Abstract
Background
Here, we aimed to evaluate and compare the anti-Helicobacter pylori activity of potential probiotic Lactiplantibacillus pentosus SLC13 to Lactobacillus gasseri BCRC 14619 T and Lacticaseibacillus rhamnosus LGG. Phenotypic assays including growth curve, cell adhesion, and cellular cytotoxicity were performed to characterize SLC13. Anti-H. pylori activity of lactobacilli was determined by the disk diffusion method and co-culture assay. Exopolysaccharide (EPS) was extracted from lactobacilli to test its immune modulation activity, and IL-8 expression in AGS and GES-1 was determined by RT-qPCR.
Results
All three lactobacilli strains were tolerant to the simulated gastrointestinal conditions. SLC13 showed the highest adhesion ability to AGS and GES-1 cells, compared to LGG and BCRC 14619 T. The coculture assays of SLC13, LGG, and BCRC 14619 T with cells for 4 h showed no significant cytotoxic effects on cells. All tested strains exhibited an inhibitory effect against H. pylori J99. The cell-free supernatant (CFS) of three strains showed activity to inhibit H. pylori urease activity in a dose-dependent manner and the CFS of SLC13 had the highest urease inhibitory activity, compared to LGG and BCRC 14619 T. Only the treatment of AGS cells with SLC13 EPS significantly decreased the IL-8 expression induced by H. pylori infection as compared to cells treated with LGG and BCRC 14619 T EPS.
Conclusions
SLC13 possesses potent antimicrobial activity against H. pylori growth, infection, and H. pylori-induced inflammation. These results suggest that SLC13 and its derivatives have the potential as alternative agents against H. pylori infection and alleviate inflammatory response.
Similar content being viewed by others
Background
Lactic acid bacteria (LAB) are a group of gram-positive cocci or rods, which produce lactic acid as the major end product of the fermentation of carbohydrates. Lactobacillus, Lactococcus, Leuconostoc,Pediococcus, Streptococcus, and Enterococcus, are the main genera of LAB with probiotic activity [1]. The biological activity of LAB, its derivative exopolysaccharide (EPS), and bacteriocins have been shown to prevent infections, against tumor progression, modulate the immune response, relieve allergies, alter the microbiome, and alleviate lactose intolerance [2,3,4,5,6].
We previously isolated a high EPS-producing Lactiplantibacillus pentosus strain, SLC13, from mustard pickles in Taiwan [4, 7]. SLC13 showed high inhibitory activity against the growth of Staphylococcus aureus, Enterococcus faecalis, and Yersinia enterocolitica [4]. The complete genome sequence of SLC13 revealed a plantaricin gene cluster, which is responsible for bacteriocins biosynthesis, and could be associated with its broad-spectrum antimicrobial activity [7].
Helicobacter pylori is a gram-negative and microaerophilic bacterium and infects approximately 50% of the world's population [8]. Importantly, persistent H. pylori infection is associated with the development of gastrointestinal diseases, including gastric adenocarcinoma [8]. H. pylori urease activity, flagella-mediated motility, and adhesins, are required for the neutralization of hostile acidic conditions and successful colonization [9]. Triple therapies consisting of a proton pump inhibitor (PPI), clarithromycin, metronidazole, levofloxacin, or amoxicillin, are the most common treatments for H. pylori infections [10]. However, the increase in antibiotic resistance of H. pylori has been reported to reduce the cure rates of H. pylori therapies [10, 11]. Therefore, In this study, we aimed to evaluate the anti-H. pylori activity of L. pentosus SLC13 to determine potential applications of SLC13 in the future.
Results
Growth of lactobacilli under simulated gastrointestinal conditions
To test and compare the growth rate of lactobacilli strains under simulated gastrointestinal conditions, lactobacilli were cultured in MRS broth, acidic MRS broth (pH 3.0), and 0.3% bile salt in MRS broth. Our results showed that all three strains were tolerant to gastrointestinal challenges (Fig. S1B & S1C). However, SLC13 grew faster than LGG and BCRC 14619 T in MRS broth and acidic MRS broth (Fig. S1A & S1B). In addition, to test the ability of three lactobacilli strains to withstand the extremely acidic conditions of the gastric cavity, the survival of lactobacilli was determined by counting the viable cells after three hours incubation in MRS broth at pH 2. The results showed that only SLC13 had a high survival rate (90.4%) under extremely acidic conditions (Fig. S2).
Adhesion ability and cytotoxicity of lactobacilli to AGS and GES-1 cells
The adhesion rates of SLC13, LGG, and BCRC 14619 T to AGS and GES-1 cells altered depending on the tested strains which ranged from 0.33 to 2.38% (AGS cells) and from 0.14 to 4.09% (GES-1 cells), respectively (Fig. 1A). Among the three strains examined, SLC13 showed the highest adhesion ability as compared to LGG and BCRC 14619 T (Fig. 1A). Consistent with the colony counting results, we observed that more SLC13 adhered to AGS and GES-1 cells, compared to LGG and BCRC 14619 T under the microscope (Fig. S3).
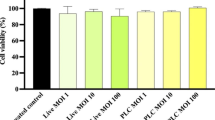
Adhesion and cytotoxicity of SLC13, LGG, and BCRC 14619 T to AGS and GES-1. A SLC13, LGG, and BCRC 14619 T adhesion to AGS and GES-1 cells. B Cell viability was determined by MTT assay after 4 h of coculture (MOI = 100). Error bars represent the standard deviation of biological triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significant difference; pc, positive control (Triton X-100 treatment); nc, negative control (AGS or GES-1 cells only, without treatment)
To test the safety of Lactobacillus in future applications, we performed the cell viability assay to determine the cytotoxicity of three lactobacilli strains to AGS and GES-1 cells (Fig. 1B). Cells treated with 0.1% Triton X-100 inhibited the proliferation of these cells significantly and thus served as a positive control in this assay (Fig. 1B). The results showed that the coculture of SLC13, LGG, and BCRC 14619 T with cells for 4 h had no significant cytotoxic effects on cells (Fig. 1B).
Antibacterial activity of lactobacilli against H. pylori
Our previous study showed that SLC13 had antibacterial activity against the growth of S. aureus, E. faecalis, and Y. enterocolitica [4]. Therefore, we performed the disk diffusion method to evaluate and compare the inhibitory activity of SLC13, LGG, and BCRC 14619 T against H. pylori reference strain J99 (Fig. 2A). Our results indicated that all tested strains exhibited an inhibitory effect against H. pylori J99, as demonstrated by forming the zones of growth inhibition (Fig. 2A).
Anti-H. pylori activity of lactobacilli. A The inhibitory effect of lactobacilli on H. pylori J99 was determined by disk diffusion method. MRS broth alone was used as a negative control. B The survival rate of H. pylori in the presence of different concentrations (10%, 20%, and 40%) of CFS or neutralized-CFS (adjusted to pH 6.5 by NaOH) at microaerophilic conditions for 4 h. The viability of H. pylori after 72 h co-incubation with CFS was evaluated by determining the viable bacterial count on Brucella agar containing 10% horse serum plates after incubation at 37 °C under microaerophilic conditions. MRS broth pH 4.0 (acid-CFS control) and pH 6.5 (neutralized-CFS control) were used as controls of the reaction. CFS, cell-free supernatant. Error bars represent the standard deviation of biological triplicates. *, p < 0.05; **, p < 0.01; ns, no significant difference
To test the anti-H. pylori activity of cell-free supernatant (CFS) collected from lactobacilli strains tested and neutralized-CFS (CFS with pH neutralization), we recorded the pH value of collected CSF from three lactobacilli strains to prevent the different acidities of CSF from affecting the phenol red indicator to determine H. pylori urease activity. The mean pH value of SLC13, LGG, and BCRC 14169 T CSF was 3.96, 3.96, and 4.12, respectively. In addition, we added ~ 80 μL NaOH (5 M) to neutralize the pH value of 10 mL collected acidic CSF to pH 6.5 to prevent the dilution effect. The survival rate of H. pylori J99 in the presence of CFS of lactobacilli was further determined, and the results showed that the CFS and neutralized-CFS (CFS with pH neutralization) of lactobacilli with various concentrations (10%, 20%, and 40%) showed different inhibition activity to H. pylori growth (Fig. 2B). In general, nearly all CFS showed higher anti-H. pylori activity compared to the neutralized-CFS (except 10 and 20% CFS of BCRC 14619 T) (Fig. 2B). Only 22%, 19%, and 13% of H. pylori survived under the treatment with 10% neutralized-CFS of SLC13, LGG, BCRC 14619 T, respectively (Fig. 2B). Moreover, the antibacterial activity of CFS from three lactobacilli strains was not significantly different (p > 0.05).
Effect of lactobacilli CFS on urease activity of H. pylori
Nearly all CFS showed higher antibacterial activity compared to neutralized-CFS (Fig. 2B). The expression level and activity of urease are critical for H. pylori survival under an acidic environment. Therefore, we further evaluated the effects of lactobacilli CFS on the urease activity of H. pylori (Fig. 3). Overall, the results showed that the urease activity of H. pylori was decreased by treating H. pylori with CFS and neutralized-CFS collected from three examined strains (Fig. 3). The anti-urease activity of H. pylori showed a dose-dependence of the lactobacilli CFS concentrations. Among three tested strains, the CFS of SLC13 showed the highest urease inhibitory activity, compared to the CFS of LGG and BCRC 14619 T. In addition, neutralized-CFS had a less potential ability to decrease urease activity compared to CFS (Fig. 3).
Inhibition of urease activity of lactobacilli cell-free supernatant. Effect of lactobacilli CFS and neutralized-CFS (10%, 20%, and 40% concentrations) on urease activity of H. pylori was determined by measuring the absorbance of ammonium at 550 nm after 4 h co-incubaction. MRS broth pH 4.0 (acid-CFS control) and pH 6.5 (neutralized-CFS control) were used as controls of the reaction. CFS, cell-free supernatant. Error bars represent the standard deviation of biological triplicates (with technical replicates). *, p < 0.05; ***, p < 0.001; ****, p < 0.0001; ns, no significant difference
Anti-H. pylori adhesion activity of lactobacilli
The adhesion ability of H. pylori to the surface of gastric epithelial cells is an important initial step towards successful colonization [9]. Therefore, we investigated whether lactobacilli or their derived CFS suppress the adhesion of H. pylori to AGS and GES-1 cells (Fig. 4A-D). The results showed that pretreatment with SLC13, LGG, and BCRC 14619 T for two hours exhibited a reduction of H. pylori adhesion to GES-1 cells by 54.2%, 52.4%, and 38.0%, respectively (Fig. 4A). Moreover, the adhesion of H. pylori to AGS cells was reduced by the pretreatment with SLC13 (67.3%), LGG (51.2%), and BCRC 14619 T (57.7%) (Fig. 4B).
Anti-H. pylori adhesion activity of lactobacilli. Adhesion of H. pylori to GES-1 (A) and AGS cells (B) after pretreatment with lactobacilli for 30 min. Adhesion of H. pylori to GES-1 (C) and AGS cells (D) in the presence of lactobacilli CFS simultaneously. Survival of H. pylori in the presence of lactobacilli CFS simultaneously in RPMI cell culture medium for GES-1 cells (E) and F12 culture medium for AGS cells (F). H. pylori J99 alone was used as a control in these assays. Error bars represent the standard deviation of biological triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significant difference
We further determined whether lactobacilli CFS has the anti-adhesion activity of H. pylori to GES-1 and AGS cells (Fig. 4C & D). The results showed that the adherence of H. pylori to GES-1 and AGS cells was dramatically reduced in the presence of lactobacilli CFS (Fig. 4C & D). The coculture of cells with CFS of SLC13, LGG, and BCRC 14619 T simultaneously caused a reduction of H. pylori adhesion to GES-1 cells by 78.7%, 73.2%, and 79.9%, respectively (Fig. 4C). Moreover, the adhesion of H. pylori to AGS cells was reduced by coculture with the CFS of SLC13 (63.7%), LGG (74.6%), and BCRC 14619 T (62.0%) (Fig. 4D).
To assess whether the inhibitory effect of lactobacilli CFS on adherence of H. pylori to GES-1 and AGS is associated with the reduction of H. pylori amount, we evaluated the survival rate of H. pylori with lactobacilli CFS treatment. Our results showed that the CFS of SLC13, LGG, and BCRC 14619 T strains killed H. pylori and resulted in a reduction of H. pylori adhesion to host gastric epithelium cells (Fig. 4E & F).
Effect of lactobacilli on H. pylori-induced Inflammation
H. pylori infection induces IL-8 production and contributes to inflammation of gastric epithelial cells [12]. We therefore determined whether the lactobacilli and their derived EPS can inhibit H. pylori-induced IL-8 gene expression of AGS and GES-1 cells by using RT-qPCR (Fig. 5A & B). The treatment of GES-1 and AGS cells with lactobacilli SLC13, LGG, and BCRC 14619 T, did not induce IL-8 mRNA expression in cells, compared to cells infected with H. pylori (Fig. 5A & B). Moreover, the pretreatment of three lactobacilli strains effectively reduced the IL-8 mRNA in GES-1 infected with H. pylori (Fig. 5A). In AGS cells, pretreatment with SLC13 and LGG effectively reduced the IL-8 mRNA which was induced by H. pylori infection (Fig. 5B). Interestingly, BCRC 14,619 exhibited a difference of IL-8 induction in GES-1 and AGS cells (Fig. 5A & B). Significant induction of IL-8 mRNA was detected in AGS cells treated with BCRC 14,619 (Fig. 5B).
Anti-H. pylori-induced inflammation of lactobacilli and their derived exopolysaccharide. GES-1 (A) and AGS (B) cells were pretreated with lactobacilli SLC13, LGG, or BCRC 14619 T for 2 h, and then cells were infected with H. pylori J99 (MOI = 100) for 2 h. C. AGS cells were treated with EPS extracted from lactobacilli SLC13, LGG, or BCRC 14619 T at a concentration of 500 ng/mL with H. pylori J99 (MOI = 100) simultaneously for 2 h. After H. pylori infection, RNA was extracted from cells to determine the il-8 gene expression. AGS and GES-1 cells alone without any treatments were used as a negative control to normalize the measurements. Error bars represent the standard deviation of biological triplicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significant difference; EPS, exopolysaccharide
Lactobacilli-derived EPS have been shown to downregulate the inflammatory cytokine expression [13]. Therefore, we next determined the EPS production in three tested lactobacilli strains, and the results showed that all three strains produced high EPS (Fig. S4). Compared to AGS cells, GES-1 infected with H. pylori induced lower IL-8 expression, therefore, we only tested the immune suppression activity of EPS extracted from three lactobacilli strains in AGS cells infected with H. pylori (Fig. 5C). The results showed that EPS derived from three strains did not induce IL-8 expression in AGS (Fig. 5C) and did not have anti-H. pylori activity (Fig. S5). Moreover, only treatment of AGS cells with SLC13 EPS significantly decreased IL-8 expression induced by H. pylori infection as compared to cells treated with LGG and BCRC 14619 T EPS (Fig. 5C).
Discussion
The increasing prevalence of antibiotic resistance in H. pylori leads to a decrease in eradication rate with standard triple therapy, including PPI, amoxicillin, and clarithromycin [14]. Moreover, antibiotic treatments have been shown to cause gut microbiota dysbiosis and subsequently alter immune cell function, which is associated with reduced synaptic transmission and gamma oscillations in the hippocampus [15]. Therefore, probiotics are considered as an alternative or adjuvant treatment to improve H. pylori eradication and reduce drug side effects [16, 17]. In this study, we characterized SLC13 and demonstrated it had the potential to against H. pylori infection and H. pylori-induced inflammation.
Although SLC13 was isolated from fermented food, our results showed that SLC13 had a high tolerance to gastrointestinal challenge (Fig. S1), colonized greatly to the gastric epithelial AGS and GES-1 cells (Fig. 1A & B), and did not cause cytotoxicity to cells (Fig. 1C). Importantly, SLC13 and its derived CFS displayed a strong anti-H. pylori J99 activity (Fig. 2). However, it remains to study whether SLC13 had broad-spectrum activity against multidrug-resistant H. pylori. Zheng et al. reported that lactobacilli LPS16 had anti-H. pylori activity depended on the secreted components and lactic acid-mediated bactericidal activity [18]. Our results showed that compared to neutralized CFS, acidic CFS had higher anti-H. pylori activity (Fig. 2B). These results suggest that SLC13 against H. pylori by acidifying culture medium in part; however, the other secreted components in the CFS also showed the antibacterial activity against H. pylori. Lactic acid causes membrane permeabilization in gram-negative bacteria, which in addition to its antimicrobial property, can promote the bactericidal effects of other compounds such as bacteriocins, which have been shown to have strong antibacterial activity against gram-negative pathogens, including H. pylori [19, 20]. The complete genome sequence of SLC13 revealed a plantaricin gene cluster, which may contribute to bacteriocins biosynthesis [7]. However, the characteristics and regulation of bacteriocins of SLC13 remain to be studied.
Urease activity is an important virulence factor for H. pylori colonization, which changes the viscoelastic nature of mucus by neutralizing acidic pH [21]. Our results revealed that CFS reduced the urease activity of H. pylori J99. In addition, the urease inhibitory effect of CFS was more efficient as compared to the neutralized-CFS (Fig. 3). We found that H. pylori treated with CFS had a lower survival rate, compared to the neutralized-CFS (Fig. 2). These results suggest that the decrease in urease activity is caused by the reduction of live bacteria. However, the treatment of H. pylori with neutralized-CFS showed a lower urease activity, compared to the control group (Fig. 3). Ryan et al. reported that the expression of 8 of 12 Cag pathogenicity island genes was downregulated by exposure to Ligilactobacillus salivarius and resulted in the loss of functionality of the Cag secretion system [22]. L. rhamnosus JB3 showed the ability to suppress the association of H. pylori to cells and its induced IL-8 levels, as well as the mRNA expression of vacA, sabA, and fucT of H. pylori [23]. Therefore, it is worth studying whether the SLC13 and its derived CFS modulate the gene expression in H. pylori and thus reduce the adhesion ability and urease activity of H. pylori.
The anti-H. pylori adhesion to cells of lactobacilli can be achieved by competition for binding sites on epithelial cells, release antimicrobial compounds to kill H. pylori, and down-regulation of adhesin gene expression. Previous results showed that lactobacilli acted directly on H. pylori by an effector molecule released into the medium and inhibited the expression of the adhesion-encoding gene sabA [24]. Our results showed that both live SLC13 and its CFS suppressed the adherence of H. pylori to GES-1 and AGS cells (Fig. 4). Furthermore, the Giemsa staining displayed that SLC13 did not adhere efficiently to cells as compared to H. pylori (data not shown). These results suggest that the bacteriocin or unknown compounds secreted by SLC13 majorly contribute to the reduction of H. pylori adherence to GES-1 and AGS cells. Jin et al. reported that a combination of L. salivarius LN12 CFS with amoxicillin and clarithromycin destroyed the H. pylori biofilm to a greater extent than when separately [25]. Therefore, the effects of SLC13 in combination with other antibiotics on H. pylori motility, biofilm, and toxin production, are worth investigating.
H. pylori can directly translocate CagA into host cells, which leads to the production of IL-8 by the epithelial cells [26]. In addition, nucleotide-binding oligomerization domain 1 (NOD1) recognizes H. pylori-derived peptidoglycan or outer membrane vesicles intracellular that trigger the pro-inflammatory signaling cascade through nuclear translocation of NF-κB subunits in gastric epithelial cells [27]. In this study, we demonstrated that SLC13 contained potential activity to inhibit H. pylori adhesion to gastric epithelial cells, which in turn, may attenuate CagA translocation and IL-8 production in cells (Fig. 5A & B). EPS have been reported to reduce TNF-α, IL-8, and MCP-1 in the gastric mucosa of H. pylori-infected mice [28]. We showed that SLC13, LGG, and BCRC 14619 T are high EPS-producing strains, however, compared to the SLC13 EPS, the EPS extracted from LGG and BCRC 14619 T did not reduce the IL-8 expression in AGS cells infected with H. pylori (Fig. 5C). The difference in biological properties of EPS extracted from three tested strains suggests that the structure and composition of their EPS are strain-specific [29]. Therefore, the mechanisms through which SLC13 and its derivatives modulate the pro-inflammatory signaling are worth investigating.
In previous clinical trial results, Chen et al. reported that the probiotics containing Lactobacillus acidophilus and L. rhamnosus can reduce the H. pylori bacterial load, but H. pylori can not be eradicated by probiotics alone [30]. However, these two lactobacilli strains used in their study are commercially available probiotics by the Taiwan Sugar Corporation. In contrast, SLC13 and previously reported JB3 with anti-H. pylori activity were isolated from fermented food and dairy food, respectively [4, 31]. At present, these two strains are not commercially available probiotics. Although we showed the in vitro anti-H. pylori activity of SLC13, an in-depth study is required to determine the characteristics (including the safety in use) of SLC13 before examining its functionality in animal models and human clinical trials. H. pylori infections were related to several extragastric diseases such as cardiovascular, hematological, and neurological conditions [32]. Probiotics generate bacteriostatic chemicals that limit H. pylori colonization and minimize treatment-related adverse effects such as antibiotic-associated diarrhea [33]. Therefore, the effects of SLC13 on H. pylori-related extragratric diseases with reduced antibiotic-causing adverse effects should be studied.
Conclusions
Our results showed that L. pentosus SLC13 had no cytotoxicity, high tolerance to acid and bile salt, and high cell adhesion ability. Moreover, SLC13 and its derivatives in CFS and EPS displayed multiactivity against colonization and pathogenesis of H. pylori through modulating inflammatory response, reducing urease activity and attachment on the cells, and killing H. pylori. Therefore, SLC13 has the potential as a preventive agent or adjuvant treatment for H. pylori infection. However, it is necessary to verify the safety and anti-H. pylori activity of SLC13 by animal models in the future.
Material and methods
Bacterial strains and culture conditions
Lactiplantibacillus pentosus SLC13, Lactobacillus gasseri BCRC 14619 T, and Lacticaseibacillus rhamnosus LGG were stored at − 80 °C in De Man, Rogosa and Sharpe (MRS) broth containing 16% glycerol (v/v) until tested. LGG and BCRC 14619 T are widely used probiotic strains and are thus used as control strains in this study [34, 35]. LGG and BCRC 14619 T were gifts from Dr. Ying-Chieh Tsai at National Yang Ming Chiao Tung University (Taiwan). SLC13 was the laboratory stock strain isolated from mustard pickles in Taiwan [4]. To activate lactobacilli, the bacteria were cultured in MRS broth at 37 °C for 24 h and then subjected to each experimental study. H. pylori J99 was stored at − 80 °C in Brucella broth containing 20% glycerol (v/v) until tested. H. pylori was grown on Brucella agar plates containing 10% (v/v) horse serum (Gibco BRL, Life Technologies, Rockville, MD) at 37 °C in microaerophilic conditions (5% O2, 10% CO2, and 85% N2).
Cell culture
AGS cells (human gastric adenocarcinoma epithelial cells, ATCC CRL 1739) and GES-1 cells (normal human gastric epithelial cells) were gifts from Dr. Hsiu-Chi Cheng at National Cheng-Kung University Hospital (Taiwan). AGS and GES-1 cells were maintained in Ham′s F12 and RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) (Gibco BRL, Life Technologies, Rockville, MD), penicillin (100 IU/mL), and streptomycin (100 µm/mL) at 37 °C with 5% CO2, respectively.
Cell adhesion assay
AGS and GES-1 cells (3 × 105 cells) were seeded into 12-well plates in F12 and RPMI-1640 media, respectively, with 10% FBS and 1% penicillin–streptomycin, and grown to a monolayer at 37 °C for 24 h. Before performing infection, the culture medium was replaced with fresh medium without antibiotics. To determine the adhesion ability of lactobacilli, cells were infected with a multiplicity of infection (MOI) of 100 of lactobacilli and incubated at 37 °C for 90 min. For lactobacilli-H. pylori co-culture, the cells were incubated with lactobacilli strains (MOI = 100) 30 min before being infected with H. pylori (MOI = 100) and then incubated at 37 °C for 1 h. For lactobacilli CFS-H. pylori co-culture, lactobacilli were cultured in cell culture medium for 4 h. After centrifugation at 3,011 xg for 10 min, the supernatant was collected and filtered by a 0.22 µm diameter filter. Sodium hydroxide (NaOH) was used as the neutralizing agent to adjust the pH of CFS to 6.5 (neutralized-CFS). The cells were then incubated with H. pylori (MOI = 100) and collected CFS at different concentrations simultaneously at 37 °C for 90 min. The cells were washed three times with PBS, lysed by incubation with 0.1% saponin at 37 °C for 15 min, and plated on MRS plates or Brucella plates with 10% horse serum. The colonies were counted to determine the number of adherent bacteria. The percentage of bacterial adhesion to cells was calculated according to the following formula: adhesion ability (%) = (adherent bacteria)/(total bacteria) × 100. Cell adhesion assay was conducted in biological triplicate to ensure reproducibility.
Cytotoxicity on AGS and GES-1 cells
The cytotoxicity effect of lactobacilli on AGS and GES-1 cells was determined by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich, St Louis, MO, USA) assay according to the previous study [36]. Cells (1.5 × 105) were grown and allowed to adhere to a 96-well plate at 37 °C for 24 h to approximately 80% confluence and then cells were treated with lactobacilli with a MOI of 100 for 4 h. The medium was removed and the cells were washed three times with PBS. Thirty µL of MTT reagent (5 mg/mL) was added to each well and the plates were incubated at 37 °C for 3 h in the dark. Two hundred μL of DMSO was added to each well and the plate was incubated with shaking (150 rpm) at room temperature for 15 min to allow the color to develop. The OD was measured at 570 nm. Cell viability (%) = (Asample/Acontrol) × 100, where Asample is the absorbance of the cells that were incubated with the medium containing lactobacilli or 0.1% Triton X-100, and Acontrol is the absorbance of the cells alone. The cytotoxicity assay was conducted in biological triplicate to ensure reproducibility.
Anti-H. pylori activity of lactobacilli
Anti-H. pylori activity of lactobacilli was determined by the disk diffusion method as previously described [37]. Briefly, H. pylori suspension (OD600 = 2 in Brucella broth) was spread on Brucella agar plates containing 10% horse serum. The overnight culture of lactobacilli was adjusted to OD600 = 1 and added to paper disks (Toyo Roshi Kaisha, Ltd, Japan) with a diameter of 6 mm. The plates were cultured under microaerophilic conditions for 72 h and the inhibition zone was determined in diameter.
For co-culture assay, H. pylori suspensions were adjusted to OD600 = 0.2 in MRS broth containing 10% horse serum and were incubated under microaerophilic conditions at 37 °C with 10%, 20%, or 40% lactobacilli CFS or neutralized-CFS for 4 h. MRS broth was used as the control. The viability of H. pylori after 4 h co-incubation with CFS was evaluated by determining the viable bacterial count on Brucella agar containing 10% horse serum plates after incubation at 37 °C under microaerophilic conditions. MRS broth (pH 4.0 and pH 6.5) was used as a control of the reaction. H. pylori survival rate (%) = [A1(Log CFU/mL)/A0(Log CFU/mL)] × 100, where A0 is the viable count of H. pylori at 0 h and A1 is the viable count of H. pylori after 4 h co-incubation with CFS. Anti-H. pylori activity of lactobacilli was conducted in biological triplicate to ensure reproducibility.
Urease activity assay
To analyze the inhibitory effects of H. pylori urease by CFS of lactobacilli, H. pylori cells were resuspended in Brucella broth (OD600 = 0.2) and mixed with 10, 20, or 40% lactobacilli CFS or neutralized-CFS, and then incubated at 37 °C for 4 h. Urease activity was determined in phosphate buffer containing urea buffer (25 mM urea and 0.012% phenol red in PBS) according to the previous study [38,39,40]. The resulting color was read at OD550 after 2 h incubation. MRS broth (pH 4.0 and pH 6.5) was used as a control of the reaction. The urease activity assay was conducted in biological triplicate along with technical triplicate to ensure reproducibility.
Exopolysaccharide extraction
Lactobacilli were inoculated in 25 mL MRS broth containing 2% sucrose at 37 °C for 24 h. Bacterial cultures were centrifuged at 3,011 xg for 30 min to remove cells and their debris. EPS were precipitated from supernatants by adding 4 volumes of 95% ethanol, and the mixture was incubated at 4 °C for 24 h [41]. After ethanol precipitation, the samples were centrifugated at 3,011 xg for 30 min, and the supernatant was removed. The precipitate of pure EPS was dried in the oven at 60 °C for 24 h. The precipitates were re-suspended in distilled water, filtered by 0.45 µm diameter filter, and stored at -80 °C until testing. The production of EPS was analyzed by the phenol–sulfuric method using glucose as a reference standard [42]. EPS production assay was conducted in biological triplicate to ensure reproducibility.
IL-8 mRNA determination by reverse-transcription quantitative PCR (RT-qPCR)
To determine the anti-inflammation ability of lactobacilli, AGS and GES-1 cells were pretreated with lactobacilli (MOI = 100) for 2 h, and then cells were infected with H. pylori (MOI = 100, 2 h). In contrast, to determine the anti-inflammation activity of EPS derived from lactobacilli, AGS cells were treated with lactobacilli EPS (500 ng/mL) and H. pylori (MOI = 100) simultaneously for 2 h. After H. pylori infection for 2 h, Trizol reagent was used for total mRNA extraction. A total of 1 μg RNA was reverse-transcribed with SuperScript II RNase H-reverse transcriptase (Bionovas biotechnology Co., Ltd, Canada) for cDNA synthesis. The amounts of IL-8 mRNA were analyzed by RT-qPCR performed in a StepOnePlus™ system (Themo Fisher Scientific Inc, USA) with the SYBR Green Master Mix (Ampliqon, Denmark) in qPCR 96 well plates (MB-Q96-LP and MB-QSM; Gunster Biotech, New Taipei City, Taiwan). The primers IL-8-forward (5′-ACTGAGAGTGATTGAGAGTGGAC-3′) and IL-8-reverse (5′- AACCCTCTGCACCCAGTTTTC − 3′) and β-actin-forward (5’-GACCTCTATGCCAACACAGT-3’) and β-actin-reverse (5'-AGTACTTGCGCTCAGGAGGA-3') were used for IL-8 and β-actin detection, respectively [43, 44]. Samples were normalized to the level of β-actin mRNA. The RT-qPCR was performed in triplicate in a total reaction volume of 25 μL containing 12.5 μL of SYBR Green PCR Master Mix, 0.5 μL of forward and reverse primers, 10.5 μL of distilled H2O, and 1 μL of cDNA from each sample. Samples were heated for 15 min at 95 °C and amplified for 40 cycles of 15 s at 95 °C and 60 s at 60 °C. Quantification was performed using the 2-ΔΔCt method [45], where the Ct value was defined as the threshold cycle of PCR at which amplified product was detected. The ΔCt was obtained by subtracting the housekeeping gene (β-actin) Ct value from the Ct value of the IL-8 gene. The fold change was calculated according to the formula 2-ΔΔCt, where ΔΔCt was the difference between ΔCt and the ΔCt calibrator value (which was assigned a value of 1 arbitrary unit). IL-8 mRNA expression level was conducted by RT-qPCR in biological triplicate to ensure reproducibility.
Statistical analysis
The data were expressed as a mean of three replicates ± SD. ANOVA one-way test was used to calculate the statistical significance of the experimental results between two groups. In the urease activity assay and anti-H. pylori activity of lactobacilli assay, ANOVA two-way test was used to assess multiple comparisons in those groups. A p-value less than 0.05 was considered as a significant difference.
Availability of data and materials
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Fijan S. Microorganisms with claimed probiotic properties: an overview of recent literature. Int J Environ Res Public Health. 2014;11(5):4745–67. https://doi.org/10.3390/ijerph110504745.
Abdalla AK, Ayyash MM, Olaimat AN, Osaili TM, Al-Nabulsi AA, Shah NP, et al. Exopolysaccharides as Antimicrobial Agents: Mechanism and Spectrum of Activity. Front Microbiol. 2021;12:664395. https://doi.org/10.3389/fmicb.2021.664395.
Balta I, Butucel E, Mohylyuk V, Criste A, Dezmirean DS, Stef L, et al. Novel Insights into the Role of Probiotics in Respiratory Infections, Allergies, Cancer, and Neurological Abnormalities. Diseases. 2021;9(3):60. https://doi.org/10.3390/diseases9030060.
Huang JY, Kao CY, Liu WS, Fang TJ. Characterization of high exopolysaccharide-producing Lactobacillus strains isolated from mustard pickles for potential probiotic applications. Int Microbiol. 2017;20(2):75–84. https://doi.org/10.2436/20.1501.01.287.
Liu Y, Wang J, Wu C. Modulation of Gut Microbiota and Immune System by Probiotics, Pre-biotics, and Post-biotics. Front Nutr. 2021;8:634897. https://doi.org/10.3389/fnut.2021.634897.
Vafaeie F. Probiotics and its effect on cancers (review). Journal of Cellular Immunotherapy. 2015;1:1–3. https://doi.org/10.1016/j.jocit.2015.10.004.
Huang ML, Huang JY, Kao CY, Fang TJ. Complete genome sequence of Lactobacillus pentosus SLC13, isolated from mustard pickles, a potential probiotic strain with antimicrobial activity against foodborne pathogenic microorganisms. Gut Pathog. 2018;10:1. https://doi.org/10.1186/s13099-018-0228-y.
Mladenova I. Clinical Relevance of Helicobacter pylori Infection. J Clin Med. 2021;10(16);3473. https://doi.org/10.3390/jcm10163473.
Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J. 2016;39(1):14–23. https://doi.org/10.1016/j.bj.2015.06.002.
Lee YC, Dore MP, Graham DY. Diagnosis and Treatment of Helicobacter pylori Infection. Annu Rev Med. 2022;73:183–95. https://doi.org/10.1146/annurev-med-042220-020814.
Alba C, Blanco A, Alarcon T. Antibiotic resistance in Helicobacter pylori. Curr Opin Infect Dis. 2017;30(5):489–97. https://doi.org/10.1097/QCO.0000000000000396.
Alzahrani S, Lina TT, Gonzalez J, Pinchuk IV, Beswick EJ. Reyes VEJWjogW. Effect of Helicobacter pylori on gastric epithelial cells. 2014;20(36):12767.
Noda M, Sultana N, Hayashi I, Fukamachi M, Sugiyama MJM. Exopolysaccharide produced by Lactobacillus paracasei IJH-SONE68 prevents and improves the picryl chloride-induced contact dermatitis. Molecules. 2019;24(16):2970.
Tshibangu-Kabamba E, Yamaoka Y. Helicobacter pylori infection and antibiotic resistance - from biology to clinical implications. Nat Rev Gastroenterol Hepatol. 2021;18(9):613–29. https://doi.org/10.1038/s41575-021-00449-x.
Caliskan G, French T, Enrile Lacalle S, Del Angel M, Steffen J, Heimesaat MM, et al. Antibiotic-induced gut dysbiosis leads to activation of microglia and impairment of cholinergic gamma oscillations in the hippocampus. Brain Behav Immun. 2022;99:203–17. https://doi.org/10.1016/j.bbi.2021.10.007.
Zhu XY. Liu FJJoDD. Probiotics as an adjuvant treatment in Helicobacter pylori eradication therapy. 2017;18(4):195–202.
Qureshi N, Li P, Gu Q. Probiotic therapy in Helicobacter pylori infection: a potential strategy against a serious pathogen? Appl Microbiol Biotechnol. 2019;103(4):1573–88.
Zheng PX, Fang HY, Yang HB, Tien NY, Wang MC, Wu JJ. Lactobacillus pentosus strain LPS16 produces lactic acid, inhibiting multidrug-resistant Helicobacter pylori. J Microbiol Immunol Infect. 2016;49(2):168–74. https://doi.org/10.1016/j.jmii.2014.04.014.
Kim TS, Hur JW, Yu MA, Cheigh CI, Kim KN, Hwang JK, et al. Antagonism of Helicobacter pylori by bacteriocins of lactic acid bacteria. J Food Prot. 2003;66(1):3–12. https://doi.org/10.4315/0362-028x-66.1.3.
Alakomi HL, Skytta E, Saarela M, Mattila-Sandholm T, Latva-Kala K, Helander IM. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl Environ Microbiol. 2000;66(5):2001–5. https://doi.org/10.1128/AEM.66.5.2001-2005.2000.
Bansil R, Celli J, Hardcastle J. Turner BJFii. The influence of mucus microstructure and rheology in Helicobacter pylori infection. 2013;4:310.
Ryan KA, O’Hara AM, van Pijkeren JP, Douillard FP, O’Toole PW. Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J Med Microbiol. 2009;58(Pt 8):996–1005. https://doi.org/10.1099/jmm.0.009407-0.
Do AD, Chang CC, Su CH, Hsu YM. Lactobacillus rhamnosus JB3 inhibits Helicobacter pylori infection through multiple molecular actions. Helicobacter. 2021;26(3):e12806. https://doi.org/10.1111/hel.12806.
de Klerk N, Maudsdotter L, Gebreegziabher H, Saroj SD, Eriksson B, Eriksson OS, et al. Lactobacilli reduce Helicobacter pylori attachment to host gastric epithelial cells by inhibiting adhesion gene expression. 2016;84(5):1526–35.
Jin F, Yang H. Effects of Lactobacillus salivarius LN12 in Combination with Amoxicillin and Clarithromycin on Helicobacter pylori Biofilm In Vitro. Microorganisms. 2021;9(8):1611. https://doi.org/10.3390/microorganisms9081611.
Fazeli Z, Alebouyeh M, Rezaei Tavirani M, Azimirad M, Yadegar A. Helicobacter pylori CagA induced interleukin-8 secretion in gastric epithelial cells. Gastroenterol Hepatol Bed Bench. 2016;9(Suppl1):S42–6.
Minaga K, Watanabe T, Kamata K, Asano N, Kudo M. Nucleotide-binding oligomerization domain 1 and Helicobacter pylori infection: A review. World J Gastroenterol. 2018;24(16):1725–33. https://doi.org/10.3748/wjg.v24.i16.1725.
Garcia-Castillo V, Marcial G, Albarracin L, Tomokiyo M, Clua P, Takahashi H, et al. The Exopolysaccharide of Lactobacillus fermentum UCO-979C Is Partially Involved in Its Immunomodulatory Effect and Its Ability to Improve the Resistance against Helicobacter pylori Infection. Microorganisms. 2020;8(4);479. https://doi.org/10.3390/microorganisms8040479.
Angelin J, Kavitha M. Exopolysaccharides from probiotic bacteria and their health potential. Int J Biol Macromol. 2020;162:853–65. https://doi.org/10.1016/j.ijbiomac.2020.06.190.
Chen MJ, Chen CC, Huang YC, Tseng CC, Hsu JT, Lin YF, et al. The efficacy of Lactobacillus acidophilus and rhamnosus in the reduction of bacterial load of Helicobacter pylori and modification of gut microbiota-a double-blind, placebo-controlled, randomized trial. Helicobacter. 2021;26(6):e12857. https://doi.org/10.1111/hel.12857.
Chen ME, Su CH, Yang JS, Lu CC, Hou YC, Wu JB, et al. Baicalin, Baicalein, and Lactobacillus Rhamnosus JB3 Alleviated Helicobacter pylori Infections in Vitro and in Vivo. J Food Sci. 2018;83(12):3118–25. https://doi.org/10.1111/1750-3841.14372.
Baj J, Forma A, Flieger W, Morawska I, Michalski A, Buszewicz G, et al. Helicobacter pylori Infection and Extragastric Diseases-A Focus on the Central Nervous System. Cells. 2021;10(9):2191. https://doi.org/10.3390/cells10092191.
Shi X, Zhang J, Mo L, Shi J, Qin M, Huang X. Efficacy and safety of probiotics in eradicating Helicobacter pylori: A network meta-analysis. Medicine (Baltimore). 2019;98(15):e15180. https://doi.org/10.1097/MD.0000000000015180.
Segers ME, Lebeer S. Towards a better understanding of Lactobacillus rhamnosus GG–host interactions. Microb Cell Fact. 2014;13(Suppl 1):S7. https://doi.org/10.1186/1475-2859-13-S1-S7.
Lin CH, Shih TW, Pan TM. A “Ct contrast”-based strain-specific real-time quantitative PCR system for Lactobacilllus paracasei subsp. paracasei NTU 101. J Microbiol Immunol Infect. 2018;51(4):535–44. https://doi.org/10.1016/j.jmii.2017.05.002.
Huang J-Y, Kao C-Y, Liu W-S, Fang TJ. Characterization of high exopolysaccharide-producing Lactobacillus strains isolated from mustard pickles for potential probiotic applications. Int Microbiol. 2017;20:75–84.
Lai C-H, Fang S-H, Rao YK, Geethangili M, Tang C-H, Lin Y-J, et al. Inhibition of Helicobacter pylori-induced inflammation in human gastric epithelial AGS cells by Phyllanthus urinaria extracts. J Ethnopharmacol. 2008;118(3):522–6.
Rezaee P, Kermanshahi RK, Falsafi F. Antibacterial activity of lactobacilli probiotics on clinical strains of Helicobacter pylori. Iran J Basic Med Sci. 2019;22(10):1118.
de Klerk N, Maudsdotter L, Gebreegziabher H, Saroj SD, Eriksson B, Eriksson OS, et al. Lactobacilli Reduce Helicobacter pylori Attachment to Host Gastric Epithelial Cells by Inhibiting Adhesion Gene Expression. Infect Immun. 2016;84(5):1526–35. https://doi.org/10.1128/IAI.00163-16.
Rezaee P, Kermanshahi RK, Falsafi T. Antibacterial activity of lactobacilli probiotics on clinical strains of Helicobacter pylori. Iran J Basic Med Sci. 2019;22(10):1118–24. https://doi.org/10.22038/ijbms.2019.33321.7953.
Chun-lei Z, Jia-qi L, Hai-tao G, Jie W, Ri-hua X. Selection of exopolysaccharide-producing lactic acid bacteria isolates from Inner Mongolian traditional yoghurt. Mljekarstvo. 2014;64(4):254–60.
Torino M, Taranto M, Sesma F, De Valdez GF. Heterofermentative pattern and exopolysaccharide production by Lactobacillus helveticus ATCC 15807 in response to environmental pH. J Appl Microbiol. 2001;91(5):846–52.
Aggarwal S, Ahuja V, Paul J. Dysregulation of GABAergic Signalling Contributes in the Pathogenesis of Diarrhea-predominant Irritable Bowel Syndrome. J Neurogastroenterol Motil. 2018;24(3):422–30. https://doi.org/10.5056/jnm17100.
Zhang M, Zhu X, Zhang Y, Cai Y, Chen J, Sivaprakasam S, et al. RCAD/Ufl1, a Ufm1 E3 ligase, is essential for hematopoietic stem cell function and murine hematopoiesis. Cell Death Differ. 2015;22(12):1922–34. https://doi.org/10.1038/cdd.2015.51.
Yeh CC, Chang HI, Chiang JK, Tsai WT, Chen LM, Wu CP, et al. Regulation of plasminogen activator inhibitor 1 expression in human osteoarthritic chondrocytes by fluid shear stress: role of protein kinase Cα. Arthritis Rheum. 2009;60(8):2350–61.
Acknowledgements
The authors thank Dr. Hsiu-Chi Cheng at National Cheng-Kung University Hospital (Taiwan) for providing the human gastric adenocarcinoma epithelial cells AGS and normal human gastric epithelial cells GES-1. Control strains LGG and BCRC 14619T were gifts from Dr. Ying-Chieh Tsai at National Yang Ming Chiao Tung University (Taiwan).
Funding
This research was funded by the Ministry of Science and Technology (Taiwan), grant numbers 109–2320-B-010–036-MY3 and 110–2320-B-A49A-524-.
Author information
Authors and Affiliations
Contributions
CYK conceived and designed the experiments. TTDT, PYK, and SML performed the experiments. CYK and TTDT analyzed the data. TTDT and CYK were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors have no conflicts of interest to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1.
Growth curves of SLC13, LGG, and BCRC 14619T incubated for 24 h in MRS broth (pH 6.5) (A), acidic MRS broth (pH 3.0) (B), and MRS broth with 3% bile salts (pH 6.5) (C). Error bars represent the standard deviation of biological triplicates. Figure S2. Survival of SLC13, LGG, and BCRC 14619T was determined by counting the viable cells after 3 hours incubation in MRS broth (pH 2.0). Error bars represent the standard deviation of biological triplicates. The survival rate of LGG and BCRC 14169T in acid MRS after 3 hours incubation was compared to SLC13 in acid MRS after 3 hours incubation. ****, p < 0.0001. Figure S3. Adhesion of SLC13, LGG, and BCRC 14619T to AGS and GES-1 cells. The microscope observation of attachment of lactobacilli to AGS and GES-1 cells. The bacteria were stained with Giemsa and thus showed purple color. Figure S4. Extraction of lactobacilli exopolysaccharide. Exopolysaccharide production in the culture medium of lactobacilli SLC13, LGG, and BCRC 14619T in MRS broth containing 2% sucrose at 37oC for 24 h. Phenol-sulfuric acid method was used to measure the content of EPS using glucose as standard. EPS content was calculated according to the regression equation based on the standard curve, and then converted with dilution ratio. EPS, exopolysaccharide. NC, negative control (detection background of phenol-sulfuric mixture). Error bars represent the standard deviation of biological triplicates. ****, p <0.0001. Figure S5. Anti-H. pylori activity of lactobacilli exopolysaccharide.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Thuy, T.T.D., Kuo, PY., Lin, SM. et al. Anti-Helicobacter pylori activity of potential probiotic Lactiplantibacillus pentosus SLC13. BMC Microbiol 22, 277 (2022). https://doi.org/10.1186/s12866-022-02701-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-022-02701-z