Abstract
Polyploidy and the microbiome are crucial factors in how a host organism responds to disease. However, little is known about how triploidization and microbiome affect the immune response and disease resistance in the fish host. Therefore, this study aims to identify the relationship between intestinal microbiota composition, transcriptome changes, and disease resistance in triploid Carassius auratus (3nCC). In China’s central Dongting lake water system, diploid (2nCC) and triploid Carassius auratus were collected, then 16S rRNA and mRNA sequencing were used to examine the microbes and gene expression in the intestines. 16S rRNA sequencing demonstrated that triploidization altered intestinal richness, as well as the diversity of commensal bacteria in 3nCC. In addition, the abundance of the genus Vibrio in 3nCC was increased compared to 2nCC (P < 0.05). Furthermore, differential expression analysis of 3nCC revealed profound up-regulation of 293 transcripts, while 324 were down-regulated. Several differentially expressed transcripts were related to the immune response pathway in 3nCC, including NLRP3, LY9, PNMA1, MR1, PELI1, NOTCH2, NFIL3, and NLRC4. Taken together, triploidization can alter bacteria composition and abundance, which can in turn result in changes in expression of genes. This study offers an opportunity for deciphering the molecular mechanism underlying disease resistance after triploidization.
Similar content being viewed by others
Background
Polyploidy has an important role in the disease resistance of fish [1]. For instance, in Atlantic salmon (Salmo salar L.), triploids may be at a disadvantage compared with their diploid siblings in their defense against bacterial infections [2]. Similarly, in response to a challenge with Vibrio anguillarum, the mortality of triploid Chinook salmon (Oncorhynchus tshawytscha) increased [3]. Dégremont et al. indicated that triploid oysters have higher disease resistance than diploids, as observed in Crassostrea virginica and Saccostrea glomerata [4]. In fertile triploid fish, normal gonadal development causes the energy needed for somatic growth to be channeled into gamete production. Consequently, this results in adverse effects, including poor physical growth and flesh quality, and can increase mortality and morbidity rates [5, 6]. Nevertheless, this phenomenon did not occur in the Carassius auratus complex in the water system of Dongting Lake, which is manifested as triploid Carassius auratus (3nCC) populations exhibited less sensitivity to environmental change than diploid Carassius auratus (2nCC) populations [7]. Consequently, it is crucial to understand how triploidy influences disease resistance in the Carassius auratus complex.
Intestinal microbial communities of animals are extremely diverse and active [8]. Microorganisms inhabiting the gastrointestinal tract, such as bacteria, archaea, viruses, fungi, and microeukaryotes, make up the intestinal microbiota [9, 10]. The intestinal microbiome is a source of key enzymes essential to food digestion and nutrient absorption for the host [11]. Technology development for next-generation sequencing (NGS) has enhanced the understanding of the diverse and complex communities of microbial symbionts residing within hosts. The diverse and dynamic microbial community of the gastrointestinal tract plays a critical role in modulating the host’s health and nutrition [12], immunity and defense against pathogens [13], growth and development [14], and behavior [15].
The intestinal microfloras are also closely associated with disease resistance in host animals and play a key role in the host’s immune system’s induction, education, and functionality [16, 17]. It has been demonstrated that Se-rich B. subtilis improves intestinal microbial changes induced by Hg, reduces Aeromonas abundance and inflammation in common carp [18]. Liu et al. found that Pb accumulated in the gut causes dysbiosis of the microbiota, affects intestinal immunity and digestion, and damages the silver carp’s intestinal barrier [19]. Shi et al. suggested antibiotics administered to grass carp would exacerbate oxidative stress, lead dysbiosis of intestinal bacteria, inhibit the immune system of the mucosa, and activate apoptosis [20]. According to Qiao et al., poly-hydroxybutyrate may have beneficial effects on immunity and disease resistance through its interaction with the gut microbiota [21]. In tilapia, NE can affect immunity indirectly by means of microbial changes as well as directly by stimulating host tissue [22]. There are potential dietary implications to altering the intestinal microbiota because the microbiota aid digestion and conversion of complex plant molecules into short-chain fatty acids are necessary for daily energy metabolism [23]. To develop and function properly in zebrafish, the immune system depends on bacteria and their products [24]. However, the role of the intestinal microbiome in the disease resistance of the triploid Carassius auratus has not been fully elucidated.
Considering the information available from previous research, we hypothesized that triploidization might change the intestine bacterial community, boost immunity, and increase disease resistance in the triploid Carassius auratus. To test this hypothesis, the composition of intestinal microbiota between diploid and triploid Carassius auratus was conducted. Additionally, mRNA-seq revealed many transcripts that differed between diploid and triploid Carassius auratus.
Results
Differences of richness and diversity of the microbiota between diploid and triploid of Carassius auratus
There were 453,811 good-quality 16S rRNA gene sequences obtained. A notable number of 400,890 sequences (88.34%) was associated with 1556 OTUs. OTUs were categorized into 34 phyla, 79 classes, 182 orders, 312 families, 594 genera, and 646 species based on the number of effective OTUs (Table 1).
For each group, we computed the Chao1 index to assess the diversity of the microbiota between diploid and triploid of Carassius auratus (Fig. 1a). The results imply that intestinal microbiota from the 3nCC group had significantly lower species diversity levels than those from the 2nCC group (P < 0.05).
PCoA distinguished the 3nCC group from the 2nCC group based on structural differences in gut microbiota from fecal samples (Fig. 1b). Triploidization changes the overall structure of the microbiota of diploid and triploid crucian carp.
Flora differences between diploid and triploid Carassius auratus
According to the LEfSe taxon, the relative abundance of bacterial phylum, class, order, family, genus, and species differed significantly between diploid and triploid crucian carp (Fig. 2). In the 3nCC groups, the proportion of Vibrionales was greater than in the 2nCC groups.
Additionally, we investigated how triploidization impacted the distribution of particular flora on 2nCC and 3nCC at the order level (Table 2), family level (Table 3), genus level (Table 4), and species level (Table 5). On an order level (Table 2), Vibrionales greatly increased in abundance in the triploid population (P < 0.05) compared to 2nCC. Rhodobacterales were observed in 3nCC at a marginally lower abundance (P < 0.1) than in 2nCC. On a family level (Table 3), a significant improvement in Vibrionaceae relative abundance was associated with triploidization (P < 0.05). In 3nCC, Rhodobacteraceae and Peptostreptococcaceae (P < 0.1) had a marginally lower abundance than in 2nCC. On a genus level (Table 4), Vibrio became more prevalent in 3nCC (P < 0.05) than in 2nCC. We observed a marginally lower abundance in uncultured_bacterium_f_Rhodobacteraceae and a higher abundance in Romboutsia (P < 0.1) were observed in 3nCC vs. 2nCC. On a species level (Table 5), the relative abundance of uncultured_bacterium_g_Vibrio was significantly increased by triploidization (P < 0.05) in comparison with 2nCC. Uncultured_bacteria_ f_Rhodobacteraceae exhibited a marginally lower abundance in 3nCC vs 2nCC, while uncultured_bacteria_g_Romboutsia exhibited a markedly higher abundance (P < 0.1).
Analysis of transcriptome sequences and sequence alignment
As measured by the OD ratio A260/A280 and RNA integrity numbers (RINs) of RNA of six samples, the RNA integrity numbers were 2.2 and 8.1–8.7, respectively (Additional file 1). Despite their high quality and lack of contamination, all samples underwent transcriptome sequencing. Illumina performed RNA sequencing on intestinal samples from 2nCC and 3nCC. Tables 6 and 7 display the RNA-Seq results. In six RNA-Seq libraries, the clean read count ranged between 40,639,622 and 64,300,602. The aligned clean reads were then aligned with the RCC reference genome (https://bigd.big.ac.cn/search?dbId=gwh&q=GWHAAIA00000000) using HISAT2. The mapped genome reads ranged from 33,237,039 to 57,311,126 sets, and genome map rates ranged from 80.77 to 89.22%.
Identification of differentially expressed transcripts (DETs) between diploid and triploid Carassius auratus
There were 293 up-regulated transcripts and 324 down-regulated transcripts observed in 3nCC and 2nCC, respectively (Fig. 3). DETs between 3nCC and 2nCC comprised osteoclast stimulatory transmembrane protein (OCSTAMP), leucine rich repeat LGI family member 2 (LGI2), upregulator of cell proliferation (URGCP), GIMAP family P-loop NTPase domain containing 1 (GIMD1), ankyrin and armadillo repeat containing (ANKAR), leucine rich repeat neuronal 2 (LRN2), G protein-coupled receptor 98 (GPR98), potassium channel tetramerization domain containing 7 (KCTD7), predicted gene 12,253 (GM12253), protein kinase superfamily protein (SNRK2.4), F-box and leucine rich repeat protein 13 (FBXL13), phosphorylase kinase regulatory subunit alpha 1 (PHKA1), leucine rich repeat containing 31 (LRRC31), etc.
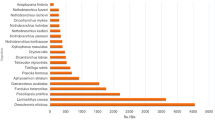
GO enrichment analysis uncovered 76, 8 and 16 terms, respectively, from the biological process, cellular component, and molecular function categories for the 3nCC group compared to 2nCC (Additional file 2). As illustrated in Fig. 4, the top 20 GO-terms were the most enriched, such as proteolysis, immune response, positive regulation of cell death, response to external biotic stimulus, response to other organism, response to biotic stimulus, response to a bacterium, positive regulation of apoptotic process, positive regulation of programmed cell death and Golgi subcompartment. Moreover, several pathways implicated in disease were identified, including inflammatory response, response to xenobiotic stimulus, activation of an innate immune response, inflammatory response to antigenic stimulus, and the detection of bacterium and cytokine production. DETs were enriched for 38 signaling pathways according to KEGG analysis (Additional file 3). As illustrated in Fig. 5, the top 20 KEGG pathways were enriched. Five of the most enriched pathways were ion channels, cellular senescence, calcium signaling pathway, human cytomegalovirus infection, and proteoglycans in cancer.
Scatter plot of enriched KEGG pathways for DETs between 3nCC and 2nCC. Pvalue is presented on the x-axis, and the top 20 pathways are shown on the y-axis. Pvalues are corrected to produce Qvalues. Color scale is used to represent Qvalues. Numbers of transcripts associated with each circle are indicated by their diameters
In conclusion, the enrichment results demonstrate that the most-enriched pathway associated with immune was immune response. The up-regulated genes included NLRP3, LY9, PNMA1, MR1, PELI1, and NOTCH2, and the down-regulated genes included NFIL3 and NLRC4.
Validation of differentially expressed transcripts (DETs) in diploid and triploid Carassius auratus by RT-qPCR
RT-qPCR was performed to validate eight DETs based on RNA-Seq data. Six DETs that were up-regulated in the 3nCC group compared with the 2nCC group, while two DETs were down-regulated in the 3nCC group. Both RT-qPCR and RNA-Seq revealed similar expression profiles for the eight DETs (Figs. 6 and 7), indicating the reliability of the RNA-Seq results.
Discussion
A prior study has indicated that environmental changes and variations can impact the population of diploid and triploid Carassius auratus [7]. In the Dongting lake water system, triploid Carassius auratus has been found to have a more generalized distribution and can adapt to many environments compared to diploid Carassius auratus [25]. This is likely because genetic and epigenetic regulation occurred in 3nCC and its adaptability to diverse environments [26, 27]. It is well-documented that the makeup of the host-microbial community is influenced by many endogenous and exogenous factors [28,29,30,31] and plays a role in regulating immune function [32]. By improving immune responses, systemic infections can be prevented [33,34,35,36]. Consequently, revealing the molecular mechanism of disease resistance from the perspective of the composition and diversity of the bacterial community in 3nCC helps us to understand its adaptability to various environments. The impact of triploidization on the microbiome and the mechanisms involved have not yet been thoroughly examined.
Triploidization raises questions about whether the immune response is influenced by the microbiota or directly by the host immune system, so we identified microbiota and gene expression in the intestines between diploid and triploid Carassius auratus. A shift or change in the structure and diversity of intestine microbiota was evident in this study. Moreover, 3nCC was significantly more abundant than 2nCC in the Vibrio genus. Numerous studies have revealed that Vibrio is associated with immune responses. For instance, grouper control of immunity and protection from Vibrio anguillarum infections can be achieved using a probiotic lactic acid bacterium Pediococcus pentosaceus strain 4012 [37]. An entire cDNA for a clip domain serine proteinase gene could bond to Aeromonas hydrophila, Vibro anguillarum, and Vibro alginolyticus, which thus reduced the pathogen-induced mortality rate [38]. The consumption of 1.0 and 2.0% Siegesbeckia glabrescens extract enriched diet significantly improved immune activity, improved the disease resistance of Epinephelus bruneus to Vibro parahaemolyticus, and reduced its cumulative mortality [39]. Several prior works suggested that Vibro is a harmful bacteria population. However, one indicates that the bacteria of the Vibrionaceae family (Vibro) are the key component of bivalve microbiota, which can cope with infectious diseases [40]. Our results suggest that triploidization can potentially alter intestine microbiota. It can be speculated that the higher abundance of Vibro in 3nCC may be more resistant to infections through immune response.
In this study, NLRP3, LY9, PNMA1, MR1, PELI1, and NOTCH2 were the up-regulated genes, and NFIL3 and NLRC4 were the down-regulated genes as a result of triploidization. NLRP3 has been demonstrated to have an important role in the inflammatory response in numerous studies. Christ et al. depicted that NLRP3-deficient mice were not affected by western diet-induced systemic inflammation, myeloid progenitor proliferation, and re-programming, which might have contributed to mediating the negative effects of trained immunity in inflammatory diseases [41]. Deng et al. uncovered that NLRP3 inflammasome in Tetraodon nigroviridis may contribute to the antibacterial immune response and produce mature TnIL-1β after activation [42]. In zebrafish, NLRP3 inflammasome has functional roles in anti-bacterial immunity [43]. In the case of LY9, it is independent of genetic background that LY9-deficient mice spontaneously developed antinuclear antibodies (ANA), anti-dsDNA, and anti-nucleosome autoantibodies, which are typical markers of systemic autoimmunity [44].
PNMA1 falls under the family of proteins implicated in an autoimmune disorder called paraneoplastic neurological syndrome [45]. The high expression of PNMA1 in mice is possibly a risk factor for neurodegenerative disorders [46]. Mammals possess a non-classical class I molecule, MR1, which serves as a sensor of microbial metabolomes and should be able to detect intracellular infection early on [47]. Many studies suggest MR1 restricted T cells have an important role in immune contexts, ranging from cancer to autoimmunity and infection [48]. As a transcriptional regulator of immune cell differentiation, NFIL3 is well-known [49]. According to Geiger et al., NFIL3 plays an important cell-intrinsic role in developing gut-associated ILC3s. Additionally, NFIL3 deficiency significantly reduces the intestinal innate immune response against acute bacterial infections such as Citrobacter rodentium and Clostridium difficile [50]. In grass carp, NFIL3 participates in host immunity against pathogen infection and can activate various gene expressions [51]. NLRC4 belongs to the Nod-like receptor family (NLRs), a group of cytosolic receptors that sense bacterial molecules [52]. By inhibiting the NLR pathway, Wang et al. suggested that NLRC4 silencing alleviates lung injury and inflammation induced by septic shock [53]. Ubiquitin E3 ligase PELI1 facilitates innate immunity and is activated by receptor signals [54]. The experimental autoimmune encephalomyelitis with mice lacking PELI1 found that antigen presentation was enhanced, adaptive and innate immune cells were more active, and proteins involved in iron metabolism were altered [55]. NOTCH2 is a single-pass transmembrane receptor that responds to ligands from the DSL (Delta-like family) receptors [56]. This transcription factor is expressed by various tissues and cells within the hematolymphatic compartment. It has a crucial role in the differentiation and functionality of various immunity cells [57]. Maekawa et al. demonstrated the impaired differentiation of Notch2-deficient T cells into cytotoxic T lymphocytes [58]. Triploidization may influence immune function by modulating the expression levels of these genes. However, it remains unclear whether changes in immune-related genes are caused either by triploidization or by an increase in Vibrio. We speculate that these immune-related genes could potentially modulate the immune response and may have a role in immune tolerance to commensal microbes, enabling 3nCC to exhibit stronger resistance to disease than 2nCC.
Conclusions
Triploidization could modify the intestine microbiota and significantly increase the relative abundance of Vibro, which may, in turn, enhance disease resistance in 3nRR. We also found that eight immunity-related genes have important implications in regulating immune response and may function in immune tolerance to commensal microbes, enabling 3nCC to exhibit a stronger resistance to disease than 2nCC. The observations provide clues to decipher disease resistance in 3nCC in future work.
Methods
Sample preparation
In 2021, three specimens of both diploid (2nCC) and triploid (3nCC) Carassius auratus were sampled from the same area of Dongting Lake in Hunan Province. A flow cytometer (BD Biosciences, San Jose, California, USA) was employed to determine the ploidy type of each sample. Each fish was injected with heparinized syringes to obtain its red blood cells from its caudal vein. Staining solution (NIM-DAPI 731085) (NPE systems, Pembroke Pines, FL, USA) was added to the blood samples for 10 min. Against the DNA of red crucian carp, the total DNA content of each of the fish was compared.
After euthanizing the fish with MS-222 (100 mg/L, Western Chemical, Inc., Ferndale, Washington), the intestinal tracts were dissected with sterile scissors. Then the intestinal contents were carefully collected into sterile tubes and stored at − 80 °C for further sequencing of the 16 s rRNA. And RNA latter (Thermo Fisher Scientific, USA) was used to store the intestinal tissue shortly after collection, clearing out the contents, and storing it at − 80 °C.
Sequencing of the 16S rRNA gene
To extract the gut microbiome DNA, MoBio’s PowerSoil DNA Isolation Kit (Carlsbad CA) was used; the quality and quantity of the resulting DNA were measured using a NanoDrop analysis method. The Illumina HiSeq 2500 library was constructed at the Biomarker Technologies Company (Beijing, China). These primers targeting the V4 and V5 regions of the 16S rRNA region were used: 338-Forward (5′-ACTCCTACGGGGGAGGCCAG) and 806-Reverse (5′-GGACTACHVGGGTWTCTAAT).
NCBI has uploaded the raw 16S rRNA sequences under BioProject ID PRJNA856111. With the aid of Trimmomatic (version 0.33), the original data were qualified, and the primer sequences were removed with Cutadapt (version 1.9.1). Following merging the paired-end reads, chimeras were removed through UCHIME (version 8.1), resulting in effective reads. Using USEARCH version 10.0, sequences with > 97% homology were classified into multiple operational taxonomic units (OTUs). Based on the SILVA reference database (version 132) and QIIME2 (version 2020.6), naïve Bayesian classifiers were employed to assign OTU sequences to SILVA representative sequences. To analyze differences in community structure between different groups, principal coordinate analysis (PCoA) was performed. Through QIIME2, the beta diversity parameters (Chao1) were calculated. At the phylum, order, class, family, genus, and species taxonomic levels, histograms were created with the R software (version 3.5.3). LEfSe (http://huttenhower.sph.harvard.edu/galaxy/) applied the nonparametric factorial Kruskal-Wallis and Wilcoxon rank-sum tests to detect significantly different species at a level of 0.05 to determine the significant difference between different groups.
mRNA sequencing
Intestinal tissue RNA was isolated with Trizol reagent (Invitrogen) after treatment with RNase Free DNase I (Dalian Takara Co. Limited, China). A NanoDrop-2000 spectrophotometer (Implant, Westlake Village, USA) was used to measure RNA concentration and quality, and agarose (1%) gel electrophoresis was used to determine RNA integrity. cDNA synthesis and sequencing were performed using high-quality RNA from each sample. Under the manufacturer’s protocol, we constructed paired-end libraries with the TruSeqTM RNA library prep kit (Illumina, San Diego, CA, USA). Six cDNA libraries were generated by combining end-repair, 3′ end adenylation, and adapter ligation and enrichment (3 2nCC, 3 3nCC). High-throughput sequencing was performed on an Illumina sequencing platform (Illumina HiSeqTM 2500). The public repository of sequenced data at the NCBI (PRJNA857759) was considered.
Using the fastp software (version 0.20.0), adapters and low-quality reads were removed after sequencing. A quality assessment of the clean reads was performed using FastQC software (version 0.11.9), and alignments of the libraries to the RCC reference genome (https://bigd.big.ac.cn/search?dbId=gwh&q=GWHAAIA00000000) were performed with the HISAT2 tool (version 2.1.0). Fragments per kilobase per million mapped fragments (FPKMs) were used to calculate the gene expression level. DEGSeq2 R package (Version 1.28.1) was used to analyze the differentially expressed transcripts (DETs) of 3nCC versus 2nCC. DETs were genes with a fold change (FC) > 2 and false discovery rate (FDR) < 0.05. ClusterProfiler (version 3.6.0) with p < 0.05 was used to perform Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on these DETs.
Verification of quantitative real-time PCR
A set of eight DETs (six up-regulated DETs and two down-regulated DETs) was tested using quantitative real-time (qRT) PCR to assess the reliability of 3nCC sequencing results compared with 2nCC sequencing results. Following manufacturer’s instructions, cDNA synthesis was performed using the PrimeScriptTM RT reagent kit (Takara, Dalian, China). Listed in Additional file 4 are the primer sequences for β-actin (the internal control gene) and these DETs. qRT-PCR reactions were conducted in a 10 μL volume using 5 μL of the SYBR Green qPCR Master Mix, 0.5 μL of 20 μM of each primer, 1 μL of cDNA (1:10 dilution), and 3 μL of nuclease-free water. As a general rule, the thermal cycle for qRT-PCR was 95 °C for 2 min, 40 cycles at 95 °C for 15 s, and annealing at 60 °C for 30 s. qRT-PCR comprised three replications per biological sample. By using the 2−ΔΔCt method, we computed the relative mRNA expression levels. The data were analyzed using SPSS (v22.0) software (SPSS Inc., Chicago, IL, USA). We analyzed Students’t-tests to determine whether the results were statistically significant.
Availability of data and materials
Raw RNA-Seq reads are available from the NCBI (PRJNA857759). And the obtained raw 16S rRNA gene sequences are available from the NCBI (PRJNA856111).
Abbreviations
- 2nCC:
-
Diploid Carassius auratus
- 3nCC:
-
Triploid Carassius auratus
- DETs:
-
Differentially expressed transcripts
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- NLRP3 :
-
NLR family pyrin domain containing 3
- LY9 :
-
Lymphocyte antigen 9
- PNMA1 :
-
PNMA family member 1
- MR1 :
-
Major histocompatibility complex, class I-related
- PELI1 :
-
Pellino E3 ubiquitin protein ligase 1
- NOTCH2 :
-
Notch receptor 2
- NFIL3 :
-
Nuclear factor, interleukin 3 regulated
- NLRC4 :
-
NLR family CARD domain containing 4
References
Fraser TW, Rønneseth A, Haugland GT, Fjelldal PG, Mayer I, Wergeland HI. The effect of triploidy and vaccination on neutrophils and B-cells in the peripheral blood and head kidney of 0+ and 1+ Atlantic salmon (Salmo salar L.) post-smolts. Fish Shellfish Immunol. 2012;33(1):60–6.
Moore LJ, Nilsen TO, Jarungsriapisit J, Fjelldal PG, Stefansson SO, Taranger GL, et al. Triploid Atlantic salmon (Salmo salar L.) post-smolts accumulate prevalence more slowly than diploid salmon following bath challenge with salmonid alphavirus subtype 3. PLoS One. 2017;12(4):e0175468.
Ching B, Jamieson S, Heath JW, Heath DD, Hubberstey A. Transcriptional differences between triploid and diploid Chinook salmon (Oncorhynchus tshawytscha) during live Vibrio anguillarum challenge. Heredity (Edinb). 2010;104(2):224–34.
Dégremont L, Garcia C, Allen SK Jr. Genetic improvement for disease resistance in oysters: a review. J Invertebr Pathol. 2015;131:226–41.
Ojolick EJ, Cusack R, Benfey TJ, Kerr SR. Survival and growth of all-female diploid and triploid rainbow trout (Oncorhynchus mykiss) reared at chronic high temperature. Aquaculture. 1995;131:177–87.
Leclercq E, Taylor JF, Fison D, Fjelldal PG, Diez-Padrisa M, Hansen T, et al. Comparative seawater performance and deformity prevalence in out-of-season diploid and triploid Atlantic salmon (Salmo salar) post-smolts. Comp Biochem Physiol A Mol Integr Physiol. 2011;158(1):116–25.
Xiao J, Zou T, Chen Y, Chen L, Liu S, Tao M, et al. Coexistence of diploid, triploid and tetraploid crucian carp (Carassius auratus) in natural waters. BMC Genet. 2011;12:20.
Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–52.
Górski A, Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cell Mol Life Sci. 2005;62(5):511–9.
Abeles SR, Pride DT. Molecular bases and role of viruses in the human microbiome. J Mol Biol. 2014;426(23):3892–906.
Liang Z, Yuan Z, Guo J, Wu J, Yi J, Deng J, et al. Ganoderma lucidum polysaccharides prevent palmitic acid-evoked apoptosis and autophagy in intestinal porcine epithelial cell line via restoration of mitochondrial function and regulation of MAPK and AMPK/Akt/mTOR signaling pathway. Int J Mol Sci. 2019;20(3):478.
Chen Y, Qin N, Guo J, Qian G, Fang D, Shi D, et al. Functional gene arrays-based analysis of fecal microbiomes in patients with liver cirrhosis. BMC Genomics. 2014;15(1):753.
Briskey D, Tucker PS, Johnson DW, Coombes JS. Microbiota and the nitrogen cycle: implications in the development and progression of CVD and CKD. Nitric Oxide. 2016;57:64–70.
Renz H, Brandtzaeg P, Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2011;12(1):9–23.
Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–12.
Tran NT, Zhang J, Xiong F, Wang GT, Li WX, Wu SG. Altered gut microbiota associated with intestinal disease in grass carp (Ctenopharyngodon idellus). World J Microbiol Biotechnol. 2018;34(6):71.
Pérez T, Balcázar JL, Ruiz-Zarzuela I, Halaihel N, Vendrell D, de Blas I, et al. Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol. 2010;3(4):355–60.
Shang X, Wang B, Sun Q, Zhang Y, Lu Y, Liu S, et al. Selenium-enriched Bacillus subtilis reduces the effects of mercury-induced on inflammation and intestinal microbes in carp (Cyprinus carpio var. specularis). Fish Physiol Biochem. 2022;48(1):215–26.
Liu H, Qian K, Zhang S, Yu Q, Du Y, Fu S. Lead exposure induces structural damage, digestive stress, immune response and microbiota dysbiosis in the intestine of silver carp (Hypophthalmichthys molitrix). Comp Biochem Physiol C Toxicol Pharmacol. 2022;262:109464.
Shi F, Huang Y, Yang M, Lu Z, Li Y, Zhan F, et al. Antibiotic-induced alternations in gut microflora are associated with the suppression of immune-related pathways in grass carp (Ctenopharyngodon idellus). Front Immunol. 2022;13:970125.
Qiao G, Chen P, Sun Q, Zhang M, Zhang J, Li Z, et al. Poly-β-hydroxybutyrate (PHB) in bioflocs alters intestinal microbial community structure, immune-related gene expression and early cyprinid herpesvirus 2 replication in gibel carp (Carassius auratus gibelio). Fish Shellfish Immunol. 2020;97:72–82.
Ran C, Hu J, Liu W, Liu Z, He S, Dan BC, et al. Thymol and carvacrol affect hybrid Tilapia through the combination of direct stimulation and an intestinal microbiota-mediated effect: insights from a germ-free zebrafish model. J Nutr. 2016;146(5):1132–40.
Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65(2):330–9.
Murdoch CC, Rawls JF. Commensal microbiota regulate vertebrate innate immunity-insights from the zebrafish. Front Immunol. 2019;10:2100.
Liu XL, Jiang FF, Wang ZW, Li XY, Li Z, Zhang XJ, et al. Wider geographic distribution and higher diversity of hexaploids than tetraploids in Carassius species complex reveal recurrent polyploidy effects on adaptive evolution. Sci Rep. 2017;7(1):5395.
Luo J, Gao Y, Ma W, Bi XY, Wang SY, Wang J, et al. Tempo and mode of recurrent polyploidization in the Carassius auratus species complex (Cypriniformes, Cyprinidae). Heredity (Edinb). 2014;112(4):415–27.
Ren L, Gao X, Yang C, Tan H, Cui J, Wang S, et al. Comparison of diploid and triploid Carassius auratus provides insights into adaptation to environmental change. Sci China Life Sci. 2018;61(11):1407–19.
Org E, Parks BW, Joo JW, Emert B, Schwartzman W, Kang EY, et al. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 2015;25(10):1558–69.
Butt RL, Volkoff H. Gut microbiota and energy homeostasis in fish. Front Endocrinol (Lausanne). 2019;10:9.
Liu C, Zhao LP, Shen YQ. A systematic review of advances in intestinal microflora of fish. Fish Physiol Biochem. 2021;47(6):2041–53.
Cheutin MC, Villéger S, Hicks CC, Robinson J, Graham N, Marconnet C, et al. Microbial shift in the enteric bacteriome of coral reef fish following climate-driven regime shifts. Microorganisms. 2021;9(8):1711.
El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11(7):497–504.
Lee PT, Yamamoto FY, Low CF, Loh JY, Chong CM. Gut immune system and the implications of oral-administered immunoprophylaxis in finfish aquaculture. Front Immunol. 2021;12:773193.
Bricknell I, Dalmo RA. The use of immunostimulants in fish larval aquaculture. Fish Shellfish Immunol. 2005;19(5):457–72.
Li H, Qiu D, Yang H, Yuan Y, Wu L, Chu L, et al. Therapeutic efficacy of excretory-secretory products of trichinella spiralis adult worms on sepsis-induced acute lung injury in a mouse model. Front Cell Infect Microbiol. 2021;11:653843.
Huo X, Wang Z, Xiao X, Yang C, Su J. Oral administration of nanopeptide CMCS-20H conspicuously boosts immunity and precautionary effect against bacterial infection in fish. Front Immunol. 2022;12:811616.
Huang JB, Wu YC, Chi SC. Dietary supplementation of Pediococcus pentosaceus enhances innate immunity, physiological health and resistance to Vibrio anguillarum in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 2014;39(2):196–205.
Jia Z, Wang M, Zhang H, Wang X, Lv Z, Wang L, et al. Identification of a clip domain serine proteinase involved in immune defense in Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2018;74:332–40.
Kim JS, Harikrishnan R, Kim MC, Jang IS, Kim DH, Hong SH, et al. Enhancement of Eriobotrya japonica extracts on non-specific immune response and disease resistance in kelp grouper Epinephelus bruneus against Vibrio carchariae. Fish Shellfish Immunol. 2011;31(6):1193–200.
Destoumieux-Garzón D, Canesi L, Oyanedel D, Travers MA, Charrière GM, Pruzzo C, et al. Vibrio-bivalve interactions in health and disease. Environ Microbiol. 2020;22(10):4323–41.
Christ A, Günther P, Lauterbach MAR, Duewell P, Biswas D, Pelka K, et al. Western diet triggers NLRP3-dependent innate immune reprogramming. Cell. 2018;172(1–2):162–175.e14.
Deng N, Zhao Y, Xu J, Ouyang H, Wu Z, Lai W, et al. Molecular characterization and functional study of the NLRP3 inflammasome genes in Tetraodon nigroviridis. Fish Shellfish Immunol. 2022;131:570–81.
Li JY, Wang YY, Shao T, Fan DD, Lin AF, Xiang LX, et al. The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E-mediated pyroptosis. J Biol Chem. 2020;295(4):1120–41.
de Salort J, Cuenca M, Terhorst C, Engel P, Romero X. Ly9 (CD229) cell-surface receptor is crucial for the development of spontaneous autoantibody production to nuclear antigens. Front Immunol. 2013;4:225.
Schüller M, Jenne D, Voltz R. The human PNMA family: novel neuronal proteins implicated in paraneoplastic neurological disease. J Neuroimmunol. 2005;169(1–2):172–6.
Chen HL, D'Mello SR. Induction of neuronal cell death by paraneoplastic Ma1 antigen. J Neurosci Res. 2010;88(16):3508–19.
Karamooz E, Harriff MJ, Lewinsohn DM. MR1-dependent antigen presentation. Semin Cell Dev Biol. 2018;84:58–64.
McWilliam HEG, Salio M. Understanding and modulating the MR1 metabolite antigen presentation pathway. Mol Immunol. 2021;129:121–6.
Schlenner S, Pasciuto E, Lagou V, Burton O, Prezzemolo T, Junius S, et al. NFIL3 mutations alter immune homeostasis and sensitise for arthritis pathology. Ann Rheum Dis. 2019;78(3):342–9.
Geiger TL, Abt MC, Gasteiger G, Firth MA, O'Connor MH, Geary CD, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med. 2014;211(9):1723–31.
Yu H, Shen Y, Sun J, Xu X, Wang R, Xuan Y, et al. Molecular cloning and functional characterization of the NFIL3/E4BP4 transcription factor of grass carp. Dev Comp Immunol. 2014;47(2):215–22.
Abdelaziz DH, Amr K, Amer AO. Nlrc4/Ipaf/CLAN/CARD12: more than a flagellin sensor. Int J Biochem Cell Biol. 2010;42(6):789–91.
Wang SS, Yan CS, Luo JM. NLRC4 gene silencing-dependent blockade of NOD-like receptor pathway inhibits inflammation, reduces proliferation and increases apoptosis of dendritic cells in mice with septic shock. Aging (Albany NY). 2021;13(1):1440–57.
Park J, Lee SY, Jeon Y, Kim KM, Lee JK, Ko J, et al. The Pellino1-PKCθ signaling axis is an essential target for improving antitumor CD8+ T-lymphocyte function. Cancer Immunol Res. 2022;10(3):327–42.
Lereim RR, Oveland E, Xiao Y, Torkildsen Ø, Wergeland S, Myhr KM, et al. The brain proteome of the ubiquitin ligase peli1 knock-out mouse during experimental autoimmune encephalomyelitis. J Proteomics Bioinform. 2016;9(9):209–19.
Lin X, Wang S, Sun M, Zhang C, Wei C, Yang C, et al. miR-195-5p/NOTCH2-mediated EMT modulates IL-4 secretion in colorectal cancer to affect M2-like TAM polarization. J Hematol Oncol. 2019;12(1):20.
Sakata-Yanagimoto M, Chiba S. Notch2 and immune function. Curr Top Microbiol Immunol. 2012;360:151–61.
Maekawa Y, Minato Y, Ishifune C, Kurihara T, Kitamura A, Kojima H, et al. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat Immunol. 2008;9(10):1140–7.
Acknowledgements
Not applicable.
Funding
This research was supported by grants from the Natural Science Foundation of Hunan Province (2021JJ30508), the Hunan Province Chinese Medicine Research Program Project (2021055), the Hunan Provincial Education Department(21B0387).
Author information
Authors and Affiliations
Contributions
KW have designed of the work. YC has contributed to this study for the design, in executing experiments and in writing manuscript. YC and KW have made substantial contributions to the acquisition and analysis of data. YC and KW have substantively revised the work. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by Ethics Committee of Hunan University of Chinese Medicine. All experiments were performed in accordance with relevant guidelines and regulations. The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org/).
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing financial interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cai, Y., Wei, K. Comparative analysis of intestinal microbiota composition and transcriptome in diploid and triploid Carassius auratus. BMC Microbiol 23, 1 (2023). https://doi.org/10.1186/s12866-022-02709-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-022-02709-5