Abstract
Background
The burden of non-tuberculous mycobacterial (NTM) disease is increasing worldwide but still its diagnosis is delayed and it is mistaken as multidrug-resistant tuberculosis (MDR-TB).The present study was performed to develop a multiplex PCR assay for detection and identification of clinically most common NTM to the species level from pulmonary samples.
Results
Out of 50 isolates, 26 were identified as Mycobacterium kansasii (MK), 20 were identified as Mycobacterium abscessus (MA) and 4 were identified as Mycobacterium avium complex (MAC) through multiplex PCR and further confirmed by sequencing.
Conclusion
Our study showed that multiplex PCR assay is a simple, convenient, and reliable technique for detection and differential identification of major NTM species.
Similar content being viewed by others
Background
Tuberculosis (TB), caused by Mycobacterium tuberculosis complex (MTBC), persists as the principal killer disease worldwide, notably in the developing countries and has been a major public health problem in spite of considerable progress in diagnosis and treatment [1]. The genus Mycobacterium comprises several species that are divided into three groups, the MTBC, Mycobacterium leprae and atypical or non-tuberculous mycobacteria (NTM) [2]. NTM, also known as environmental mycobacteria or mycobacteria other than tuberculosis (MOTT), are mycobacteria which are generally free-living organisms and found ubiquitously in the environment [3]. There has been approximately 200 NTM species identified to-date [4]. They can cause a wide range of infections, with pulmonary infections being the most frequent (65–90%) [5].
Nowadays NTM have become important human pathogens as the incidence and prevalence of disease caused by them continue to increase worldwide [6]. The disease causing agents among NTM differ geographically, but the most common species that are frequently isolated from patients with NTM infection are Mycobacterium avium complex (MAC) (Mycobacterium avium, Mycobacterium intracellulare and Mycobacterium chimaera), Mycobacterium abscessus complex (Mycobacterium abscessus subspecies bolletii, subspecies massiliense and Mycobacterium chelonae) and Mycobacterium kansasii [7, 8]. The identification and differentiation of NTM from MTBC is of important diagnostic value as the pathogenesis and treatment regimens for these diseases are different [6, 9, 10].The varying pattern of susceptibility towards anti-TB drugs imposes need of different treatment strategies even among the NTM of same species complex [11,12,13]. Thus, rapid differentiation of MTBC from NTM and species-specific identification of NTM is crucial for proper treatment and appropriate patient management.
Usually, the prevalence of NTM infections has been notified from TB non-endemic countries and rarely from TB endemic countries because the chances of missing NTM infections are higher in TB endemic countries [14, 15]. The current standard of care for diagnostic tests does not include bacterial characterization leading to smear positive NTM cases being misclassified as MTBC. So, majority of NTM infections remain either undetected or receive chemotherapy commonly used for tuberculosis causing evolution of drug resistant NTM strains. Besides cases of mixed infection have also been reported by a few workers [16, 17].Mostly these NTM species are identified by phenotypic methods which are very cumbersome and time taking. Thus the study was designed to develop a multiplex PCR assay for detection and identification of clinically most common NTM to the species level from pulmonary samples.
Result
In this study, there were a total of 50 cultures which were smear positive but negative by capilia and GenoType MTBDR plus Assay. The multiplex PCR using four sets of primers for three unrelated mycobacterial species was successfully developed. At first multiplex PCR was tested with control strains of Mycobacterium avium complex (MAC), Mycobacterium kansasii (MK) and Mycobacterium abscessus (MA). In each strain of MAC, MK and MA we found two bands, one genus specific 688 bp and another band of 169 bp, 218 bp and 271 bp respectively for each species as shown in Fig. 1.In Mycobacterium tuberculosis(H37Rv), clinically confirmed MTBC and known strain of Mycobacterium fortuitum only one genus specific (688 bp) band was found as shown in Fig. 2. Out of 50 isolates, 26 were identified as Mycobacterium kansasii, 20 were identified as Mycobacterium abscessus and 4 were identified as Mycobacterium avium complex through multiplex PCR and further confirmed by sequencing. The sequencing data of control strains and other isolates were analyzed with the help of Basic Local Alignment Search Tool (BLAST) which finds regions of local similarity between the Internal transcribed spacer (ITS) region sequences of reference strains such as Mycobacterium avium complex: accession no.CP040255.1, Mycobacterium abscessus: accession no.CP030860.1, Mycobacterium kansasii: accession no.LR031424.1.
Discussion
In the current study, we identified the most common NTM species from pulmonary samples. Out of these NTM species 26 (52%) were identified as Mycobacterium kansasii, 20 (40%) Mycobacterium abscessus and 4(8%) as Mycobacterium avium complex by multiplex PCR and further confirmed by sequencing.
Based on only phenotypic characterization or combined with hybridization with DNA probes, the commonest species isolated from pulmonary specimens in earlier studies has been MAC. The other species were Mycobacterium abscessus, Mycobacterium xenopi, Mycobacterium kanasii, Mycobacterium chelonae and Mycobacterium fortuitum although their prevalence varied [18,19,20,21,22,23]. In contrast Wang HX, et al., [24] reported Mycobacterium chelonae (26.7%), followed by Mycobacterium fortuitum (15.4%), Mycobacterium kansasii (14.2%), Mycobacterium avium-intracellulare complex (13.1%) and Mycobacterium terrae (6.9%) which were identified with conventional biochemical tests and 16S rRNA gene sequencing in suspected pulmonary and extra pulmonary tuberculosis [24].
A study from Singapore reported the identification and differentiation of clinically relevant NTM species with two sets of multiplex PCR targeting the ITS region. They found Mycobacterium abscessus (26.3%) followed by Mycobacterium fortuitum (24.5%) and Mycobacterium avium–intracellulare complex (18.2%) [25]. According to Ito Y, et al., [26], MAC was most frequently isolated (85.9%), followed by Mycobacterium abscessus (2.8%) and Mycobacterium kansasii (1.2%) with the help of molecular techniques like COBAS Amplicor PCR assay, COBAS TaqMan MAI test (Roche Diagnostics, Basel, Switzerland) and DNA-DNA hybridization with the DDH Mycobacteria Kit (Kyokuto Pharmaceutical Industrial Co., Tokyo, Japan) [26]. A study from Saudi Arabia showed that the incidences of NTM causing pulmonary and extrapulmonary diseases were reportedly increasing and most prominent species were Mycobacterium simiae (22.6%), Mycobacterium fortuitum (18.1%), Mycobacterium abscessus (17.8%), MAC (11.2%) and Mycobacterium kansasii (3.7%). Primary species identification was carried out by line probe assays followed by sequencing [27]. Hu C, et al., [28] observed five different species of NTM causing pulmonary disease with the help of Mycobacterium Species Identification kit (PCR-reverse dot blot) of DaAn Gene company, such as Mycobacterium intracellulare (70.1%), Mycobacterium abscessus (11.5%) and Mycobacterium avium (11.5%) of isolates. A small number of cases were due to Mycobacterium kansasii, (7.5%) and Mycobacterium gordonae (1.1%) [28].
In a study from India NTM often associated with pulmonary and extrapulmonary disease, which are identified by PCR restriction analysis (PRA) of the hsp65 gene, that included Mycobacterium chelonae (28.97%), Mycobacterium fortuitum (19.62%), Mycobacterium avium complex (17.75%), Mycobacterium gordonae (11.21%), Mycobacterium terrae complex (8.41%), Mycobacterium kansasii (3.73%), Mycobacterium scrofulaceum (2.80%), Mycobacterium simiae (2.80%), Mycobacterium ulcerans (1.86%), Mycobacterium abscessus (0.93%), Mycobacterium malmoense (0.93%) and Mycobacterium phlei (0.93%) [29]. Jain S, et al., [30] found that the most common NTM species from pulmonary and extrapulmonary samples were Mycobacterium kansasii (30.1%) Mycobacterium chelonae (23.3%), Mycobacterium xenopi (15.4%), Mycobacterium scrofulaceum (7.8%), Mycobacterium avium (7.8%), Mycobacterium asiaticum (7.8%), and Mycobacterium fortuitum (7.8%) based on biochemical tests. Further they performed multiplex PCR using different primers for Mycobacterium genus (targeting hsp65), Mycobacterium tuberculosis complex (targetingESAT6), and Mycobacterium avium complex (targeting MAC) specific genes. So MAC was the only NTM which was identified by multiplex PCR [30]. A study by Sharma P, et al., [31] reported that NTM isolated from pulmonary samples were Mycobacterium intracellulare (62.5%), Mycobacterium flavescens (12.5%), Mycobacterium genavense (12.5%), and Mycobacterium gordonae (12.5%) whereas extrapulmonary NTM isolates included Mycobacterium intracellulare (6.5%), Mycobacterium abscessus (2.6%), Mycobacterium avium (1.3%), Mycobacterium mucogenicum (1.3%), Mycobacterium austroafricanum (1.3%), and Mycobacterium gordonae (10.4%). These were identified by PRA and gene sequencing [31].
Thus it can be seen that spectrum of NTM varied in different geographical regions. Molecular tools like multiplex PCR or PRA can help in rapid identification. However, PRA is costlier compared to multiplex PCR. The limitation of this study is that it includes only fifty isolates. Multiplex PCR has disadvantages that it couldn’t include more number of primers because primers inhibit each other. However, all fifty isolates could be identified by this multiplex PCR.
Conclusion
In conclusion, multiplex PCR is a simple, fast, convenient and reliable technique for identification of NTM species in the routine laboratory. This method can be used in developing countries for identification of most common NTM from pulmonary samples. To the best of our knowledge this is the first type of study conducted in India.
Methods
Study design and identification of isolates
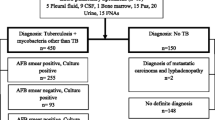
This study was undertaken in the Department of Microbiology, Institute of Medical Sciences, Banaras Hindu University, at Varanasi. It is the extension of our previous work [32] where out of 60 positive cultures, 10 (16.7%) were found positive by both GenoType MTBDR plus assay (LPA) and PCR but remaining 50 which were liquid culture (MGIT 960) positive, but capilia and LPA negative were included in this study. Control strains of Mycobacterium avium complex (MAC), Mycobacterium kansasii (MK) and Mycobacterium abscessus (MA) were obtained from National Reference Laboratory, National Institute of Tuberculosis and Respiratory Diseases, New Delhi, India.
DNA extraction
DNA isolation from the positive MGIT cultures as well as solid culture was done by CTAB-chloroform method with some modifications in BSL-3 laboratory [33, 34]. The quality and quantity of DNA were analyzed by a spectrophotometer (Thermo Scientific NanoDrop 2000).
Primer designing for the study
At first sequence of Mycobacterium tuberculosis, Mycobacterium avium complex, Mycobacterium kansasii and Mycobacterium abscessus was downloaded from National Center for Biotechnology Information (NCBI) data base. Then the genus specific primer was designed from 16S rRNA region which is specific for Mycobacterium genus. The species specific primer from the Internal transcribed spacer (ITS) region of Mycobacterium avium complex, Mycobacterium abscessus and Mycobacterium kansasii were designed. Annealing temperature and GC content were calculated for both forward and reverse complementary primer using Tm calculator Thermo Fisher Scientific software. The detail of prepared primers is enlisted in Table 1.
Development of multiplex PCR for identification of different NTM species
Initially, all fifty isolates were screened individually by targeting ITS region (MAC, MA and MK) sequences which were species-specific. This helped us to identify all the isolates at species level. Further gradient multiplex PCR was performed with 16S rRNA genus-specific primer and ITS (MAC, MA and MK) species-specific primers to know the primer working conditions. On the basis of results obtained in gradient multiplex PCR, the in house multiplex PCR was developed. This multiplex PCR targeted 16S rRNA (genus specific) and ITS (MAC, MA and MK) sequences as shown in Table 1. Different reaction mixtures were added in the PCR tube and run in thermal cycler at the amplifying conditions as shown in Table 2. The reference strain MAC, MK and MA were used as positive controls and PCR grade water was used as a negative control. The multiplex PCR was also tested against standard strain of Mycobacterium tuberculosis (H37Rv), clinically confirmed MTBC and known strain of Mycobacterium fortuitum.
Multiplex PCR running conditions
Initial denaturation step at 95 °C for 15 min followed by following parameters:
Final extension step at 72 °C for 10 min.
Sequencing
The ITS region was amplified with the help of primers for MAC, MK and MA. Product size was confirmed by agarose (2%) gel electrophoresis. Four representative strains from each species were sent for sequencing.
Sequence data analysis
The sequencing data of control strains and other isolates were analyzed with the help of Basic Local Alignment Search Tool (BLAST). BLAST finds regions of local similarity between sequences [35].
Availability of data and materials
The datasets generated and/or analyzed during the current study are not publicly available due confidentiality agreement at the department of microbiology, Institute of Medical Sciences, Banaras Hindu University but are available from the corresponding author on reasonable request.
Abbreviations
- NTM:
-
Non-tuberculous mycobacteria
- MGIT:
-
Mycobacterium Growth Indicator Tube
- TB:
-
Tuberculosis
- MTBC:
-
Mycobacterium tuberculosis complex
- MOTT:
-
Mycobacterium other than tuberculosis
- MAC:
-
Mycobacterium avium complex
- MK:
-
Mycobacterium kansasii
- MA:
-
Mycobacterium abscessus
- LPA:
-
Line Probe Assay
- NCBI:
-
National center for biotechnology information
- ITS:
-
Internal transcribed spacer
- BLAST:
-
Basic local alignment search tool
- PRA:
-
PCR restriction analysis
References
Stop TB. Partnership: a partnership hosted by United Nations at UNOPS; 2015. http://stoptb.org/assets/documents/global/plan/GlobalPlanToEndTB_TheParadigmShift_2016-2020_StopTBPartnership.pdf.
Park SC, Kang MJ, Han CH, Lee SM, Kim CJ, Lee JM, et al. Prevalence, incidence, and mortality of nontuberculous mycobacterial infection in Korea: a nationwide population-based study. BMC Pulm Med. 2019;19(1):140.
Sharma S, Dhar R. Nontuberculous mycobacterial diseases: current diagnosis and treatment. Astrocyte. 2017;4(1):67.
Johansen MD, Herrmann J-L, Kremer LJNRM. Non-tuberculous mycobacteria and the rise of mycobacterium abscessus; 2020. p. 1–16.
Porvaznik I, Solovič I, Mokrý J. Non-Tuberculous Mycobacteria: Classification, Diagnostics, and Therapy. Adv Experimental Med Biol. 2017;944:19-25.
Stout JE, Koh W-J, Yew WW. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis. 2016;45:123–34.
Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36(1):13–34.
Johnson MM, Odell JAJJotd. Nontuberculous mycobacterial pulmonary infections. J Thorac Dis. 2014;6(3):210.
Ryu YJ, Koh W-J, Daley CL. Diagnosis and treatment of nontuberculous mycobacterial lung disease: clinicians' perspectives. Tuberc Respir Dis. 2016;79(2):74–84.
Chae H, Han SJ, Kim S-Y, Ki C-S, Huh HJ, Yong D, et al. Development of a one-step multiplex PCR assay for differential detection of major mycobacterium species. J Clin Microbiol. 2017;55(9):2736–51.
Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med. 2007;175(4):367–416.
Tortoli E. Clinical manifestations of nontuberculous mycobacteria infections. Clin Microbiol Infect. 2009;15(10):906–10.
Griffith DE. Nontuberculous mycobacterial lung disease. Curr Opin Infect Dis. 2010;23(2):185–90.
Gopinath K, Singh S. Non-tuberculous mycobacteria in TB-endemic countries: are we neglecting the danger? PLoS Negl Trop Dis. 2010;4(4):e615.
Kimerling ME, Schuchter J, Chanthol E, Kunthy T, Stuer F, Glaziou P, et al. Prevalence of pulmonary tuberculosis among HIV-infected persons in a home care program in Phnom Penh, Cambodia. Int J Tuberc Lung Dis. 2002;11(6):988–94 PMID: 12475145.
Wolinsky E. Mycobacterial diseases other than tuberculosis. Clin Infect Dis. 1992;15(1):1–12.
Aliyu G, El-Kamary SS, Abimiku Al, Brown C, Tracy K, Hungerford L, Blattner W. Prevalence of non-tuberculous mycobacterial infections among tuberculosis suspects in Nigeria. PloS one. 2013;8(5):e63170.
Koh W-J, Kwon OJ, Jeon K, Kim TS, Lee KS, Park YK, et al. Clinical significance of nontuberculous mycobacteria isolated from respiratory specimens in Korea. Chest. 2006;129(2):341–8.
Dailloux M, Abalain M, Laurain C, Lebrun L, Loos-Ayav C, Lozniewski A, et al. Respiratory infections associated with nontuberculous mycobacteria in non-HIV patients. Eur Respir J. 2006;28(6):1211–5.
Marras TK, Chedore P, Ying AM, Jamieson F. Isolation prevalence of pulmonary non-tuberculous mycobacteria in Ontario, 1997–2003. Thorax. 2007;62(8):661–6.
Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182(7):970–6.
Lai C-C, Tan C-K, Chou C-H, Hsu H-L, Liao C-H, Huang Y-T, et al. Increasing incidence of nontuberculous mycobacteria, Taiwan, 2000–2008. Emerg Infect Dis. 2010;16(2):294.
Chen C-Y, Chen H-Y, Chou C-H, Huang C-T, Lai C-C, Hsueh P-R. Pulmonary infection caused by nontuberculous mycobacteria in a medical center in Taiwan, 2005–2008. Diagn Microbiol Infect Dis. 2012;72(1):47–51.
Wang HX, Jun Y, Min HA, Yang JH, Gao RL, Jing LJ, et al. Nontuberculous mycobacteria: susceptibility pattern and prevalence rate in Shanghai from 2005 to 2008. Chin Med J (Engl). 2010;123(2):184–7.
Ngan G, Ng L, Jureen R, Lin R, Teo J. Development of multiplex PCR assays based on the 16S–23S rRNA internal transcribed spacer for the detection of clinically relevant nontuberculous mycobacteria. Lett Appl Microbiol. 2011;52(5):546–54.
Ito Y, Hirai T, Fujita K, Maekawa K, Niimi A, Ichiyama S, et al. Increasing patients with pulmonary Mycobacterium avium complex disease and associated underlying diseases in Japan. J Infect Chemother. 2015;21(5):352–6.
Varghese B, Enani M, Shoukri M, AlJohani S, Al Ghafli H, AlThawadi S, et al. The first Saudi Arabian national inventory study revealed the upcoming challenges of highly diverse non-tuberculous mycobacterial diseases. PLoS Negl Trop Dis. 2018;12(5):e0006515.
Hu C, Huang L, Cai M, Wang W, Shi X, Chen W. Characterization of non-tuberculous mycobacterial pulmonary disease in Nanjing district of China. BMC Infect Dis. 2019;19(1):764.
Verma AK, Kumar G, Arora J, Singh P, Arora VK, Myneedu VP, et al. Identification of mycobacterial species by PCR restriction enzyme analysis of the hsp65 gene—an Indian experience. Can J Microbiol. 2015;61(4):293–6.
Jain S, Sankar MM, Sharma N, Singh S, Chugh T. High prevalence of non-tuberculous mycobacterial disease among non-HIV infected individuals in a TB endemic country–experience from a tertiary center in Delhi, India. Pathogens Global Health. 2014;108(2):118–22.
Sharma P, Singh D, Sharma K, Verma S, Mahajan S, Kanga A. Are we neglecting nontuberculous mycobacteria just as laboratory contaminants? Time to reevaluate things. J Pathogens. 2018;21:2018.
Singh K, Kumari R, Tripathi R, Gupta A, Anupurba S. Mutation in MPT64 gene influencing diagnostic accuracy of SD bioline assay (capilia). BMC Infect Dis. 2019;19(1):1048.
Van Embden J, Cave MD, Crawford JT, Dale J, Eisenach K, Gicquel B, et al. Strain identification of mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31(2):406–9.
Kumari R, Banerjee T, Anupurba S. Molecular detection of drug resistance to ofloxacin and kanamycin in mycobacterium tuberculosis by using multiplex allele-specific PCR. J Infect Public Health. 2018;11(1):54–8.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10.
Acknowledgements
Authors acknowledge Dr. Vithal Prasad Myneedu, National Reference Laboratory, National Institute of Tuberculosis and Respiratory Diseases, New Delhi, India for providing control strains and also Foundation of Innovative New Diagnostics (FIND) India for logistic support.
Funding
The current study is not supported by any funding agency.
Author information
Authors and Affiliations
Contributions
KS and SA designed the study. KS, RK, and RT collected and analyzed data. KS, RK, SG and SA interpreted the results. KS primarily wrote the manuscript. KS, RK, RT, SG and SA provided valuable insight for revising the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study has been ethically approved by the Institute ethical committee of Institute of Medical Sciences (Ethical committee No-ECR/Bhu/Inst/UP/2013/Re-registration-2017 dt. 31.01.2017 and Approval No- Dean/2018/EC/322), Banaras Hindu University, Varanasi.
Consent for publication
Not applicable.
Competing interests
The author(s) declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Singh, K., Kumari, R., Tripathi, R. et al. Detection of clinically important non tuberculous mycobacteria (NTM) from pulmonary samples through one-step multiplex PCR assay. BMC Microbiol 20, 267 (2020). https://doi.org/10.1186/s12866-020-01952-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-020-01952-y