Abstract
Background
Raw milk may contain pathogenic microorganism that can sometimes fatally affect the health of consumers. However, risks related to raw milk consumption in Southwest Ethiopia are not well studied. The aim of this study was to evaluate the presence of five target pathogenic bacteria including Escherichia coli O157:H7, Salmonella enterica Typhimurium, Staphylococcus aureus, Listeria monocytogenes, and Campylobacter jejuni in raw milk and to assess exposure associated with the consumption of raw milk.
Method
A cross-sectional study was carried out between November 2019 and June 2020 to in Jimma zone, Southwest Ethiopia. Laboratory analysis was conducted on milk samples collected from Seven Woreda towns, including, Agaro, Yebu, Sekoru, Serbo, Shebe, Seka, Sheki and Jimma town administration. Semi-structured interview questions were administered to collect data on the amount and frequency of consumption. Descriptive statistics were used to summarize laboratory results and questionnaire survey data.
Result
Among 150 total raw milk samples, about 61.3% were found contaminated by one or more types of pathogens along the dairy value chain. The highest and the least bacterial counts recorded were 4.88 log10cfu/ml and 3.45 log10cfu/ml from E. coli and L. monocytogenes respectively. The mean concentrations of pathogens demonstrated significant statistical difference (p < 0.05) using 95% confidence interval where the prevalence percentage of isolates increased as the milk was transported from farms to the retail outlets. Except for C. jejuni; all other pathogens were detected in the range of unsatisfactory level of milk microbiological quality along the chain. The estimated mean annual risk of acquiring intoxication of E. coli across retailer outlets is 100% whereas salmonellosis, S. aureus intoxication, and listeriosis are 84%, 65% and 63% respectively.
Conclusion
The study highlights the significant health risks associated with the consumption of raw milk due to its unacceptable microbiological quality. The traditional production and consumption patterns of raw milk are the primary reasons for the high annual probability of infection. Therefore, regular monitoring and implementation of hazard identification and critical control point principles are necessary from raw milk production to retail points to ensure the safety of consumers.
Similar content being viewed by others
Introduction
The increasing magnitude of microbial contamination of food is among the most important challenges associated to public health, since the hazard existence has been recognized [34]. Animal products are generally considered as high risk food items with respect to pathogen contents, natural toxins and other unavoidable pre-processing and post-processing activities [12]. Milk and milk-based food products are highly susceptible to microbial contamination because of their rich composition, which provides a favorable medium for growth of spoilage agents [4].
The most predominant zoonotic bacterial contaminants of raw milk and other dairy products include Staphylococcus aureus, Escherichia coli, Salmonella enterica typhimurium, Shigella spp., Listeria monocytogenes, Yersinia enterocolitica, Campylobacter jejuni, Streptococcus spp., and Pseudomonas spp. [27, 37, 38]. Foods contaminated with such biological hazards and ready for consumption are one of the health concerns in the world causing food-born health problems. Ethiopia shares the same global health concerns associated to raw milk consumption, mainly gastroenteritis and typhoid fever as the rest of the world. Food-borne gastrointestinal disease can be either due to infection by vegetative bacterial cells or by intoxication from toxigenic bacteria such as S. aureus and E. coli. Food-borne illnesses create an enormous burden on the country’s economy such as consumer costs including medical, legal, and other expenses, as well as absenteeism at work and school [2]. In relation to food-borne illness, World Health Organization (WHO) and the Food and Agriculture Organization (FAO) have set standards for acceptable daily intake of microbial load in foods in order to safeguard human health [12].
Even though raw milk is considered as relatively free from contaminants when it leaves the udder, it is highly prone to the invasion of various exogenous pathogenic bacteria [22, 26, 27, 36, 37]. It has been reported between 2006 and 2012 that twenty outbreaks of illness have been associated with the consumption of raw milk in New Zealand [27].
Milk is an important source of nutrition in developing countries like Ethiopia. Although its milk yield is found to be low, Ethiopia ranks first in the number of livestock in Africa [25]. It is estimated that Jimma zone has 456,893 heads of cattle, which is relatively the largest cattle population in Oromia region [3]. Hence, the people are highly dependent on the foods of animal source, mainly on dairy products. Despite the preference of consuming raw milk and other dairy products in the community, there is no formal standard followed to maintain the hygienic conditions of milk along the production and transportation line. Hand milking, unsafe transportation under ambient temperatures and collection of milk in unsterile containers are the common practices among the community that lead to poor milk hygiene. Microorganisms take advantages of time for proliferation all the way through such unhygienic production, transportation and storage [6, 13].
In spite of these unhygienic practices that could pose potential health problems in our community, there are very few related studies assessing biological hazards and public exposures. In addition, Ethiopia has no systematic surveillance and management systems that help in identifying, quantifying and controlling food safety hazards and potential adverse health effects resulting from human exposure [2]. It is necessary,however, to quantify biological hazards in readily consumable foods and to have baseline information on the risk factors related to food contamination. Therefore, the objective of this study is to identify and characterize hazards and to assess exposure associated with the consumption of raw milk in Jimma zone.
Methods
The study area, sampling and design
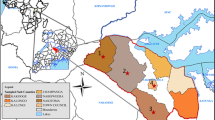
A cross sectional study was carried out in Jimma zone, Southwest Ethiopia located about 350 km from Addis Ababa, the capital of the country. The elevation of Jimma Zone varies from 1000 to 3360 m above sea level. According to the 2007 national population and housing census, the zone has a total population of 2.6 million, of which 88.7% are rural residents. A total of 521,506 households were counted in this zone, which results in an average of 4.77 persons to a household, and 500,374 housing units. Jimma zone has estimates of 456,893 heads of cattle [5, 7]. It is estimated that about 203L of milk is produced in Jimma Zone per one lactation period of a cow [19]. Seven Woreda (a geo-political sub division of a zone) towns including Agaro, Yebu, Sekoru, Serbo, Shebe, Seka, Sheki and Jimma town administration were purposely selected for this study.
Sample collection
A total of 150 milk samples were collected from sampling points; 50 from dairy farms (a sample from each Woreda center and 43—samples from Jimma town administration), 50 from collection and distribution centers of Jimma town administration, and 50 from retail outlets of selected Woreda towns and Jimma town administration (a sample from each Woreda center and 43—samples from Jimma town administration). The samples were collected from all small to large scale urban dairy farms, examining the value chain from farm to table; nearly all farms within the selected study area were included in the analysis (Fig. 1).
About 25 ml of raw milk samples were collected aseptically with sterile universal screw capped plastic bottles using sterilized 50 ml syringe. The samples were collected at dairy farms immediately after milking from the storage containers ready to be transported. At distribution centers and retailer outlets, the samples were taken directly from milk storage tanks immediately after the milk had been transported and collected in the tanks. At each stage of dairy supply chain, a sample was taken from two to three randomly selected containers within the same sterile bottle. All collected samples were labeled, placed in cold ice boxes and transported within 4 h of sampling to the Microbiology Laboratory School of Medical Laboratory Sciences in Jimma University for microbiological analysis.
Questionnaire and interview survey
As there is no reliable estimate on the exposure status of inhabitants of Jimma zone for microbial contamination through raw milk consumption, a 50% proportion which leads to the highest possible sample size was used as recommended by Daniel [8]. The estimate is desired to be with 5% margin of error and 95% confidence interval. Therefore the required sample size for this study was determined using the following statistical formula:
where “N” is the total number of households in towns of Jimma zone which is 60,107, “n” is minimum number of sample size, “Z” is standard normal score, “p” is the prevalence value, “q” = 1-p and “d” is marginal error. At 95% confidence interval Z = 1.96 and marginal error is 5%. Since no report is yet recorded for prevalence of exposure the p-value is considered to be 50%. Considering 10% non-response rate, the final sample size for raw milk exposure assessment is calculated to be 420. Interview questions were administered to collect data on the amount and frequency of consumption of raw milk so as to determine the exposure status. Trained data collectors who can understand both English and the local languages of the study area (mainly Oromifa) interviewed volunteer respondents. A total of four hundred and twenty milk consumers were interviewed on the spot of consumption from randomly selected 50 milk retail outlets (restaurants, cafeterias, hotels) along the chain of dairy clients.
Microbial isolation and quantification
The bacterial load estimation per 1 ml of milk was done by mixing 25 ml of milk samples into 225 ml of sterilized buffered peptone water and were thoroughly shaken to make one-in-ten initial dilution of the sample; the stock solution. Ten-folds of serial dilutions were made from the homogenates up to 10–6 with three replicates each except for Salmonella. Appropriate spread plates were made with 0.1 ml aliquots from all serial dilution tubes and incubated at appropriate temperature for the pathogen of an interest. Bacterial colonies were counted using colony counter to determine colony forming units (cfu)/ml. Dilutions with the total number of colonies on a plate were used for cfu computation according to the following formula.
Salmonella spp. were detected from milk samples by the conventional microbiological analysis according to International Standards Organization ISO-6579–2002 methods using buffered peptone water as pre-enrichment medium. Selenite Fraser Broth (SFB) was used as both selective enrichment and enumeration medium of salmonella with Most Probable Number (MPN) tubes. The probable numbers of colony forming units of salmonella was determined by turbidity of the MPN tubes. For isolation of Salmonella, Salmonella-Shigella agar (SSA) was used as selective plating. One milliliter of a stock solution was transferred into SFB and incubated for 24 h followed by which a loop full of SFB was streaked on SSA and incubated for 24 h at 37 °C. Finally, black colonies on SSA were identified as presumptive colonies of Salmonella. Finally, Indole Methyl-red Voges-Proskauer Citrate IMVC test was applied for biochemical confirmation of salmonella. Staphylococcus spp. were isolated and enumerated by spread plate on Mannitol-salt agar (MSA) using American Public Health Association-2001 method. After 24 h incubation of spread plates at 37 °C, golden yellow colonies were considered as presumptive S. aureus. Grams staining and coagulase tests using plasma [28, 31] were used as confirmation of the presumptive colonies. Gram positive and coagulase-positive colonies were considered as S. aureus. Buffered peptone water was used as primary enrichment followed by Buffered Listeria Enrichment Broth (BLEB) as secondary enrichment to isolate Listeria spp. The preparations were incubated at 30 0C for 48 h. Listeria selective agar base (LSA) enriched with sheep blood and Listeria selective supplements were used as a selective differential medium for L. monocytogenes. The spread plate cultures were incubated for 37 0C for 48 h and β-hemolytic colonies on LSA were identified as L. monocytogenes. Gram staining, microscopy and catalase tests were performed for morphological characterization and confirmation. Isolation and enumeration of presumptive E. coli in meat sample was done by plating dilutions on MacConkey Sorbitol Agar (MacSA, Oxoid, UK). The characteristic colonies that are identified as presumptive E. coli O157:H7 were sub cultured on Eosin Methylene Blue Agar (EMBA, Oxoid, UK) and aerobically incubated at 37 °C for 24 h after incubation which the colonies with distinct metallic sheen were counted. IMViC test was also applied for biochemical confirmation. Campylobacter Selective Agar base (CSA) enriched with sheep blood and Campylobacter selective supplement was used as a selective differential medium for isolation of Campylobacter spp. Because of cellular sensitivity outside the gastrointestinal environment resulting in possible sub-lethal effects on the cells, a loop full milk sample were streaked directly from the stock solution onto enriched CSA. The cultures were incubated in moist anaerobic jar at 400C. Grayish flat colonies growing on the medium were considered as presumptive C. jejuni. Further morphological (microscopic) examination and biochemical tests (Gram’s, catalase, H2S and oxidase tests) were done to confirm C. jejuni.
Microbial hazards and exposure assessment
The exponential model \(\mathrm{P}\left(\mathrm{d}\right)=1-{\mathrm{e}}^{-\mathrm{rd}}\) [33] was used to estimate the probability of infection through consumption of contaminated milk,where, “P(d)” is probability of infection at dose (d) per serving, “d” is the mean dose (cfu) of pathogens consumed per person per a day (microbial infection, “r” is dimensionless infectivity constant. The model parameter “r” was 2.18 × 10–4 for E. coli, 3.97 × 10–6 for Salmonella, 7.64 × 10–8 for Staphylococcus, 3.29 × 10–7 for Listeria, and 2.44 × 10–8 for Campylobacter [9, 10, 12, 14, 32]. The duration, number of times of raw milk consumption practice (frequency) and the amount of milk consumed in a given period (quantity) were identified and the information was combined to estimate the level of exposure of study subjects to the microbiological hazards. The annual probability of infection was calculated from the probability of infection and the number of days of exposure within the year according to the relation: P(ann) = 1-(1-P(d))n [33],where “n” is the number of days of exposure within the year.
Quality assurance
Sample collection and laboratory analyses were carried out under close supervision. All media and reagents used were up to date, and all microbiological analysis was carried out inside level II biosafety cabinet (BDK, Genkingen, Germany). All culture media and materials were sterilized using autoclave (Astell, England). The adequacy of sterilization was also assured using sterilization indicator.
Data management and statistical analysis
Laboratory results and questionnaire survey data were stored in Microsoft Excel spreadsheet. The statistical data analysis was done by using Statistical Package for Social Science (SPSS) version 23. Descriptive measures including mean, percentage and frequency were used to quantify the level of contamination of milk by the photogenic bacteria. The microbial load measurement was normalized to cfu/ml converting into log 10 values. Statistical differences among the mean concentrations of pathogens along the dairy value chain were assessed using p-values by which values less than 0.05 were considered as significant.
Results
Hazard identification and milk microbial quality
Frequency of milk contamination along the dairy value chain
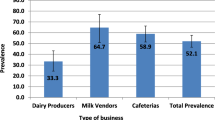
Among the total of 150 raw milk samples collected, 92 (≈61.3%) were found contaminated along the dairy value chain. One or more types of pathogens were found to be in a milk sample analyzed in the laboratory. A total of 42 (28%) samples contained at least one type of pathogens, whereas 2(1.3%) samples contained four different types of pathogens of interest in this study (Fig. 2).
Incidence of specific pathogens in milk samples along the value chain
Out of the total of 150 raw milk samples analyzed in the laboratory, 54 (36%) were positive for E. coli; 30 (20%), 58 (38.7%), 8 (5.3%), and 2 (1.3%) were positive for Salmonella, S. aureus, Listeria, and Campylobacter isolates respectively. In all cases, the microbial contamination incidence (percentage) of the samples appeared to increase from dairy farms to the retail outlets except for Campylobacter isolates (Table 1).
The logarithmic means of pathogenic bacteria counts in all milk samples ranged from 1.48 log10cfu/g (30 cfu/ml) to 4.88 log10cfu/ml (7.50 × 105 cfu/ml). The highest bacterial count was recorded from E. coli contamination ranging from 2.64 log10cfu/ml (4.36 × 103 cfu/ml)—4.88 log10cfu/ml (7.50 × 105 cfu/ml) whereas the lowest from Campylobacter which was 1.48 log10cfu/ml (30 cfu/ml) found only at distributer points. The counts of Salmonlla, S. aureus, and L. monocytogenes ranged from 2.53 log10cfu/ml (3.40 × 103 cfu/ml)—4.04 log10cfu/ml (1.10 × 105 cfu/ml), 2.89 log10cfu/ml (7.68 × 103 cfu/ml)—4.65 log10cfu/ml (4.43 × 105 cfu/ml), and 2.32 log10cfu/ml (2.10 × 103 cfu/ml)—3.45 log10cfu/ml (2.80 × 104 cfu/ml) respectively. Samples from retail outlets showed the highest bacterial counts, where as those from farms where the least; except for Campylobacter counts, in which the case appeared only at distributer points (Table 2).
Microbial quality of milk samples along the dairy value chain
The pathogens were detected in the range which unfit for human consumption (unsatisfactory level) at each point along the dairy value chain except for C. jejuni; which were isolated only from distributors (Table 3). However, the incidence of pathogens was varied that 48% of samples were found contaminated with S. aureus in the range of unsatisfactory level among consumable samples at retailers. But none of the samples was recorded from C. jejuni contamination. E. coli, Salmonella and L. monocytogenes were 44%, 24%, and 8% respectively.
Dose–response and exposure assessment
Raw milk consumption and exposure practice
A socio-demographic report of 420 raw milk consumers revealed that 254 were male and 166 were female. In terms of age, 6 were less than or equal to 18, 353 were greater than up to less than or equal to thirty, and 61 were greater than 30. Educational status showed that 13 were illiterate, 29 were primary, 71 were secondary, and 307 were tertiary. Religion-wise, 235 were Christian, 176 were Muslim, and the rest 9 were others. Over two-third (75.7%) of respondents had the custom of drinking raw milk. Majority (45.7%) of them experienced long period raw milk consumption, at least once in a week, and chiefly 300 ml per serving (69.5%). It was reported that most respondents prefer raw milk, mainly in the form of homemade yoghurt, for its taste (69.5%), minority (2.9%) for it is tradition of their community, and some others for both reasons (3.3%). Contrastingly, respondents also reported their experience of disease symptoms due to consumption of raw milk (Table 4).
Dose response to pathogen exposure
Estimates of most of the pathogens in the serving units of raw milk were too large that the highest (7.50 × 105 cfu) recorded from E. coli and the least (2.80 × 104 cfu) from L monocytogenes. Probability of infection at the consumption dose shows very high that the annual probability of infection is perfectly-1 in the cases of four pathogens. But, none of samples from retail outlets contained C. jejuni (Table 5).
Discussion
Results of this study revealed the contamination of raw milk by either one or a combination of pathogenic bacteria. The number of contaminated raw milk samples was found to be significantly high (61.3%). This figure is virtually comparable to the 62.5% contamination in the distribution center milk containers in Mekelle, Ethiopia [11] and 65.5% of contamination reported from parallel study conducted in selected street foods in Gondar Town [1]. The chance of contact of raw milk to different types of bacterial pathogens increased from farms to the retail outlets along the dairy value chain. This can be associated to the advantage that microorganisms take, having different contact surfaces and time through the milk production chain, from farm-to-table.
Except for the isolates of C. jejuni, which appeared only in the samples from distributers, all other pathogenic bacteria were identified in the milk samples from every source; dairy farms, distributers and retail outlets. The load of E. coli was ranged from 2.64–4.88 log10 cfu/ml, where the mean concentration was 3.76 ± 1.12 log10 cfu/ml (mean ± SD). This is the highest incidence recorded in our study as shown in Table 2. The result from E. coli contamination in this study is comparable to 2.00–6.07 log10 cfug−1 reported from the rural household served foods in Malawi by Taulo et al., [35]. Sabuj et al., [32] also reported 4.10–4.58 log10 cfug−1 E. coli from street-vended ready-to-eat foods in Bangladesh Agricultural University. Next to the eminent raw milk contaminant pathogens considered in this study, E. coli, were S. aureus, S. enterica Typhimurium, L. monocytogenes, C. jejuni that appeared to be with mean concentrations of 3.74 ± 0.88 log10 cfu/ml, 3.42 ± 0.79 log10 cfu/ml, 2.92 ± 0.56 log10 cfu/ml, and 1.48 log10 cfu/ml respectively. The total absence of C. jejuni from the retail outlets in our study directly coincides with its zero occurrences reported by Taulo et al. [35] in ready-to-eat foods,which can be ascribed to the fastidious nature of the organism.
S. aureus was the second prevalent pathogen determined in our study followed by E. coli. The mean concentration of S. aureus surpassed 105 cfu/ml in the samples from retailers, which could have posed potential risk among raw milk consumers. Studies conducted by Heidinger et al., [17] and Giacometti et al., [15] in raw milk also revealed parallel prevalence of S. aureus. S. enterica Typhimurium and L. monocytogenes had relatively lower mean concentration, each having 3.42 ± 0.79 log10 cfu/ml and 2.92 ± 0.56 log10 cfu/ml respectively. Studies also revealed corresponding results for high Salmonella contamination [16, 29]. However, the figures of prevalence of L. monocytogenes in this study are considerably large in relation to its very low (0.08%) to zero occurrences [21, 24].
Generally, the prevalence and percentages of most isolates of pathogens considered in our study increased as milk transported from farms to the retail outlets (Table 1). This could be connected with the contamination of bacteria through milking, collection, and transportation processes and the proliferation of bacteria using the opportunity of time from production and processing to the table for consumption. Except for C. jejuni, the mean concentrations of all pathogens demonstrated significant statistical difference (p < 0.05) using 95% confidence interval along the dairy value chain.
Except for the C. jejuni, all pathogens considered in this study were detected in the unsatisfactory level along the dairy value chain, particularly at retail outlets, where raw milk is ready-to-eat. It is alike the result in the study conducted by Manguiat and Fang [24] on street vended foods where most of the pathogens found in unsatisfactory levels. The laboratory analysis of samples (48%) showed that S. aureus were present in the ranges of 105 cfu/ml,which is certainly large number compared to the unsatisfactory level (> 104 cfu/ml) rated in guide lines [6]. Odu and Assor [30] also obtained corresponding results (42.1%) from S. aureus contamination. E. coli were detected in 44% of milk samples from retailers, against the limits of guidelines, in which their presence in > 102 cfu/ml considered as potentially hazardous. S. enterica Typhimurium (24%) and L. monocytogenes (8%) were present in the ranges of 105 cfu/ml and 104 cfu/ml respectively. According to the standard limits of guidelines [6], however, the ranges determined for both pathogens is classified as unsatisfactory.
In this present study, the survey of raw milk consumption data on the amount, frequency, and duration was translated for the estimates of the number of pathogens present in the milk. These variables were significantly associates with the annual probability of infection (Table 5) among the study population. An exponential dose–response model was used, assuming quantitative estimates of pathogens at the contaminated milk portion size (serving units) ingested by the presumptive population at risk [20, 32, 33]. The mean amount of raw milk consumed (0.31 ± 0.05 L) had evidently direct relation to the large number of pathogens in the serving units in addition to the chance of proliferation that the pathogens could have along the dairy chain to the retailer outlets. In this study, therefore, the probable dose of pathogens assumed was the product of cfu/ml in raw milk and the mean amount of milk consumed at once. The results of the current study, accordingly, demonstrated ingestion of very high number of pathogens with serving units of raw milk,2.37 × 108 E. coli, 1.41 × 108 S. aureus, 3.49 × 107 S. enterica T., and 8.89 × 106 L. monocytogenes, (Table 5). Similarly, [32], Manguiat and Fang 2013, [18, 29] reported high ranges of pathogen contamination in their findings. In general, our study reflected that the microbiological quality of raw milk in the study area was considerably unsatisfactory with respect to pathogens treated. Mamun et al., [23] also reported high levels of potential hazards to the public health threats especially due to high number of coliforms,like S. aureus, E. coli, and S. enterica T in foods vended by school-based streets.
Except for C. jejuni, the annual probability of infection by all other pathogens is perfectly one; i.e. 100% (Table 5). This finding coincides with the 100% chance of being infected by potential pathogens as reported in previous studies [39] and [32]. Raw milk consumers had experience and/or information of disease symptoms like nausea (26.7%), abdominal discomfort (24.5%), vomiting (20.5%) and diarrhea (4.0%). This could be associated to either one or combination of intoxications of E. coli and S. aureus, salmonellosis, and listeriosis. The probability of infection at consumption dose was also high regardless of the category of consumers varied in experience of duration, amount and reason for raw milk consumption. Generally, the estimated risk at consumption dose of acquiring intoxications of E. coli across retailer outlets is 100%, whereas salmonellosis, S. aureus intoxication and listeriosis is 84%, 65% and 63% respectively.
Conclusion
Our study specifically found that raw milk in the dairy value chain in Jimma, Southwest Ethiopia, is contaminated with almost all types of bacterial hazards. The prevalence of frequent bacterial contaminants and the deterioration of raw milk across retail outlets were also demonstrated. Over half of the total samples had high level of pathogenic bacteria, most of which were found to be in the range of unsatisfactory limits that could pose health risks to consumers. Therefore, regular monitoring and implementing hazard identification and critical control point principles along the dairy value chain are necessary to ensure the safety of raw milk. These findings highlight the need for immediate action to prevent the spread of foodborne diseases caused by bacterial hazards in food.
Availability of data and materials
All data and materials are available for this work and can be accessed from the corresponding author.
References
Adimasu A, Mekonnen B, Guadu T, Giaw Z, Adane T. Bacteriological quality assessment of selected street foods and their public health importance in Gondar Town. North West Ethiopia Glob Vet. 2016;17(3):255–64.
Ayalew H, Birhanu A, Asrade B. Review on food safety system: Ethiopian perspective. African J Food Sci. 2013;7:431–40.
Brandsma, W., Mengistu, D., Kassa, B., Yohannes, M., Van der Lee, J. The major Ethiopian milk sheds: an assessment of development potential. In: Wageningen, Wageningen UR (University & Research center) Livestock Research Report. 2013: 735, 247 blz.
Buyser ML, Dufour B, Maire M, Lafarge V. Implication of milk and milk products in food-borne diseases in France and in different industrialized countries. Int J of Food Microbiol. 2001;67:1–17.
CACC (Central Agricultural Census Commission). Paperback. 2 vols (labeled Part I and Part II). folding forms, viii, 200, iv, 502p. Addis Ababa, Ethiopia: Central Agricultural Census; 2002.
CFS (Center for Food Safety),. Microbiological guideline for food: for ready-to-eat foods in general and specific food items. Hong Kong: Center for Food safety; 2014.
CSA (Central Statistical Agency). Central Statistical Agency of the Federal Democratic Republic of Ethiopia. Agricultural Sample Survey of 2011/12 (2004 E.C). Volume II. Report on Livestock and Livestock Characteristics (Private Peasant Holdings). Addis Ababa: Central Statistical Agency; 2012.
Daniel WW. Biostatistics: a foundation for analysis in the health sciences. 7th ed. New York: John Wiley and Sons; 1999. pp.606-11.
EFSA (European Food Safety Authority). Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. European Food Safety Authority. Journal published by John Wiley and Sons Ltd on behalf of European Food Safety Authority. 2018.
EPA (Environmental Protection Agency. Microbial Risk Assessment Guideline: Pathogenic Organisms with Focus on Food and Water. FSIS Publication No. USDA/FSIS/2012–001; EPA Publication No. EPA/100/J12/001. 2012.
Faisal SD, Ahmed AF. Bacteriological Quality Assessment of Milk in College of Veterinary Medicine (Cvm) Dairy Farm and Kalamino Dairy Farm in Mekelle, Tigray, Ethiopia. Dairy Vet Sci J. 2018;8(2):555734. https://doi.org/10.19080/JDVS.2018.08.555734.
FAO/WHO (Food and Agriculture Organization, and World Health Organization). Pilot of new approaches to estimate dietary exposure to veterinary drug residues. Seventy-eighth meeting of the Joint FAO/WHO Expert Committee on food additives. 2014.
FAO/WHO (Food and Agriculture Organization, and World Health Organization). Risk Assessment of Microbiological Hazards in Food: report of a Joint FAO/WHO Expert Consultation, Geneva, Switzerland, 15-19 March 1999. World Health Organization; 1999. https://apps.who.int/iris/handle/10665/65973.
FSANZ (Food Standards Australia New Zealand). Guidelines for the Microbiological Examination of Ready-to-eat Foods. 2015. Available at: http://www.foodstandards.gov.au/education/publications/guidelinesformicrobi1306.cfm (Accessed 19 Feb 2021).
Giacometti F, Serraino A, Bonilauri P, Ostanello F, Daminelli P, Finazzi G, Losio MN, Marchetti G, Liuzzo G, Zanoni RG, Rosmini R. Quantitative microbial risk assessment of verocytotoxin-producing Escherichia coli O157 and Campylobacter jejuni related to consumption of raw milk in a province in northern Italy. J of Food Protection. 2012;75:2031–8.
Gurtler BJ, Hinton JA, Bailey BR, Cray JC, Meinersmann JR, Ball AT, Jin ZT. Salmonella isolated from ready-to-eat pasteurized liquid egg products: Thermal resistance, biochemical profile, and fatty acid analysis. Int J of Food Microbiology. 2015;206:109–17.
Heidinger JC, Winter CK, Kullor JS. Quantitative microbial risk assessment for Staphylococcus aureus and Staphylococcus enterotoxin A in raw milk. J of Food Protection. 2009;72:1641–53.
Hughes AF, Gyamfi AA, Appiah. Microbiological and parasitological quality of local beef retailed in Accra and radiation sensitivity of Salmonella spp. Int J Curr Microbiol App Sci. 2015;4(4):86–96.
Hussen M, Kechero Y, Molla M. Productive and reproductive performances of ruminant livestock in Jimma Zone Southwest Ethiopia. J Reprod Infertil. 2015;6(2):27–34.
Lammerding AM, Fazil A. Hazard identification and exposure assessment for microbial food safety risk assessment. Int J of Food Microbiol. 2000;58:147–57.
Latha C, Anu CJ, Ajaykumar VJ, Sunil B. Prevalence of Listeria monocytogenes, Yersinia enterocolitica, Staphylococcus aureus, and Salmonella enterica Typhimurium in meat and meat products using multiplex polymerase chain reaction. Vet World. 2017;10(8):927–31.
Leykun B, Beje G, Tesfaye K, Lelisa SD, Dechassa T, Masrie G, Habib B, Assegid G, Sultan S, Seid TM. Microbial quality of raw cow milk and its predictors along the dairy value chain in Southwest Ethiopia. Int J of Food Microbiology. 2021;350:1–7.
Mamun AM, Rahman MS, Turin CT. Microbbiological uality of selected street food items vended by school-based street food vendors in Dhaka Bangladesh. Int J of Food Microbiol. 2013;166:413–6.
Manguiat SL, Fang JT. Microbiological quality of chicken-and pork-based street-vended foods from Taichung Taiwan and Laguna, Philippines. Food Microbiol. 2013;36:57–62.
Misganaw G, Hailemariam F, Mamo D, Tajebe S, Nigussie Y. Production potential, challenges, and prospects of dairy cooperatives in Aksum and Adwa towns. Ethiopian J Dairy Vet Anim Res. 2017;5(6):221–6.
Mohamed FS, Farah AA. Bacteriological quality assessment of milk in College of Veterinary Medicine (Cvm) dairy farm and Kalamino dairy farm in Mekelle, Tigray, Ethiopia. J of Dairy and Vet Sci. 2018;8(2):1–8.
MPI (Ministry for Primary Industries). Technical paper No: 2014/12 on Assessment of the microbiological risks associated with the consumption of raw milk. 2013. Available at: http://www.foodstandards.gov.au/education/publications/guidelinesformicrobi1306.cfm (Accessed 17 Mar 2022).
Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH. Manual of Clinical Microbiology. 8th ed. Washington DC: American Society of Microbiology; 2003.
Ngo TH, Thanh NL, Duc PP, Xuan DS, Hang L-T, Robichaud DJ, Viet NH, Le HT, Grace D, Unger F. Microbial contamination and associated risk factors in retailed pork from key value chains in Northern Vietnam. Int J of Food Microbiol. 2021;346:1–8.
Odu NN, Assor P. Microbiological analysis of ready to eat food (cooked rice and beans) sold among different restaurants in University of Port Harcourt, Port Harcourt Nigeria. Academia Arena. 2013;5(1):62–6.
Pumipuntu N, Kulpeanprasit S, Santajit S, Tunyong W, Kong-ngoen T, Hinthong W, Indrawattana N. Screening methods for Staphylococcus aureus identification in subclinical bovine mastitis from dairy farms. Vet World. 2017;10(7):721–6.
Sabuj AAM, Haque ZF, Younus MI, Pondit A, Barua N, Hossain MG, Islam MA, Saha S. Microbial risk assessment of ready-to-eat fast foods from different street-vended restaurants. Int J One Health. 2020;6(1):41–8.
Soller JA. Use of microbial risk assessment to inform the national estimate of acute gastrointestinal illness attributable to microbes in drinking water. J Wat Health. 2006;4(2):165–86.
Steven T, Anne W, Roger A, Grant K, Rajab M, Anthony G. Microbiological hazard identification and exposure assessment of food prepared and served in rural households of Lungwena Malawi. Int J of Food Microbiol. 2008;125:111–6.
Taulo S, Wetlesen A, Abrahamsen R, Kululanga G, Mkakosya R, Grimason A. Microbiological hazard identification and exposure assessment of food prepared and served in rural households of Lungwena Malawi. Int J of Food Microbiol. 2008;125:111–6.
Tegegne B, Tesfaye S. Bacteriological milk quality: possible hygienic factors and the role of Staphylococcus aureus in raw bovine milk in and around Gondar Ethiopia. Int J Food Contam. 2017;4(1):1–9.
Weldeabezgi LT, Atsbha TW, Kassegn HH, Gebremichael TF, Berhe MH. A quantitative risk assessment model for Staphylococcus aureus and Salmonella associated with consumption of informally marketed milk products in Tigray Ethiopia. J Food Saf. 2019;2019(e12749):1–11.
Yohannes G. Antimicrobial susceptibility testing of Escherichia coli isolated from selected dairy farms in and around Mekelle, Ethiopia. J Bacteriol Mycol Open Access. 2018;6(1):47–51. https://doi.org/10.15406/jbmoa.2018.06.00176.
Younus MI, Sabuj AAM, Hague ZZF, Sayem SM, Majumder S, Parvin MS, Islam MA, Saha S. Microbial risk assessment of ready-to-eat mixed vegetable salads from different restaurants of Bangladesh Agricultural University campus. J Adv Vet Anim Res. 2020;7(1):34–41.
Acknowledgements
Authors would lie to acknowledge Jimma University for funding this research work. We are grateful to the Medical Microbiology and Environmental Health Science and Technology Laboratories of Jimma University for their provision of facilities and technical assistance.
Funding
This work was supported by Jimma University.
Author information
Authors and Affiliations
Contributions
Beje Gume wrote the original manuscript text draft including figures and tables. Beje Gume and Seid Tiku Mereta involved in the conceptualization, investigation methodology of the manuscript. Project and resource administration, supervision and validation were done by Seid Tiku Mereta. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval of research protocol letter was obtained from ethical review board of Jimma University College of Health Sciences. All methods were carried out according to relevant guidelines and regulations. Written informed consent was also obtained from zonal and district administrations; dairy farms, milk collection and distributor shops and retailer outlets to inform all participants in the research. Informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest towards the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gume, B., Berhanu, L., Kassa, T. et al. Bacterial hazard identification and exposure assessment of raw milk consumption in Jimma zone, South West Ethiopia. BMC Microbiol 23, 166 (2023). https://doi.org/10.1186/s12866-023-02910-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-023-02910-0