Abstract
Background
In Ethiopia, the distribution of bovine tuberculosis (BTB) has long been known and documented as a major problem of animal health. However, the burden of circulating M. bovis strains is poorly understood in the country. Therefore; this study aimed to identify and characterize the mycobacterial isolates responsible for BTB in Northwest Ethiopia.
Methods
A cross-sectional study was conducted on tuberculous lesions that had been collected from slaughtered cattle between September 2018 to June 2019. Collected lesions were cultured and tested for tuberculous bacilli. The MPT64 assay and Genotype line probe assay (LPA) were used for identification of mycobacterial isolates, and region of deletion 4 (RD4) typing and spoligotyping were used to characterize the M. bovis strains.
Results
Of the total 1458 examined slaughtered cattle, only 62 (4.3, 95%CI; 0.0328–0.0542) had tuberculous lesions. The highest number of gross tuberculous lesions were observed from the lymph nodes of the thoracic cavity; at the mediastinal (40.3%, 25/62) and bronchial (22.6%, 14/62) lymph nodes. Of the 62 collected tuberculous lesions; 18 (29.0%) were culture positive for mycobacterium isolates, and only five isolates were confirmed for M. tuberculosis complex (MTBc) by the MPT64 assay and LPA. All the five MTBc isolates were positive for RD4 typing of M. bovis with a PCR product size of 446 bp, and no isolate was noticed to have M. tuberculosis. The detected M. bovis strains displayed five spoligotypes; with the common SB1176 and SB0133 M. bovis strains, although the two spoligotypes had not been previously reported.
Conclusion
The present study shows that BTB in North Gondar, Ethiopia, is caused by M. bovis strains SB1176 and SB0033, with low frequency. Thus, the finding highlights the importance of continuous surveillance for mycobacterial strains in cattle populations.
Similar content being viewed by others
Introduction
Bovine tuberculosis (BTB), caused by Mycobacterium bovis (M. bovis), is an infectious, chronic disease of cattle, which are known to be the primary hosts for this organism [1], although the disease has also been reported in different mammalian hosts, including humans [2]. The disease has high economic relevance within the context of livestock farming; directly affects animal productivity and also influences international trade of animal products [3, 4].
The World Organization for Animal Health (OIE) recognizes BTB as an important animal disease [1], and it presents enormous economic and public health problems in developing countries, including Ethiopia, mainly amongst the vulnerable area in rural populations [5]. The risk is supposed to be high in communities where close and frequent contact between humans and animals is common [5, 6]. According to reports [7], an estimated 68,900 new cases of zoonotic TB, due to M. bovis, occurred in Africa where pasteurization of milk is rare and BTB in cattle is common; however, M. bovis infection might be higher than the presently estimated.
In Ethiopia, the distribution of BTB in animal populations has long been described [8] and most studies also showed that BTB is prevalent mainly in cattle in wide areas of the country [9,10,11]. Absence of regular surveillance or any control measure strategies in the country aggravates the widespread and endemicity of BTB and being a threat to public health [12, 13]. In Ethiopia, different studies conducted so far on animals are based mostly on slaughterhouse reports made by researchers and veterinarians investigating gross tuberculous lesions of BTB, and also through the use of skin tuberculin testing [14], with a prevalence ranging from 4.7 to 10.2% for gross tuberculous lesions [10, 15, 16] and 9.7 to 50% when using tuberculin skin testing [13, 17]. Various studies have also described the importance of the slaughterhouse BTB surveillance in endemic situations; particularly, in countries with a low prevalence of BTB, between 5 and 15% of BTB infected herds were detected through the slaughterhouse surveillance [18,19,20,21]. However, these diagnostic tools cannot distinguish among members of the mycobacterial species, including M. bovis. A definitive examination can only be accomplished by isolation of M. bovis from clinical or post mortem samples [22, 23].
Several molecular methods, including RD4 deletion typing [24], spoligotyping [25], and other polymerase chain reaction (PCR)-based techniques [26], have been applied to genotype strains of the mycobacterial diseases and used to complement epidemiological studies on BTB. Although, in Ethiopia, few molecular studies have been used to genotype strains of the mycobacterial species and for molecular epidemiological studies on BTB [10, 27, 28]; there is still a lack of data on the strain diversity of mycobacterial species from slaughter cattle in the northwest of Amhara region. It was; therefore, aimed at molecular characterization of mycobacterial species (M. bovis strains) responsible for BTB in northwest Ethiopia.
Materials and methods
Study area
The study was conducted at Gondar ELFORA abattoir, which is located 740 km away from the capital city, Addis Ababa, in the Northwest of Ethiopia, and located at the GPS coordinates of 12° 36′ 10.8648″ N and 37° 27′ 7.6752″ E, Fig. 1. The abattoir delivers animal slaughter services for Gondar town and the surrounding community; butcher houses, restaurants, hotels, and supermarkets. Cattle; brought to the abattoir for slaughter service, were purchased at various local markets in Gondar town and surrounding livestock rich districts, and transported to the abattoir. In this area, several people are involved in smallholder businesses, supplying food animals, and animal products to the communities and the slaughterhouses. According to Central Statistical Agency [29] report, North Gondar has an estimated 3.2 million cattle, 1.3 million sheep, and 1.8 goat population, and ranked first of the total 11 zones of the Amhara regional state in its livestock population.
Study design and animals
A cross-sectional study design with a systematic random sampling method was performed between September 2018 to June 2019. For this study, all cattle that were presented for slaughter at the abattoir were enrolled as study animals. All of the cattle slaughtered at the abattoir were local zebu breed, and the sex of examined animals was male; while female animals were not slaughtered in an abattoir during this study. Subsequently, a total of 1458 cattle were examined for tuberculous lesions suspected of TB. Additionally, epidemiological data including breed, age, sex, and body condition score (BCS) of the individual animal were recorded during the study period. Body condition scoring of cattle was measured using a numerical scale ranging from 1 to 9 as described by Nicholson and Butterworth [30]; and BCS was categorized as poor (2 or 3 measurement scale), medium (4 to 6), and good (> 6).
Antemortem and postmortem inspection
Cattle were examined physically before slaughter according to the antemortem inspection protocol [31]; particularly body conditions, condition of regional lymph nodes, and visible mucous membranes were examined for the individual animal.
Postmortem examination was carried out as described in the World Organization for Animal Health [32]. Briefly, lymph nodes of the head (retropharyngeal and mandibular), lymph nodes of the thoracic cavity (mediastinal and bronchial), and mesenteric lymph nodes were visualized, palpated, and incised into small size sections and then be inspected under bright light for the presence of tuberculous lesions. Organs including lungs, liver, kidneys, and intestines were also examined for visible lesions. Additional examinations of other lymph nodes and organs or body systems were considered whenever lesions are detected. If gross lesions suggestive of BTB are found in any of the examined tissues, the slaughtered animal was classified as tuberculous lesioned. Slaughter animals without any suggestive tubercular lesions were classified as non-lesioned [33].
Sample collection and transportation
All tuberculous suspected tissue samples were collected in sterile screw-capped universal bottles with phosphate buffer saline (PBS) according to the World Health Organization (WHO) guidelines [34]. Following collection, the samples were transported by maintaining a cold chain to TB laboratory, University of Gondar Comprehensive Specialized Hospital, and kept stored at − 20 °C (a maximum of 3–4 weeks) until mycobacteriological analysis.
Isolation of mycobacteria
Samples from suspected tuberculous lesions were processed and cultured following the protocol in World Organization for Animal Disease [32]. Briefly, the specimens were further sectioned into pieces using a sterile scalpel blade, and then ground and homogenized by pestle and mortar with the gradual addition of PBS. The homogenate was then transferred into a centrifuge tube and decontaminated by adding an equal volume of 4% NaOH, followed by centrifugation at 3000 rpm for 15 min. The supernatant was discarded, and the sediment was neutralized by 1% (0.1 N) HCl using phenol red as an indicator. Neutralization was achieved when the color of the solution changed from purple to yellow. Subsequently, 0.1 ml of the sediment from each sample was spread onto a slope of Lowenstein-Jensen (LJ) medium in duplicates; one enriched with sodium pyruvate while the other enriched with glycerol. Inoculated LJ media were incubated aerobically at 37 °C for about 8 weeks with weekly observation for mycobacterial growth.
Identification of mycobacterial isolates
Preliminary identification of tubercle bacilli was done primarily with typical mycobacterial colony morphological characteristics on LJ media, and observation of acid-fast bacilli (AFB) using Ziehl–Neelsen (ZN) smear microscopy [34]. A rapid immunochromatographic MPT64 antigen test kit (SD Bioline, BD Diagnostic System, Sparks, MD) was used to identify mycobacterial species between M. tuberculosis complex (MTBc) isolates and nontuberculous mycobacteria (NTM). Strain identification was further confirmed by the Hain Genotype line probe assay (LPA) (Hain Lifescience GmbH, Nehren, Germany) [35] and Region of difference (RD4) deletion typing [24].
DNA extraction
Deoxyribonucleic acid (DNA) extraction was done from culture-positive isolates by resuspending two loopful of bacteria in 200 μl 1 × TE buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA), vortexed and boiled the mixture in a water bath for 5–10 min based on the template DNA isolation methods described elsewhere [24]. The extracted DNA suspension was stored at a − 20 °C freezer, and then later transported to Aklilu Lemma Institute of Pathobiology (ALIPB), Addis Ababa (Ethiopia) by keeping the cold chain in an icebox with an icepack for RD4-deletion typing and spoligotyping. Five μl of the DNA supernatant was used for PCR amplification.
Region of difference (RD4) deletion typing
Region of difference (RD4) deletion typing was performed following the methods described previously [24] in a final volume of 20 μl for PCR amplification. In brief, the PCR products were electrophoresed on 1.5% agarose gel, and the product size was visualized using Syngene BioImaging System (Syoptics Group) for the presence or absence of RD4. Both M. tuberculosis H37Rv and M. bovis BCG DNAs were used as positive controls and distilled water as a negative control.
Spoligotyping
Spoligotyping for MTBc strain typing was conducted following the protocol of the previous study [25]. Briefly, the amplified PCR products were hybridized to a set of 43 immobilized oligonucleotides at a spoligotype membrane for 60 min at 60 °C and followed by incubation (42 °C) in streptavidin-peroxidase conjugate for 45 min. Detection of hybridized DNA was performed by using chemiluminescent detection liquid and exposed to X-ray film. Both M. tuberculosis H37Rv and M. bovis BCG reference strains DNAs were included in each run as positive controls, and also distilled water as a negative control. The results were interpreted for the presence or absence of spacers from spoligotype images using the consensus of binary digits and an octal numbering system [25, 36].
Genotype analysis
For genotype analysis, the publicly available spoligotype database (Mbovis.org) [37] was used to assign the M. bovis genotypes. An SB number was assigned when one or more isolates in the database shared the identical spoligotype-patterns, while spoligotype-patterns that had not been designated before were named as “New pattern or unknown”.
Statistical analysis
Data analysis was performed using STATA® version 15.1 (StataCorp, USA). Descriptive statistics were used to analyze the overall proportion of tuberculous lesions and lesion frequency in different anatomical sites. Chi-square was used to evaluate the statistical significance of the associations of age and body conditions of cattle with the occurrence of tuberculous lesions. Binary logistic regressions were also used to investigate possible associations between the prevalence and the descriptive variables. A p-value of < 0.05 was considered statistically significant.
Results
Slaughter cattle characteristics and prevalence of BTB
A total of 1458 slaughtered cattle were examined in this study; all of which were male and local zebu breeds, with a higher proportion of the cattle (734, 50.3%) were grouped in the age category of 5–8 years old. The BCS showed 862 (59.1%) medium and 232 (15.9%) poor body conditioned, Table 1.
Of the total 1458 examined slaughtered heads of cattle, only 62 (4.3, 95%CI; 0.0328–0.0542) had suspected tuberculous lesions. Of these, 39 (62.9%) of the tuberculous lesions were detected in cattle age between 5 and 8 years old, Table 1. In the overall statistical analysis, suspected tuberculous lesions were most likely to be observed in cattle age category of 5–8 years old (OR = 4.23; 95%CI (1.783–10.110); p = 0.001), and in age greater than 8 years old (OR = 5.21; 95%CI (2.027–13.379); p = 0.001). Likewise, suspected tuberculous lesions were significantly higher in medium body conditioned cattle (OR = 4.42; 95%CI (1.748–11.179); p = 0.002), Table 1.
Distribution of tuberculous lesions among tissues and organs
The highest number of suspected gross tuberculous lesions were observed from the lymph nodes of the thoracic cavity; particularly at the mediastinal (40.3%, 25/62) and bronchial (22.6%, 14/62) lymph nodes, and followed by the lung tissue (17.7%, 11/62). While the least distribution of tuberculous lesions was observed from lymph nodes of the head (retropharyngeal (5, 8.1%) and submandibular (5, 8.1%) lymph nodes), Table 2. The number of lesions per inspected cattle had only a single lesion in the examined anatomical sites of the slaughtered cattle.
Isolation and identification of Mycobacterium bovis
In the present study, all 62 suspected tuberculous lesions were cultured on LJ media. Culture-positive isolates were identified in 29.0% (18/62) of the tuberculous suggestive lesions, Table 2, between 6 and 8 weeks of incubation with the majority on pyruvate enriched LJ media, 77.8% (14/18), and had flat, smooth, and non-pigmented colony characteristics. Forty samples (64.5%) had no growth in culture, and four samples were contaminated.
Among the 18 isolates, 10 (55.6%) were grouped as acid-fast bacilli (AFB) by smear microscopy (Ziehl–Neelsen) and colony characteristics; of which only five (27.8%) isolates were confirmed as members of the MTBc by the MPT64 TBAg test, Fig. 2, and Hain GenoType LPA, Table 3 and Supplement Fig. 1, while the remained 13 isolates were assumed to be non-tuberculosis mycobacteria (NTM).
RD4 deletion typing
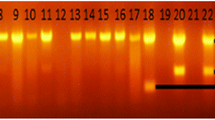
RD4 deletion typing of 18 isolates produced only five (27.8%) interpretable bands, Fig. 3. All the five isolates generated a PCR product of 446 bp, a characteristic of RD4 signal, and confirmed that they were M. bovis strains, while the remaining 13 (72.2%) isolates did not produce the band, and were identified as negative results.
Gel electrophoresis of PCR products by RD4 typing of AFB and culture-positive isolates. (Lane 1: M. tuberculosis H37Rv DNA positive control; Lane 2: Distilled water as a negative control; Lane 3: M. bovis BCG DNA positive control; Lanes 4 to 21: PCR amplicons of culture-positive isolates DNAs; Lane MW: 100 bp molecular weight marker)
Spoligotyping of MTBc strains
Spoligotyping patterns were effectively detected in the five (27.8%) M. bovis strains among 18 isolates, while 13 (72.2%) were missed all spacers; of which five isolates were microscopy smear-positive. Besides, for the five MTBc strains with both MPT64 TBAg test, Hain Genotype LPA, and RD4 typing results available, spoligotyping also produced concordant MTBc strains in 100% (5/5).
Five spoligotypes of M. bovis were identified; three of them had been previously reported in the MBovis.com database, Table 4. The common spoligotypes were SB1176 (n = 2, 40%) and SB0133 (n = 1, 20%) of the M. bovis strains, while the remained two spoligotypes were not documented in the database “unknown patterns of the M. bovis strains”. One spoligotype (SB1176) was clustered and the other three strains were unique spoligotype patterns.
Discussion
In the present study, we described the M. bovis strains among slaughtered cattle in northwest Ethiopia. The study showed that tuberculous lesions were detected in 4.3% of slaughtered cattle, which was consistent with other studies in Ethiopia [28, 38, 39], but lower than the reports elsewhere [15, 40]. Indeed, the prevalence of BTB varies from country to country, or from one place to another place within a country [14]. This variation might be linked to the type of animal production system [41, 42], and cattle breed [9, 43], while the extensive farming of zebu cattle breeds is commonly experienced in our study area that might not show the real picture of BTB prevalence. Studies have also supported that indigenous zebu cattle are relatively resistant to M. bovis infections, BTB [9, 41, 43, 44]. Our study animals were indigenous zebu cattle.
In our findings, culture-positive isolates were identified in 29% (18/62) of the tuberculous lesions, of which M. bovis was detected only in five isolates, all confirmed by MPT64 Ag assay and molecular tests, such as Hain Genotype LPA, RD4 typing, and spoligotyping. This agrees with the findings that explained M. bovis is isolated among slaughtered cattle in Ethiopia [10, 27, 28, 45]. However, our study displays a lower prevalence of M. bovis than other reports elsewhere [46,47,48,49,50]. In general, lower rates of M. bovis isolation from tuberculous lesions have been documented in the country [10], although the diagnosis of slaughter animals through abattoir-based inspection is the common practice in Ethiopia. Misdiagnosis of tuberculous lesions due to non-tuberculous mycobacteria and other granuloma-causing organisms might be the reason for a fewer number of M. bovis in our study; this is also in agreement with other studies [10, 51,52,53]. Moreover, the paucibacillary nature of calcified lesions, the influence of decontamination, and culture growth conditions in practice could also be the probable cause for the low efficiency of bacteriological isolation of M. bovis [23, 43, 51]. Nevertheless, our findings are consistent with the studies in other African countries [48,49,50], and hence revealed the distribution of M. bovis infection in cattle.
In this study, molecular analysis of the mycobacterial strains identified five spoligotypes in the Mbovis.org database [37], with the common SB1176 and SB0133 M. bovis genotypes. These M. bovis genotypes have also been reported as the most prevalent M. bovis strains in Ethiopia [10, 27, 28], and implicating the major cause of BTB circulating in Ethiopian cattle. Both M. bovis genotypes; SB1176 and SB0133, are members of the African 2 (Af2) clonal complex identified only in cattle adapted in East African countries, and characterized by the absence of spacers 3–7, 16, and 39–43 in their patterns [54]. The predominance of these M. bovis strains in cattle over this geographical area might certainly be explained by the uncontrolled free transboundary movement of the animal, probably with infected cattle or wild animals, within and between countries and neighboring areas [55, 56]. Also, the poor BTB control measures, including the absence of test and slaughter policy and insufficient quarantine measures might be the possible reasons for the spread and endemicity nature of the disease, BTB. Nevertheless, the two M. bovis spoligotypes (Octal number: 412773777000200) without assigned SB number (Mbovis.org) might therefore be emerged more recently and be distributed in our study area. It is, thus, important to note that further studies using highly polymorphic markers might need for a better knowledge of the circulating M. bovis strains in the region.
In the present study, no M. tuberculosis was isolated, however, various studies in Ethiopia have been reported both M. bovis as well as M. tuberculosis strains in cattle and humans [6, 10, 57, 58]; implicating the possible risk of M. bovis infection as zoonotic TB. In the country, due to the consumption habit of raw meat and unpasteurized milk, M. bovis should be considered to be a potential threat for human tuberculosis in a setting with a high prevalence of BTB and no control strategies.
This study had limitations. Because of the resource constraints and the limited investigation period, the study only included cattle slaughtered at the Gondar abattoir; the genotypes of M. bovis observed might not reflect the overall prevalence of these strains in the animal population of the region. In addition, we did not perform the statistical analysis for M. bovis spoligotypes distribution among slaughtered cattle, in which all cattle that were presented for slaughter were local zebu breed and male sex purchased from multiple markets in this study area.
Conclusion
The current findings show that tuberculous lesions were common in cattle slaughtered in North Gondar, Ethiopia, and present the first insight of the strains of M. bovis, confirmed by the MPT64 antigen test, Hain Genotype LPA, RD4 typing, and spoligotyping. The study also reveals that M. bovis spoligotypes; SB1176 and SB0133, were the common mycobacterial strains identified from the tuberculous lesions collected among slaughtered cattle. Although the frequency is relatively low, the identified M. bovis strain suggests circulation of the pathogen in the cattle population of the study area and continues as a public health threat. Thus, the findings highlight the importance of continuous bacteriological and molecular investigations for mycobacterial strains in animal populations, and to assist the control strategies of BTB.
Availability of data and materials
Data are available from the authors upon reasonable request.
Abbreviations
- AFB:
-
Acid-fast bacilli
- BCS:
-
Body condition score
- BTB:
-
Bovine tuberculosis
- LJ:
-
Lowenstein-Jensen
- LPA:
-
Line probe assay
- M. bovis :
-
Mycobacterium bovis
- MTBc:
-
Mycobacterium tuberculosis complex
- NTM:
-
Nontuberculous mycobacteria
- PCR:
-
Polymerase chain reaction
- RD4:
-
Region of deletion 4
- ZN:
-
Ziehl–Neelsen
References
OIE. Bovine tuberculosis. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2019. In: OIE Terrestrial Manual; 2018. p. 1058–74.
Thoen CO, Steele JH, Gilsdorf MJ. Mycobacterium Bovis Infection in Animals and Humans: Second Edition. Mycobacterium Bovis Infection in Animals and Humans: Second Edition; 2008. p. 1–329.
Tschopp R, Hattendorf J, Roth F, Choudhoury A, Shaw A, Aseffa A, et al. Cost estimate of bovine tuberculosis to Ethiopia. Curr Top Microbiol Immunol. 2013;365:249–68.
Amanfu W. The situation of tuberculosis and tuberculosis control in animals of economic interest. Tuberculosis (Edinb). 2006;86(3–4):330–5.
Olea-Popelka F, Muwonge A, Perera A, Dean AS, Mumford E, Erlacher-Vindel E, et al. Zoonotic tuberculosis in human beings caused by Mycobacterium bovis—a call for action. Lancet Infect Dis. 2017;17:e21–5.
Shitaye JE, Tsegaye W, Pavlik I. Bovine tuberculosis infection in animal and human populations in Ethiopia: a review. Vet Med (Praha). 2007;52(8):317–32.
World Health Organization. Global tuberculosis report 2019. Geneva: WHO/CDS/TB/2019.15; 2019. p. 1–283.
Hailemariam S. A brief analysis of the activities of the meat inspection and quarantine division. Addis Ababa: Ministry of Agriculture; 1975. p. 1–57.
Ameni G, Aseffa A, Engers H, Young D, Gordon S, Hewinson G, et al. High prevalence and increased severity of pathology of bovine tuberculosis in holsteins compared to zebu breeds under field cattle husbandry in Central Ethiopia. Clin Vaccine Immunol. 2007;14(10):1356–61.
Berg S, Firdessa R, Habtamu M, Gadisa E, Mengistu A, Yamuah L, et al. The burden of mycobacterial disease in ethiopian cattle: implications for public health. PLoS One. 2009;4(4):e5068.
Biffa D, Bogale A, Skjerve E. Diagnostic efficiency of abattoir meat inspection service in Ethiopia to detect carcasses infected with Mycobacterium bovis: implications for public health. BMC Public Health. 2010;10:462.
Bogale A, Tameru B, Habtemariam T. Status and control of bovine tuberculosis in Ethiopia. In: Zoonotic Tuberculosis: Mycobacterium bovis and Other Pathogenic Mycobacteria. 3rd ed; 2014. p. 109–32.
Firdessa R, Tschopp R, Wubete A, Sombo M, Hailu E, Erenso G, et al. High prevalence of bovine tuberculosis in dairy cattle in Central Ethiopia: implications for the dairy industry and public health. PLoS One. 2012;7(12):e52851.
Sibhat B, Asmare K, Demissie K, Ayelet G, Mamo G, Ameni G. Bovine tuberculosis in Ethiopia: a systematic review and meta-analysis. Prev Vet Med. 2017;147:149–57.
Demelash B, Inangolet F, Oloya J, Asseged B, Badaso M, Yilkal A, et al. Prevalence of bovine tuberculosis in Ethiopian slaughter cattle based on post-mortem examination. Trop Anim Health Prod. 2009;41(5):755–65.
Mekibeb A, Fulasa TT, Firdessa R, Hailu E. Prevalence study on bovine tuberculosis and molecular characterization of its causative agents in cattle slaughtered at Addis Ababa municipal abattoir, Central Ethiopia. Trop Anim Health Prod. 2013;45(3):763–9.
Fetene T, Kebede N. Bovine tuberculosis of cattle in three districts of northwestern Ethiopia. Trop Anim Health Prod. 2009;41(2):273–7.
Olea-Popelka FJ, Costello E, White P, McGrath G, Collins JD, O’Keeffe J, et al. Risk factors for disclosure of additional tuberculous cattle in attested-clear herds that had one animal with a confirmed lesion of tuberculosis at slaughter during 2003 in Ireland. Prev Vet Med. 2008;85(1–2):81–91.
Abernethy DA, Upton P, Higgins IM, McGrath G, Goodchild AV, Rolfe SJ, et al. Bovine tuberculosis trends in the UK and the Republic of Ireland, 1995-2010. Vet Rec. 2013;172(12):312.
Probst C, Freuling C, Moser I, Geue L, Köhler H, Conraths FJ, et al. Bovine tuberculosis: making a case for effective surveillance. Epidemiol Infect. 2011;139(1):105–12.
Guta S, Casal J, Napp S, Saez JL, Garcia-Saenz A, Perez De Val B, et al. Epidemiological investigation of bovine tuberculosis herd breakdowns in Spain 2009/2011. PLoS One. 2014;9(8):e104383.
Gormley E, Corner LAL, Costello E, Rodriguez-Campos S. Bacteriological diagnosis and molecular strain typing of Mycobacterium bovis and Mycobacterium caprae. Res Vet Sci. 2014;97(S):S30–43.
Corner LAL, Gormley E, Pfeiffer DU. Primary isolation of Mycobacterium bovis from bovine tissues: conditions for maximising the number of positive cultures. Vet Microbiol 2012;156(1–2):162–71.
Huard RC, De Oliveira Lazzarini LC, Butler WR, Van Soolingen D, Ho JL. PCR-based method to differentiate the subspecies of the mycobacterium tuberculosis complex on the basis of genomic deletions. J Clin Microbiol. 2003;41(4):1637–50.
Kamerbeek J, Schouls L, Kolk A, Van Agterveld M, Van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–14.
Warren RM. Gey Van Pittius NC, Barnard M, Hesseling a, Engelke E, De Kock M, et al. differentiation of mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int J Tuberc Lung Dis. 2006;10(7):818–22.
Biffa D, Skjerve E, Oloya J, Bogale A, Abebe F, Dahle U, et al. Molecular characterization of mycobacterium bovisisolates from Ethiopian cattle. BMC Vet Res. 2010;6(1):28.
Ameni G, Desta F, Firdessa R. Molecular typing of Mycobacterium bovis isolated from tuberculosis lesions of cattle in north eastern Ethiopia. Vet Rec. 2010;167(4):138–41.
CSA. Federal Democratic Republic of Ethiopia. Agricultural Sample Survey 2016/17, Volume II report on livestock and livestock characteristics (private). CSA. 2017.
Nichlson JM, Butherworth MH. A Guide to Body Condition Scoring of Zebu Cattle. ILCA; A monograph International Livestock Center for Africa; 1986. p. 1–30.
Ninios T, Lundén J, Korkeala H, Fredriksson-Ahomaa M. Meat inspection and control in the slaughterhouse. Ante-Mortem Inspection. Chapter 3. UK: Wiley; 2014. p. 19–28.
OIE. OIE Manual of diagnostic tests and vaccines for terrestrial animals. Paris: World Organization for Animal Health; 2012. p. 1058–74. https://www.oie.int.
Corner LA. Post mortem diagnosis of Mycobacterium bovis infection in cattle. Vet Microbiol. 1994;112:303–12.
Global Laboratory Initiative. Mycobacteriology laboratory manual. Geneva: Global Laboratory Initiative. World Health Organization; 2014. p. 1–146.
Hain Lifescience. GenoType MTBDRplus products. Mycobacteria Diagnostics. Nehren: Hain Lifesciences. GmbH; 2013.
Dale JW, Brittain D, Cataldi AA, Cousins D, Crawford JT, Driscoll J, et al. Spacer oligonucleotide typing of bacteria of the mycobacterium tuberculosis complex: recommendations for standardised nomenclature. Int J Tuberc Lung Dis. 2001;5:216–9.
Smith NH, Upton P. Naming spoligotype patterns for the RD9-deleted lineage of the mycobacterium tuberculosis complex; www.Mbovis.org. Infect Genet Evol 2012;12:873–6.
Tekle Y, Mamo G, Ameni G, Mulugeta F. Assessment of bovine tuberculosis like lesions and its risk factors in cattle slaughtered at Hawassa University and municipal abattoirs, Southern Ethiopia. J Vet Sci Med. 2017;5(2):1–9.
Aylate A, Shah SN, Aleme H, Gizaw TT. Bovine tuberculosis: prevalence and diagnostic efficacy of routine meat inspection procedure in Woldiya municipality abattoir north Wollo zone, Ethiopia. Trop Anim Health Prod. 2013;45(3):855–64.
Nemomsa B, Gebrezgabiher G, Birhanu T, Tadelle H, Tadesse G, Getachew B. Epidemiology of bovine tuberculosis in Butajira, southern Ethiopia: a cross-sectional abattoir-based study. Afr J Microbiol Res. 2014;7(33):3112–7.
Ameni G, Aseffa A, Engers H, Young D, Hewinson G, Vordermeier M. Cattle husbandry in Ethiopia is a predominant factor affecting the pathology of bovine tuberculosis and gamma interferon responses to mycobacterial antigens. Clin Vaccine Immunol. 2006;13(9):1030–6.
Tschopp R, Schelling E, Hattendorf J, Aseffa A, Zinsstag J. Risk factors of bovine tuberculosis in cattle in rural livestock production systems of Ethiopia. Prev Vet Med. 2009;89(3–4):205–11.
Diguimbaye-Djaibé C, Hilty M, Ngandolo R, Mahamat HH, Pfyffer GE, Baggi F, et al. Mycobacterium bovis isolates from tuberculous lesions in Chadian zebu carcasses. Emerg Infect Dis. 2006;12(5):769–71.
Allen AR, Minozzi G, Glass EJ, Skuce RA, Mcdowell SWJ, Woolliams JA, et al. Bovine tuberculosis: the genetic basis of host susceptibility. Proc R Soc B. 2010;277:2737–45.
Mekonnen GA, Mihret A, Tamiru M, Hailu E, Olani A, Aliy A, et al. Genotype diversity of Mycobacterium bovis and pathology of bovine tuberculosis in selected emerging dairy regions of Ethiopia. Front Vet Sci. 2020;7:5539–40.
Romha G, Ameni G, Berhe G, Mamo G. Epidemiology of mycobacterial infections in cattle in two districts of Western Tigray zone, northern Ethiopia. Afr J Microbiol Res. 2013;7(31):4031–8.
Tsegayea W, Aseffab A, Machec A, Mengistuc Y, Stefan B, Amenib G. Conventional and molecular epidemiology of bovine tuberculosis in dairy farms in Addis Ababa city, the capital of Ethiopia. Int J Appl Res Vet Med. 2010;8(2):143–51.
Müller B, Steiner B, Bonfoh B, Fané A, Smith NH, Zinsstag J. Molecular characterisation of Mycobacterium bovis isolated from cattle slaughtered at the Bamako abattoir in Mali. BMC Vet Res. 2008;4:26.
Asiimwe BB, Asiimwe J, Kallenius G, Ashaba FK, Ghebremichael S, Joloba M, et al. Molecular characterisation of mycobacterium bovis isolates from cattle carcases at a city slaughterhouse in Uganda. Vet Rec. 2009;164(21):655–8.
Ghebremariam MK, Hlokwe T, Rutten VPMG, Allepuz A, Cadmus S, Muwonge A, et al. Genetic profiling of Mycobacterium bovis strains from slaughtered cattle in Eritrea. PLoS Negl Trop Dis. 2018;12(4):e0006406.
Teklu A, Asseged B, Yimer E, Gebeyehu M, Woldesenbet Z. Tuberculous lesions not detected by routine abattoir inspection: the experience of the Hossana municipal abattoir, southern Ethiopia. OIE Rev Sci Tech. 2004;23(3):957–64.
Asseged B, Woldesenbet Z, Yimer E, Lemma E. Evaluation of abattoir inspection for the diagnosis of Mycobacterium bovis infection in cattle at Addis Ababa abattoir. Trop Anim Health Prod. 2004;36(6):537–46.
Nuru A, Zewude A, Mohammed T, Wondale B, Teshome L, Getahun M, et al. Nontuberculosis mycobacteria are the major causes of tuberculosis like lesions in cattle slaughtered at Bahir Dar abattoir, northwestern Ethiopia. BMC Vet Res. 2017;13(1):237.
Berg S, Garcia-Pelayo MC, Müller B, Hailu E, Asiimwe B, Kremer K, et al. African 2, a clonal complex of Mycobacterium bovis epidemiologically important in East Africa. J Bacteriol. 2011;193(3):670–8.
Katale BZ, Mbugi EV, Siame KK, Keyyu JD, Kendall S, Kazwala RR, et al. Isolation and potential for transmission of Mycobacterium bovis at human–livestock–wildlife Interface of the Serengeti ecosystem, Northern Tanzania. Transbound Emerg Dis. 2017;64(3):815–25.
Mekonnen GA, Conlan AJK, Berg S, Ayele BT, Alemu A, Guta S, et al. Prevalence of bovine tuberculosis and its associated risk factors in the emerging dairy belts of regional cities in Ethiopia. Prev Vet Med. 2019;168:81–9.
Ameni G, Tadesse K, Hailu E, Deresse Y, Medhin G, Aseffa A, et al. Transmission of mycobacterium tuberculosis between farmers and cattle in Central Ethiopia. PLoS One. 2013;8(10):e76891.
Gumi B, Schelling E, Berg S, Firdessa R, Erenso G, Mekonnen W, et al. Zoonotic transmission of tuberculosis between pastoralists and their livestock in south-East Ethiopia. Ecohealth. 2012;9(2):139–49.
Acknowledgments
The authors would like to acknowledge the University of Gondar (UoG), Ethiopia for financial support of this research work. We would also be grateful to the staff members of the TB culture lab at the University of Gondar and Aklilu Lemma Institute of Pathobiology, Ethiopia. In addition, we are very thankful to the veterinarians and animal health experts in the Gondar ELFORA slaughterhouse for their assistance and teamwork. Special thanks to Emeritus Professor Fusao Ota at Tokushima University for his small financial grant support through collaborative Scholarship Scientific Research program with the University of Gondar.
Funding
This study was funded by the University of Gondar, as part of the Mega Research Project with the grant number of VP/RCS/05/442/2017.
Author information
Authors and Affiliations
Contributions
Mebrat Ejo, Elias Kebede and Seleshe Nigatu conceived and designed the experiments. Mebrat Ejo, Belete Haile, and Anwar Nuru supervised the bacteriological and molecular work. Tsegaye Tariku, Belete Haile, and Mussie Girma carried out the bacteriological and molecular work (RD4 deletion typing and spoligotyping). Mebrat Ejo, Belete Haile, Tsegaye Tariku, and Abebe Belete: analyzed the data and wrote the manuscript. Seleshe Nigatu, Anwar Nuru, Yitayew Demessie, Gashaw Getaneh, and Atnaf Alebie supported the data analysis and reviewed the manuscript. Fusao Ota provided small grant support for laboratory reagents and facilities. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Research ethical support was obtained from the ethical clearance review committees of the University of Gondar before the commencement of the research activities.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no conflicts of interest
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplement Figure 1.
The Hain Genotype LPA result from 18 culture-positive isolates obtained from LJ culture.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ejo, M., Haile, B., Tariku, T. et al. Bacteriological and molecular characterization of Mycobacterium bovis isolates from tuberculous lesions collected among slaughtered cattle, Northwest Ethiopia. BMC Microbiol 21, 286 (2021). https://doi.org/10.1186/s12866-021-02349-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-021-02349-1