsSNF7 caused 64.4% mortality, which was 17.7% more compared to dsStaufen + dsSNF7. This is again indicative that knockdown of Staufen hindered the RNAi of SNF7, which in turn affected the mortality caused by dsSNF feeding in thrips Fig. 14. Similarly, simultaneous knockdown of Dicer-2 and AQP resulted in 17.3% reduction in mortality of thrips compared to the feeding of dsGFP + dsAQP. This may also be attributed to hampering of AQP RNAi due to knockdown of key RNAi pathway gene i.e. Dicer-2. The results of simultaneous feeding of dsRNA against the target and RNAi pathway genes confirmed the presence of well-functioning RNAi pathway in T. tabaci. These data also showed that the mortality observed in dsRNA treated insects is indeed due to RNAi effect. The results have given the clue for role of Dicer-2 and Aubergine in RNAi of T. tabaci. The Thrips Staufen analyzed for protein domains using SMART (Simple Modular Architecture Research Tool) [15] showed presence of 4 dsRBDs and one staufen domain at C-terminal, however we could find only 5 dsRBDs without Staufen C-terminal domains in insect orders other than coleoptera (Additional file 5: Fig. S1). The phylogenetic analysis revealed that the Staufen from T. tabaci shows evolutionary relationship with Staufen and StauC from coleopterans (Additional file 5: Fig. S2). The phylogenetic analysis also predicts that the StauC is specific only to coleopteran insects, however some unique protein motifs were predicted in T. tabaci using SALAD (Surveyed conserved motif ALignment diagram and the Associating Dendrogram) [16] (Additional file 5: Fig. S3).
a RNAi of RNAi pathway genes genes in Thrips tabaci: a AQP expression levels in Dicer Knockdown. b Dicer Knockdown (66.7%). c AQP expression levels in Aubergine knockdown. d Aubergine knockdown (56.8%). The expression level has been normalized with GSTD2. Error bar represents standard deviation (n = 6) and *indicates significant levels of mRNA transcripts of target genes compared to GFP (P ≤ 0.05, Student’s t-test)
a SNF expression levels in Staufen knockown Thrips tabaci. b Staufen knockdown (78.5%). The expression level has been normalized with GSTD2. Error bar represents standard deviation (n = 6) and *indicates significant levels of mRNA transcripts of target genes compared to GFP- Control (P ≤ 0.05, Student’s t-test)
Percent mortality of Thrips tabaci post 48 h feeding of dsRNA incorporated in synthetic diet through membrane feeding assay. Error bars represents SE ± mean based on three replication. a Bioassay with dsSNF7 and dsAQP as compared to dsGFP (N = 75–100). b Reduction in dsSNF7 mortality due to simultaneous feeding of dsStaufen + dsSNF7 compared to dsGFP + dsSNF7 (N = 45). c Reduction in mortality of dsDicer + dsAQP compared to dsGFP + dsAQP (N = 75). *Indicates significant percent mortality compared to dsGFP-control and #indicates significant difference between first two treatments in B and C (P ≤ 0.05, Student’s t-test)
Discussion
The lack of genomic/transcriptomic information for Thysanoptera insect T. tabaci motivated us to sequence its RNA and assembly de novo transcriptome. This sequence resource was used to identify stable reference genes for RT-qPCR studies in T. tabaci. The data resource was also used to identify the existence of RNAi machinery and demonstration of feeding RNAi in this insect. A respectable number of CDS (27,836) successfully identified from transcripts indicated the completeness of transcriptome, which was further validated by BUSCO analysis. The quality of our assembled transcripts seems on par when compared to assemblies listed in Simao et al. [17, 18]. Previous studies with another thysanopteran insect western flower thrip, F. occidentalis also reported comparable number (24,262) of assembled CDS from de novo transcriptome [19]. This may be an indication of gene conservation among thysanopteran insects. The RNA-seq approach sometimes fails to include sequences of rare transcripts. However, the number of CDS identified in the current study is comparable to those reported from previously sequenced transcriptomes of other insect species including 14,797 CDS in Drosophila melanogaster [20]; 18,501 in Bombyx mori [21] and 18,071 in Plutella xylostella [22]. The assembled and annotated transcriptome is the first open source sequence data available for T. tabaci, which can be used for initiating molecular studies in this insect.
The reference gene evaluation using BestKeeper, NormFinder, geNorm and RefFinder revealed GSTD2, UbiCE, 28s, SOD and TUB as stable genes. The previous reports in Bactrocera minax [23] and Lucilia cuprina [24] also showed GSTD2 and GST1 as the most stable reference genes, respectively. The NormFinder results, as well as overall analysis with RefFinder, showed GSTD2 to be the most stably expressed gene in thrips nymphs and adults. Present studies also revealed UbiCE in adults as a highly stable gene, however overall RefFinder analysis across stages and starvation stress designated it as the least stable gene. Previous reports in different insect species with various ubiquitins, i.e. ubiquitin conjugation factor [25], polyubiquitin [26] and ubiquitin [27] have shown their suitability as a reference gene. Our previous studies in A. biguttula also identified UbiCF as a stable gene under starvation stress [28]. SOD gene expression levels have been found variable in Caenorhabditis elegans [29]. However, our studies reveal it to be the most stable gene in late nymph based on geNorm and RefFinder analysis. Thus it is clear that stability of a particular gene varies from organism to organism and even within organism under a different set of experimental conditions.
Similarly, Actin has been widely used as a reference gene across insect species in various expression studies [30]. However, its use as universal internal control has been questioned in many instances due to uncertainty in its expression [31, 32]. It is actually, impossible to discover a gene that is stable across different developmental stages. To reach a final decision on stable reference genes, we have done successful evaluation and validation of fourteen genes identified in the de novo transcriptome of T. tabaci. The results of each algorithm are variable; however, this study gives first-hand information on possible reference genes in T. tabaci, which may be helpful for gene expression studies to the community working on this insect. So, rather than identifying a single stable gene for each developmental stage, we have identified the top 3 to 5 genes for each developmental stage based on comprehended RefFinder analysis. Moreover, we also suggest that prior to conducting any expression analysis studies, it will be worthwhile to test and validate a few of the reported reference genes under respective experimental conditions.
To authenticate our results on reference genes and their significance in expression studies, we studied the impact of normalization with most stable (GSTD2, SOD) and least stable (TUB, EF) genes on relative expression of RNAi knockdown genes. It is evident that there was a significant difference in the knockdown percentage of AQP and SNF7 when normalized individually with GSTD2 and SOD in comparison to TUB and EF. For RT-qPCR studies use of two reference genes is always advisable as it ensures greater accuracy and improves the results in the expression analysis [33, 34]. We also observed that use of two reference genes significantly improves the expression results by reducing variation within the housekeeping genes. Our previous studies with RNAi in mealybug also revealed that the gene expression data normalized with unstable genes varied significantly compared to those normalized with most stable genes [28]. Thus, the use of appropriate reference genes is the key factor for interpretation of right mRNA levels in RNAi studies. The normalization with stable vs. unstable reference genes showed significant knockdown in target gene compared to control. Present study reflects that using the least stable genes for normalization may misinterpret the actual knockdown efficiency in RNAi experiments.
Transcriptome sequence resource may be helpful in generating first-hand information on genes involved in various physiological and biochemical pathways in insects. One such process, RNAi has come up as one of the important functional genomics tools and is now being exploited to identify novel targets in insects for their management [35, 36]. Sequence-specific knockdown of target genes has been so far demonstrated in many insect species [37, 38]. There is no previous report of RNAi in T. tabaci, so this sequence data resource revealed the presence of a good number of genes involved in RNAi. We were successful in identifying 21 RNAi pathway genes from the current transcriptome. Previous studies used the transcriptome and genome sequence data for identifying RNAi pathway genes in Tuta absoluta [39] and Anoplophora glabripennis [40] and identified a comparable number of genes associated with this pathway. These core RNAi pathways genes from T. tabaci, when blasted in NCBI database, showed significant similarity with their homologs from other insect species. This was further supported by the phylogenetic analysis of RNAi pathway genes from T. tabaci and other insect species. The phylogeny at an average bootstarp of > 60 confirmed the significant evolutionary relationship among the various gene families associated with RNAi pathway. The SID-1 gene (systemic RNA interference defective-I) identified in T. tabaci, showed high sequence similarity with that in Z. nevadensis. The SID proteins are essential for dsRNA uptake and spreading systemic RNAi across cells [35, 41] and have been reported in many insect species [42]. Two dicer paralogues from T. tabaci, i.e., Dicer-1 (miRNA precursors) and Dicer-2 (long dsRNA endonuclease) were also identified, which are closely related to those reported from Blattella germanica [43]. Earlier studies in Drosophila suggested the role of Dicer-2 in siRNA/miRNA silencing pathways as it is an important component of RISC activation complex [44]. Moreover, the knockdown of Dicer-2 in L. decemlineata cell line partially blocked RNAi in these cells [45]. Drosha predicted in T. tabaci, is involved in the processing of miRNA precursors and has shown significant match with that of Z. nevadensis. However, we could not identify dsRNA binding proteins such as RDRP and Loquacious in T. tabaci transcriptome. The RDRP was also not found in Tuta absoluta [39]. It is possible that the expression of these genes may be induced in response to dsRNA feeding in these insects [46]. Furthermore, the homologs of Ago-1, Ago-2, Alg-2, Alg-4, Alg-5, Alg-6, Alg-7, Alg-10, piwi, Aubergine and SpindleE predicted in T. tabaci have shown high similarity to those reported from Z. nevadensis and Locusta migratoria [47]. The Clathrin heavy chain protein known to function in endocytosis pathway in Leptinotarsa decemlineata is present in T. tabaci, this gene exhibits high sequence similarity to that reported from Z. nevadensis. A single transcript of dsRNase has been identified from T. tabaci. dsRNases have been known to degrade long dsRNA in the gut of hemipteran insects, thus posing a major bottleneck in successful RNAi in this group [48, 49]. Further studies with dsRNases may be helpful in better understanding of RNAi in this insect. We have also identified Staufen in T. tabaci, which is a dsRNA-binding protein composed of four or five dsRBDs and initially identified from Drosophila [50]. Staufen from C. elegans has been known to play a role in RNAi. However recently robust RNAi efficiency in beetles has been attributed to coleopteran-specific Staufen (StauC), which is not present in insects of other orders [51, 52]. Thus transcriptome mining of T. tabaci reveals the existence of well-defined RNAi machinery in T. tabaci, which was further, validated through sequence-specific knockdown of SNF7 and AQP, as well as core RNAi pathway genes such as Dicer-2, Aubergine and Staufen. The knockdown efficiency through feeding of dsSNF7 and dsAQP was about 93% in T. tabaci. This may be attributed to robust RNAi machinery in this insect or higher concentration of dsRNA (500 ng/µl of diet) used for feeding bioassay in the current study compared to the previous studies that reported 20 to 27% reduction in vATPase transcripts through injection of 80 ng dsRNA directly into the females of a thysanopteran insect, F. occidentalis [53]. The mortality of T. tabaci observed in dsSNF and dsAQP was high compared to dsGFP control and this further support the knockdown assay results of dsRNA feeding. Knockdown of RNAi pathway genes Dicer-2, Aubergene and Staufen reduced RNAi effect caused by dsSNF7 and dsAQP demonstrating the function of these genes in RNAi and cofirming that the mortality induced by dsSNF7 and dsAQP is indeed due to RNAi effect.
The simultaneous knockdown of Dicer-2, Aubergine and Staufen and target gene (SNF7 and AQP) confirmed their role in RNAi pathway by hampering the knockdown efficiency of latter two genes. This was further supported by low mortality of T. tabaci observed in dsDicer + dsAQP and dsStaufen + dsSNF compared to dsAQP and dsSNF7 alone. The studies with coleopterans cell line of L. decemlineata showed that knockdown of Dicer-2 and Aubgerine partially blocked RNAi in these cells [45]. Earlier studies have shown that Staufen in coleopterans does not play any role in RNAi, however StauC (StaufenC) predominantly contributes to this process [51]. StauC specific to beetles is a key player for robust RNAi response in coleopteran insects, additionally lower expression of StauC in RNAi resistant L. decemlineata cell lines (Lepd-SL1RR) showed poor RNAi response [51]. StauC domain is missing in lepidopterans which are poor processors of dsRNA to siRNA [54]. It may be possible that in other insect species like thrips, Staufen may be contributing to RNAi in place of StauC. Thrips Staufen have four dsRNA binding domains (dsRBD) similar to what has been reported in coleopteran specific StauC protein, however in most of the other insect species Staufen has five dsRBD [51]. The Staufen from T. tabaci was in close resemblance to coleopteran Staufen and additionally it showed some unique protein motifs, which were absent in other insect species. Thus Staufen from T. tabaci needs further characterization to understand its exact role in RNAi. Our studies preliminarily elucidate the role of Dicer-2, Aubergine and Staufen in RNAi of T. tabaci, which needs to be further validated through indepth studies. The validation of in silico data through mRNA expression of some predicted RNAi pathway genes, phylogenetic analysis as well dsRNA feeding assay confirm the existence of well-functioning RNAi machinery in T. tabaci.
Conclusion
This study has generated good quality assembled and annotated transcriptome with 27,836 predicted CDS from T. tabaci. This sequence resource will be useful for molecular studies in T. tabaci. Fourteen putative reference genes have been identified in the transcriptome and evaluated for their stability through expression studies. Based on analysis using different algorithms, GSTD2 has been predicted as the best reference gene in nymphs, adults and starvation stress. The top three stable genes are UbiCE, 28s, TATA in adults, SOD, GSTD2 and 18s in nymphs, and TUB, 18s, UbiCE under starvation conditions. Both in silico and in vivo experiments demonstrated the presence of robust RNAi machinery in T. tabaci. The efficient RNAi in T. tabaci may hold potential for developing novel strategies for the management of this pest.
Materials and methods
Insect culture
Thrips tabaci culture was maintained on Gossypium hirsutum variety HS 6 raised in the walk-in environmental chamber (RH 65%, Temperature 30 ± 2 °C). Staggered sowing of the cotton plants was done to ensure the year-round availability of cotton plants for the rearing of T. tabaci.
Total RNA isolation and sequencing
Individuals representing each developmental stage were collected using a soft paintbrush (# 000) and pooled for total RNA isolation and transcriptome sequencing. For validation of reference genes (HKGs), a mixture of 1st and 2nd nymphal stage individuals (200 per pool) and adults (150 per pool) was collected in three replicates. In addition to this, 200 adult thrips in three replicates were kept under starvation stress for 5 h. The total RNA was isolated from these samples using Tri Reagent® (Sigma-Aldrich) as per the manufacturer’s protocol. For RNA sequencing, the quality and quantity of the isolated RNAs were determined using denatured RNA agarose gels (Additional file 6: Fig. S1) and Qubit Fluorometer (ThermoFisher Scientific) spectrophotometer (Additional file 6: Fig. S2), respectively. For other studies the quantification of the isolated RNAs was done using Eppendorf BioSpectrometer® basic and quality of RNA was checked by denaturing agarose gel electrophoresis.
RNA sequencing libraries were prepared using TruSeq stranded mRNA sample preparation kit (Illumina). mRNA was enriched from the total RNA using poly-T magnetic beads. The mRNA was fragmented enzymatically and reverse transcribed to cDNA. The cDNA was purified using XP beads (Ampure) followed by A-tailing, adapter ligation, and enrichment by PCR. The quality and quantity of the libraries were determined by 4200 Tape Station system (Agilent Technologies) using High sensitivity D1000 Screen tape (Agilent Technologies). The libraries were sequenced using the NextSeq 500 (Illumina).
Transcriptome assembly
The sequenced raw data were processed to obtain high-quality clean reads using Trimmomatic V0.35 [55] to remove adapter sequences, ambiguous reads with unknown nucleotides “N” and more than 5% and low-quality sequences with PHRED score (QV) < 20. These high-quality reads (QV > 20), obtained were used for de novo assembly using default parameters. Adapter trimming, the Sliding window, Leading Trailing were performed at a threshold quality of below 25. Additionally, the reads were eliminated if its length was below 100 bp using Minlength setting [56]. The high-quality filtered reads were assembled into transcripts using Velvet V1.2.10 [57] and OasesV0.2.09 [58] on optimized K mer 57 and 47, respectively. To check for misassembled, erroneous and poorly supported transcripts that arose during the assembly, all high-quality reads were mapped back to their respective assembled transcripts using Burrows-Wheeler Aligner BWAV0.7.12 for validation. Open reading frames (CDS) of all assembled transcripts were predicted using TransDecoder (http://transdecoder.sourceforge.net).
Gene ontology and functional annotation
Gene ontology and functional annotation were performed for all predicted CDS by Blast2GO program. GO mapping was carried out to retrieve GO terms for all the BLASTX functionally annotated CDS. To identify the potential involvement of the predicted CDS in biological pathways, the CDS were mapped to reference canonical pathways in KEGG. All the CDS were classified mainly into four categories: Metabolism, Cellular processes, Genetic information processing, Environmental information processing. The output of KEGG analysis included KEGG Orthology (KO) assignments, Corresponding Enzyme Commission (EC) numbers and metabolic pathways of predicted CDS using KEGG automated annotation server KASS (http://www.genome.jp/kaas-bin/kaas_main). The completeness and contiguity of the assembled transcriptome was further validated using BUSCO v1.1b1 (Benchmarking Universal Single-Copy Orthologs) 15. This analysis was performed using insect (1658) and metazoan (978) BUSCO dataset, which includes BUSCO lineages of 42 insects and 65 metazoan species.
Identification of stable reference genes of T. tabaci
Selection of genes and primer design
Primer sequences and the associated amplicon characteristics for fourteen commonly used reference genes are summarized in Table 2. Based on reference genes described in the literature for insects [25, 28, 59], transcriptome data was searched for identification of following homologous T. tabaci sequences: Actin, 18s (ribosomal protein 18), 28s (ribosomal protein 28), TUB (tubulin), GAPDH (Glyceraldehyde 3-phosphate dehydrogenase), EF (elongation factor alpha), RPL17 (ribosomal protein component L17), HIST3 (Histone 3), UbiCE (ubiquitin-conjugating factor), TATA (TATA box-binding protein-like protein 1), E2F (transcription factor E2F1), vATPase (V-ATPase holoenzyme subunit B), SOD (Mn superoxide dismutase) and GSTD2 (glutathione s-transferase D2) which were amplified using RT-qPCR (Table 3). The Primer3 software was used to design the primers [60] with predefined parameters such as amplicon length 100–150 bp, melting temperature 60–62 °C, primer length 20–22 bp, and GC content 45–50%.
Quantitative real-time PCR analysis
Total RNA (1 µg) isolated from 2nd instar, adults and starved insects was used for cDNA synthesis using First Strand cDNA Synthesis Kit (ThermoFisher Scientific). Real-time PCR reactions were performed using three biological replicates and three technical replicates for each treatment. The reaction contained 10 µl total reaction volume (5 μl SYBR Premix Ex Taq II (Clontech Takara, USA), 1 µl (1:10) of cDNA template and 0.2 μl each of gene-specific primers (Tables 1, 2). Reactions were carried out in a 96 Lightcycler (Roche, USA) with PCR parameters as follows: 95 °C for 3 min, 40 cycles of 95 °C for 5 s, 30 s at the Tm value of primer pairs. The melting curve analysis was performed to verify the specificity of amplified PCR products.
Statistical analysis
To evaluate the expression stability of the reference genes in both developmental stages and starvation stress, four algorithms, geNorm [61], NormFinder [62], Bestkeeper [63] and RefFinder (http://leonxie.esy.es/RefFinder/?type=reference) were used. Ct values were obtained from the experiment run in the Lightcycler software (Roche). Comparative Ct method was used to convert these into linear values by assuming the lowest relative quantity for each gene as 1 and further using these values as input figures in NormFinder and geNorm algorithms. geNorm determines the stability score (M) by calculating the average of mean pairwise variation of each reference gene. Thus the genes with M value > 0.5 were excluded from studies. The NormFinder analysis was based on the expression stability of genes within inter and intragroup and standard deviation calculated by advanced analysis. BestKeeper is an open source MS Excel-based algorithm, which uses Ct values of genes to infer their stability taking into consideration standard deviation, P-values and correlation coefficient of each gene. Consequently, lower SD signifies the suitability of a particular gene to serve as a better internal control. RefFinder (http://leonxie.esy.es/RefFinder/?type=reference) is an online tool, which combines the outcome of all three above algorithms along with Delta CT method and calculates the geometric mean (comprehensive analysis) for each gene to rank best reference gene.
Validation of reference genes through RNAi studies
To evaluate the impact of selection of reference genes in RNAi studies, two most stable and two least stable genes as per results obtained were used for normalization of mRNA levels in dsSNF7 (double-stranded RNA of SNF7) and dsGFP (double-stranded RNA of GFP-control) fed T. tabaci adults. The gene-specific primers having T7 promoter sequence (TAATACGACTCACTATAG) at 5′ end of both primers were used to amplify the template for dsSNF7 i.e. (Fwd: 5′GGTTTGCGAGGAGAGCTTATGG3′; Rev.:5′GGGAGGGCAGCAATTCCTACTT3′). The amplified product was purified using Nucleospin Gel and PCR Cleanup kit (Macherey–Nagel) as per instruction manual and used for dsRNA synthesis using MEGAscript™ RNAi Kit (Thermo Fisher Scientific). Biological samples in triplicate each comprising of 200 adults thrips were released in 1.5 ml tube (lower 1/4th part of the tube was cut and covered with 2–3 folds of muslin cloth). The dsSNF7 and dsAQP (500 ng/ul) were fed along with sucrose diet stretched between two layers of parafilm as per earlier described methodology used for insecticide bioassay [64]. Live insects were collected 48 h post-feeding, and total RNA was isolated using Tri-Reagant® (Sigma-Aldrich). For relative expression studies, cDNA synthesis was done using First Strand cDNA Synthesis Kit (ThermoFisher Scientific). The cDNA from different samples was used to quantitate mRNA level of SNF7 with RT-qPCR (as described section: Quantitative Real-time PCR analysis) using two most stable (GSTD2 and SOD) and two least stable genes (TUB and EF) individually as well as in combination.
Identification of RNAi pathway genes and their validation through RT-qPCR and RNAi feeding assay
To explore functional RNAi in T. tabaci, the sequences of major genes involved in RNAi pathway in other insects were used as query against thrips transcriptome. The E-value < e−30 was used as a cutoff and the identity of the retrieved sequences was confirmed through BlastX against NCBI nr database. A phylogenetic tree with the help of multiple sequence alignment was constructed using sequences of core RNAi genes identified from T. tabaci and their homologs reported from D. melanogaster M., Bombyx mori L., Apis mellifera L., T. castaneum Herbst, Z. nevadensis H. and Nilaparvata lugens Stål. ClustalW program and neighbor-joining analysis (bootstrap 1000) of MegaX software were used for phylogeny. To reconfirm and assess the expression levels of various RNAi pathway genes in T. tabaci, the primers were designed in Primer3 software and used in RT-qPCR (Table 4).
We chose SNF7 and AQP (Aquaporin) as target genes to confirm the RNAi functioning in T. tabaci. SNF7 functions as a constituent of ESCRT (Endosomal Sorting Complex Required for Transport) pathway, which is responsible for the cellular functions such as internalization, transport, sorting and lysosomal degradation of transmembrane proteins [65]. Aquaporins are majorly involved in water regulation in some insect species such as phloem-feeding leafhopper Cicadella viridis [66] and B. tabaci [67]. Gene-specific primers to amplify fragments of SNF7 (Fwd. 5′TAATACGACTCACTATAGGGTTTGCGAGGAGAGCTTATGG3′; Rev.:5′TAATACGACTCACTATAGGGGAGGGCAGCAATTCCTACTT3′) and AQP (Fwd: 5′TAATACGACTCACTATAGGGGAGATGAAGTACACGATGGC3′; Rev.: 5′TAATACGACTCACTATAGGGCGCAGCACATCTGGATAA3′) with T7 promoter sequence at 5′ ends of both forward and reverse primer were custom synthesized. The product amplified from these primers was purified using Nucleospin Gel and PCR Cleanup kit (Macherey–Nagel) as per instruction manual. The purified product was used as template for the dsRNA synthesis using MEGAscript™ RNAi Kit (Thermo Fisher Scientific). Biological samples in triplicate each comprising of 200 insects both nymph and adult thrips were released in 1.5 ml tube (lower 1/4th part of the tube was cut and covered with 2–3 folds of muslin cloth) (Additional file 6: Fig. S3). The dsSNF7 and dsAQP (500 ng/μl) were fed along with sucrose diet stretched between two layers of parafilm as per earlier described methodology used for insecticide bioassay [64]. Similarly synthesized dsRNA against Dicer-2, Aubergine and Staufen genes was evaluated to examine their respective knockdown impact on RNAi of AQP/SNF7 gene (Additional file 4: Table S6). For this, 500 ng/μl each of dsDicer-2 and dsAubergine mixed with dsAQP (500 ng/μl) while dsStaufen mixed with dsSNF was co-administered to thrips in sucrose diet separately for the each RNAi pathway and target gene. Live insects were collected 48 h post-feeding, and total RNA was isolated using Tri-Reagant® (Sigma-Aldrich). cDNA was synthesized from 1 µg total RNA using First Strand cDNA Synthesis Kit (ThermoFisher Scientific) followed by RT-qPCR analysis as described in the earlier section (Quantitative Real-time PCR analysis). In a separate experiment, a bioassay was conducted in three replicates (70–100 adults in each) to assess the mortality of thrips after feeding 500 ng/µl of dsSNF, dsAQP, dsDicer-2 + dsAQP, dsAubergine + dsAQP and dsStaufen + dsSNF7 and compared with dsGFP control. Further, Staufen sequences were analyzed through SMART and SALAD to study conserved domains and motifs, respectively [15, 16].
Abbreviations
- RT-qPCR:
-
real-time quantitative PCR
- RNA:
-
ribonucleic acid
- dsRNA:
-
double stranded ribonucleic acid
- RNAi:
-
RNA interference
- NCBI:
-
National Center for Biotechnology Information
- TSA:
-
Transcriptome Shotgun Assembly
- SRA:
-
Sequence Read Archives
- KASS:
-
KEGG Automatic Annotation Server
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- cDNA:
-
complementary deoxy-ribonucleic acid
- CDS:
-
coding sequence
- RNA-seq:
-
RNA sequencing
- GO:
-
gene ontology
- BUSCO:
-
Benchmarking Universal Single-Copy Orthologs
- 18S:
-
ribosomal protein 18
- 28s:
-
ribosomal protein 28
- TUB :
-
tubulin
- GAPDH :
-
glyceraldehyde 3-phosphate dehydrogenase
- EF :
-
elongation factor alpha
- RPL17 :
-
ribosomal protein component L17
- HIST3 :
-
histone 3
- UbiCE :
-
ubiquitin-conjugating factor
- TATA :
-
TATA box-binding protein-like protein 1
- E2F :
-
transcription factor E2F1
- VATPase :
-
V-ATPase holoenzyme subunit B
- SOD :
-
Mn superoxide dismutase
- GSTD2 :
-
glutathione s-transferase D2
- GFP:
-
green florescent protein
- Ct:
-
cycle threshold
- SD:
-
standard deviation
References
Mound LA, Morris DC. A new thrips pest of Myoporum cultivars in California, in a new genus of leaf-galling Australian Phlaeothripidae (Thysanoptera). Zootaxa. 2007;1495:35–45.
Heming BS. Development of the mouthparts in embryos of Haplothrips verbasci (Osborn) (Insecta, Thysanoptera, Phlaeothripidae). J Morphol. 1980;164:235–63. https://doi.org/10.1002/jmor.1051640303.
Jacobson AL, Booth W, Vargo EL, Kennedy GG. Thrips tabaci population genetic structure and polyploidy in relation to competency as a vector of tomato spotted wilt virus. PLoS ONE. 2013;8:1–10. https://doi.org/10.1371/journal.pone.0054484.
Childers C, Achor D. Thrips feeding and oviposition injuries to economic plants, subsequent damage and host responses to infestation. In: Parker BL, Skinner M, Lewis T, editors. Thrips biology and management. Boston: Springer US; 1995. p. 31–51. https://doi.org/10.1007/978-1-4899-1409-5_3.
Hunter WB, Ullman DE. Precibarial and cibarial chemosensilla in the western flower thrips, Frankliniella occidentalis (Pergande)(Thysanoptera: Thripidae). Int J Insect Morphol Embryol. 1994;23:69–83. https://doi.org/10.1016/0020-7322(94)90001-9.
Morse JG, Hoddle MS. Invasion biology of Thrips. Annu Rev Entomol. 2006;51:67–89. https://doi.org/10.1146/annurev.ento.51.110104.151044.
Reitz SR. Biology and ecology of the western flower thrips (Thysanoptera: Thripidae): the making of a pest. Fla Entomol. 2009;92:7–13. https://doi.org/10.1653/024.092.0102.
Riley DG, Joseph SV, Srinivasan R, Diffie S. Thrips vectors of tospoviruses. J Integr Pest Manag. 2011;2:I1–10. https://doi.org/10.1603/IPM10020.
Immaraju JA, Paine TD, Bethke JA, Robb KL, Newman JP. Western flower thrips (Thysanoptera: Thripidae) resistance to insecticides in coastal California greenhouses. J Econ Entomol. 1992;85:9–14.
Palli SR. RNA interference in Colorado potato beetle: steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci Insect Sci. 2014;6:1–8.
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. https://doi.org/10.1093/bioinformatics/bti610.
http://busco.ezlab.org/v2/datasets/insecta_odb9.tar.gz. Accessed 15 May 2018
http://busco.ezlab.org/v2/datasets/metazoa_odb9.tar.gz. Accessed 15 May 2018
García-Ortega LF, Martínez O. How many genes are expressed in a transcriptome? estimation and results for RNA-Seq. PLoS ONE. 2015;10:1–22. https://doi.org/10.1371/journal.pone.0130262.
Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–6. https://doi.org/10.1093/nar/gkx922.
Mihara M, Itoh T, Izawa T. SALAD database: a motif-based database of protein annotations for plant comparative genomics. Nucleic Acids Res. 2009;38:D835–42.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM, Simao FA, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. https://doi.org/10.1093/bioinformatics/btv351.
Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, et al. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2017;35:543–8. https://doi.org/10.1093/molbev/msx319.
Zhang Z, Zhang P, Li W, Zhang J, Huang F, Yang J, et al. De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics. 2013;101:296–305. https://doi.org/10.1016/j.ygeno.2013.02.005.
Daines B, Wang H, Wang L, Li Y, Han Y, Emmert D, et al. The Drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21:315–24.
Wang X, Li Y, Peng L, Chen H, Xia Q, Zhao P. Comparative transcriptome analysis of Bombyx mori spinnerets and Filippi’s glands suggests their role in silk fiber formation. Insect Biochem Mol Biol. 2016;68:89–99.
Xia X, Zheng D, Zhong H, Qin B, Gurr GM, Vasseur L, et al. DNA sequencing reveals the midgut microbiota of diamondback moth, Plutella xylostella (L.) and a possible relationship with insecticide resistance. PLoS ONE. 2013;8:e68852. https://doi.org/10.1371/journal.pone.0068852.
Fu W, Xie W, Zhang Z, Wang S, Wu Q, Liu Y, et al. Exploring valid reference genes for quantitative real-time PCR analysis in Plutella xylostella (Lepidoptera: Plutellidae). Int J Biol Sci. 2013;9:792–802.
Bagnall NH, Kotze AC. Evaluation of reference genes for real-time PCR quantification of gene expression in the Australian sheep blowfly, Lucilia cuprina. Med Vet Entomol. 2010;24:176–81. https://doi.org/10.1111/j.1365-2915.2010.00866.x.
Bansal R, Mittapelly P, Chen Y, Mamidala P, Zhao C, Michel A. Quantitative RT-PCR gene evaluation and rna interference in the brown marmorated stink bug. PLoS ONE. 2016;11:e0152730.
Maroniche GA, Sagadín M, Mongelli VC, Truol GA, Del Vas M. Reference gene selection for gene expression studies using RT-qPCR in virus-infected planthoppers. Virol J. 2011;8:308. https://doi.org/10.1186/1743-422X-8-308.
Huis R, Hawkins S, Neutelings G. Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol. 2010;10:71. https://doi.org/10.1186/1471-2229-10-71.
Singh S, Gupta M, Pandher S, Kaur G, Rathore P, Palli SR. Selection of housekeeping genes and demonstration of RNAi in cotton leafhopper, Amrasca biguttula biguttula (Ishida). PLoS ONE. 2018;13:e0191116.
Hoogewijs D, Houthoofd K, Matthijssens F, Vandesompele J, Vanfleteren JR. Selection and validation of a set of reliable reference genes for quantitative sod gene expression analysis in C. elegans. BMC Mol Biol. 2008;9:1–8.
Kreuzer KA, Lass U, Landt O, Nitsche A, Laser J, Ellerbrok H, et al. Highly sensitive and specific fluorescence reverse transcription-PCR assay for the pseudogene-free detection of β-actin transcripts as quantitative reference. Clin Chem. 1999;45:297–300.
Foss DL, Baarsch MJ, Murtaugh MP. Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and β-actin mRNA expression in porcine immune cells and tissues. Anim Biotechnol. 1998;9:67–78.
Spanakis E. Problems related to the interpretation of autoradiographic data on gene expression using common constitutive transcripts as controls. Nucleic Acids Res. 1993;21:3809–19. https://doi.org/10.1093/nar/21.16.3809.
Jiang H, Qian Z, Lu W, Ding H, Yu H, Wang H, et al. Identification and characterization of reference genes for normalizing expression data from red swamp crawfish Procambarus clarkii. Int J Mol Sci. 2015;16:21591–605.
Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre J-F, Louvet R, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008;6:609–18.
Price DRG, Gatehouse JA. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008;26:393–400.
Burand JP, Hunter WB. RNAi: future in insect management. J Invertebr Pathol. 2013;112(SUPPL. 1):9–14. https://doi.org/10.1016/j.jip.2012.07.012.
Gu L, Knipple DC. Recent advances in RNA interference research in insects: implications for future insect pest management strategies. Crop Prot. 2013;45:36–40. https://doi.org/10.1016/j.cropro.2012.10.004.
Inderkant S, Singh S, Mogilicherla K, Shukla JN, Palli SR. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci Rep. 2017;7:17059.
de Camargo RA, Herai RH, Santos LN, Bento FMM, Lima JE, Marques-Souza H, et al. De novo transcriptome assembly and analysis to identify potential gene targets for RNAi-mediated control of the tomato leafminer (Tuta absoluta). BMC Genomics. 2015;16:1–17. https://doi.org/10.1186/s12864-015-1841-5.
Rodrigues TB, Dhandapani RK, Duan JJ, Palli SR. RNA interference in the Asian Longhorned Beetle: identification of key rnai genes and reference genes for RT-qPCR. Sci Rep. 2017;7:1–10. https://doi.org/10.1038/s41598-017-08813-1.
Shih JD, Hunter CP. SID-1 is a dsRNA-selective dsRNA-gated channel. RNA. 2011;17(6):1057–65. https://doi.org/10.1261/rna.2596511.
Jose AM, Hunter CP. Transport of sequence-specific rna interference information between cells. Annu Rev Genet. 2007;41:305–30. https://doi.org/10.1146/annurev.genet.41.110306.130216.
Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–9.
Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81.
Yoon JS, Shukla JN, Gong ZJ, Mogilicherla K, Palli SR. RNA interference in the Colorado potato beetle, Leptinotarsa decemlineata: identification of key contributors. Insect Biochem Mol Biol. 2016;78:78–88. https://doi.org/10.1016/j.ibmb.2016.09.002.
Garbutt JS, Reynolds SE. Induction of RNA interference genes by double-stranded RNA; implications for susceptibility to RNA interference. Insect Biochem Mol Biol. 2012;42:621–8. https://doi.org/10.1016/j.ibmb.2012.05.001.
Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–50.
Allen ML, Walker WB. Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol. 2012;58:391–6. https://doi.org/10.1016/j.jinsphys.2011.12.014.
Gupta M, Singh H, Kaur R, Pandher S, Singh S. Improving feeding RNAi efficiency in Bemisia tabaci through knockdown of gut dsRNases to understand osmoregulatory genes. In: Curbing whitefly-plant virus pandemics: the departure from pesticides to genomic solutions. Punjab Agricultural University; 2017. p. 51.
St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63.
Yoon J-S, Mogilicherla K, Gurusamy D, Chen X, Chereddy SCRR, Palli SR. Double-stranded RNA binding protein, Staufen, is required for the initiation of RNAi in coleopteran insects. Proc Natl Acad Sci. 2018;115:8334–9.
Prentice K, Pertry I, Christiaens O, Bauters L, Bailey A, Niblett C, et al. Transcriptome analysis and systemic RNAi response in the African sweetpotato weevil (Cylas puncticollis, Coleoptera, Brentidae). PLoS ONE. 2015;10:1–18.
Badillo-Vargas I, Rotenberg D. RNA interference tools for the western flower thrips, Frankliniella occidentalis. Insect Physiol. 2015;76:36–46. https://doi.org/10.1016/j.jinsphys.2015.03.009.
Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol. 2011;57:231–45.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20.
Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci. 2009;106:12950–5. https://doi.org/10.1073/pnas.0903091106.
Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9.
Schulz C, Perdiguero EG, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, et al. A lineage of myeloid cells independent of myb and hematopoietic stem cells. Science. 2012;335:86–90.
Barros Rodrigues T, Khajuria C, Wang H, Matz N, Cunha Cardoso D, Valicente FH, et al. Validation of reference housekeeping genes for gene expression studies in western corn rootworm (Diabrotica virgifera virgifera). PLoS ONE. 2014;9:e109825.
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:9–14.
Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques. 2004;36:84–91. https://doi.org/10.1186/gb-2002-3-7-research0034.
Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50.
Wojciechowska-Durczyńska K, Krawczyk-Rusiecka K, Cyniak-Magierska A, Zygmunt A, Gałecka E, Lewiński A. Relative quantification of PIK3CA gene expression level in fine-needle aspiration biopsy thyroid specimens collected from patients with papillary thyroid carcinoma and non-toxic goitre by real-time RT-PCR. In: Thyroid research. 2010. p. 5. https://doi.org/10.1186/1756-6614-3-5.
Huseth AS, Chappell TM, Langdon K, Morsello SC, Martin S, Greene JK, et al. Frankliniella fusca resistance to neonicotinoid insecticides: an emerging challenge for cotton pest management in the eastern United States. Pest Manag Sci. 2016;72:1934–45.
Ramaseshadri P, Segers G, Flannagan R, Wiggins E, Clinton W, Ilagan O, et al. Physiological and cellular responses caused by RNAi- mediated suppression of snf7 orthologue in Western Corn Rootworm (Diabrotica virgifera virgifera) Larvae. PLoS ONE. 2013;8:e54270. https://doi.org/10.1371/journal.pone.0054270.
Le Caherec F, Guillam MT, Beuron F, Cavalier A, Thomas D, Gouranton J, et al. Aquaporin-related proteins in the filter chamber of homopteran insects. Cell Tissue Res. 1997;290:143–51.
Mathew LG, Campbell EM, Yool AJ, Fabrick JA. Identification and characterization of functional aquaporin water channel protein from alimentary tract of whitefly, Bemisia tabaci. Insect Biochem Mol Biol. 2011;41:178–90. https://doi.org/10.1016/j.ibmb.2010.12.002.
Authors’ contributions
SS, MG, SP and PR conceived the experiments; SS, MG and GK conducted the experiments; SS, MG, GK, SP and SRP analyzed the results; SS, MG and NG did the bioinformatics analysis; SS, MG, SP, PR and SRP prepared the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Punjab Agricultural University, Ludhiana for the support to conduct this study and BMC for waiving the APC charges.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data sets supporting the results of this article are deposited in the National Center for Biotechnology Information (NCBI) repository under Bioproject PRJNA392363, SRA (Sequence Read Archives) accession SRX2977733 and TSA (Transcriptome Shotgun Assembly) accession GFQQ00000000.
Funding
No formal funding has been received for this research. However, the research has been carried out through internal funds of Punjab Agricultural University.
Consent to publish
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1.
Annotations of T. tabaci CDS sequences.
Additional file 2.
Gene ontology.
Additional file 3.
KEGG Automatic Annotation Server (KASS) pathway analysis.
Additional file 4.
Reference gene amplicon characteristics, analysis with NormFinder, BestKeeper and RefFinder, list of RNAi pathway genes identified from T. tabaci and dsRNA primer details.
Additional file 5.
Staufen protein domains from different insect, phylogenetic analysis of Staufen from insect species and Unique motifs in T. tabaci Staufen compared to other insects.
Additional file 6.
RNA quality assessment for transcriptome sequencing and dsRNA feeding schematic layout.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Singh, S., Gupta, M., Pandher, S. et al. RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.) (Thysanoptera: Thripidae). BMC Molecular Biol 20, 6 (2019). https://doi.org/10.1186/s12867-019-0123-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12867-019-0123-1