Abstract
Background
Salmonella enterica serovar Enteritidis (S. Enteritidis) has emerged as one of the most important food-borne pathogens for humans. Lipopolysaccharide (LPS), as a component of the outer membrane, is responsible for the virulence and smooth-to-rough transition in S. Enteritidis. In this study, we screened S. Enteritidis signature-tagged transposon mutant library using monoclonal antibody against somatic O9 antigen (O9 MAb) and O9 factor rabbit antiserum to identify novel genes that are involved in smooth-to-rough transition.
Results
A total of 480 mutants were screened and one mutant with transposon insertion in rfbG gene had smooth-to-rough transition phenotype. In order to verify the role of rfbG gene, an rfbG insertion or deletion mutant was constructed using λ-Red recombination system. Phenotypic and biological analysis revealed that rfbG insertion or deletion mutants were similar to the wild-type strain in growth rate and biochemical properties, but the swimming motility was reduced. SE Slide Agglutination test and ELISA test showed that rfbG mutants do not stimulate animals to produce agglutinating antibody. In addition, the half-lethal dose (LD50) of the rfbG deletion mutant strain was 106.6 -fold higher than that of the parent strain in a mouse model when injected intraperitoneally.
Conclusions
These data indicate that the rfbG gene is involved in smooth-to-rough transition, swimming motility and virulence of S. Enteritidis. Furthermore, somatic O-antigen antibody-based approach to screen signature-tagged transposon mutants is feasible to clarify LPS biosynthesis and to find suitable markers in DIVA-vaccine research.
Similar content being viewed by others
Background
Salmonella enterica serovar Enteritidis (S. Enteritidis, SE) has emerged as one of the most important food-borne pathogens for humans, with poultry meat and eggs being the most common sources of human S. Enteritidis food-borne infections [1]. Young chicks showed high mortality rate when infected with S. Enteritidis. However, in adult chickens, S. Enteritidis usually leads to symptomless carriage, and is able to colonize the tissues of the ovary and oviduct of egg-laying hens which result in egg contamination [2, 3]. Above all, S. Enteritidis constitutes a risk for public health.
In Salmonella, the lipopolysaccharide (LPS), as a significant component of the outer membrane, is responsible for virulence, smoothness and for mounting cross reactivity [4]. LPS is composed of three major structures—a core polysaccharide unit; the O-antigen, a polysaccharide consisting of repeating units of sugars that extend from the cell surface; and lipid A, a potent activator of the immune response, which anchors the LPS to the outer membrane [5]. Mutations in genes that are required for the synthesis of the LPS often result in a truncated LPS [4]. Mutant strains harboring incomplete LPS due to its truncation in the polysaccharide structure may have a smooth-to-rough transition [6]. Until now, some LPS deficient mutants (for instance, rfaJ, rfaL or rfc.) have been used to prevent infection caused by fowl typhoid, Salmonella Typhimurium and Salmonella Choleraesuis [4, 7, 8]. The purpose of this study was to identify novel factors that are involved in smooth-to-rough transition in S. Enteritidis.
Signature-tagged mutagenesis (STM) is a powerful tool to identify genes that are associated with a particular phenotype. The STM technique has been applied in several pathogens to identify conditionally essential genes during infection [9–11]. In previous studies in our laboratory, Geng et al. screened an STM bank of 1800 S. Gallinarum biovar Pullorum mutants and identified the genes essential for its survival in chickens. The attenuation of 10 mutants was confirmed by in vivo and in vitro competitive index (CI) studies. One highly attenuated spiC mutant was further characterized as a candidate vaccine [11].
Different serotypes of Salmonella have different O antigens. O antigens are used for serotyping of Salmonella. O9 antigen is one of the antigens produced on Salmonella Enteritidis. Salmonella Enteritidis with smooth phenotype have O9 antigen and therefore can be agglutinated by O9 antibody [12, 13]. However, Salmonella Enteritidis without O9 antigen would show rough phenotype. In this study, we used homemade monoclonal antibody against somatic O9 antigen (O9 MAb) [14] and commercial O9 factor rabbit antiserum to screen S. Enteritidis signature-tagged transposon mutants to identify novel factors involved in smooth-to-rough transition.
Methods
Bacteria, plasmids, primers and grow media
Bacterial strains, plasmids and primers used in this study are listed in the Table 1. Wild-type Salmonella Enteritidis strain C50041 was used in this study [15]. SE C50041ΔspiC, which was constructed by suicide plasmid in previous studies in our laboratory, was used as the recipient strain to make the mutant library in this study [16]. The plasmid pUT mini-Tn5Km2 (Cm) was constructed by inserting CmR gene into pUT mini-Tn5Km2. Bacteria were grown in LB broth (Difco). When needed, this medium was supplemented with 1.5% (w/v) Bacto-agar, ampicillin (Amp, 100 μg/ml), kanamycin (Km, 50 μg/ml) and chloromycetin (Cm, 40 μg/ml).
Construction of the transposon mutant library
According to the PCR-based STM working scheme, pUT mini-Tn5Km2(Cm) was used for transformation into E. coli. χ7213-pir, which required 2,6-diaminopimelic acid (DAP) for growth. The transformants were plated on selective LB agar plates containing Amp, Km, Cm and DAP. Thus the donor strain was generated. Conjugation was performed between the donor strain and the recipient strain SE C50041ΔspiC strain as described previously [11].
Briefly, 400 μl of the donor was mixed with 400 μl of the recipient. The mixture was immobilized on a 0.45 μm pore-size membrane filter placed on LB agar at 30 °C for 24 h. Transconjugants were recovered in 2 ml phosphate-buffered saline (PBS) and a 100 μl aliquot was plated on LB agar containing Km and Cm. All the potential conjugants were analysed for exclusive pUT mini-Tn5Km2(Cm) insertion by confirmation of Amp sensitivity. Each transconjugant was grown in a 96-well plate and stored in LB containing 20% glycerol at -80 °C for further use.
Screening rough strains from mutant library
Frozen plates of the SE C50041ΔspiC transposon mutants were defrosted and subcultured by transferring 20 μl from each well to a new 96-well plate containing 180 μl of LB (containing Km and Cm). Plates were incubated overnight in a shaking incubator at 50 rpm at 37 °C. Subcultured strains were grown on LB agar containing Km and Cm at 37 °C for 16 h.
The slide agglutination tests were performed using O9 MAb developed previously in our laboratory and O9 factor rabbit antiserum (SSI®SALMONELLA ANTISERA, Denmark). AS handbook indicated, mutant culture from LB agar medium was mixed homogeneously with 1 drop of PBS and 1 drop of the O9 MAb or O9 factor rabbit antiserum on a glass slide. After the slide was tilted gently for approx. 1 min, the results were read.
Of the 480 colonies screened, a rough strain from primary screen was further verified by acriflavine agglutination test.
Identification of transposon insertion site
Chromosomal DNA was isolated from transposon mutant and completely digested with NlaIII that cut on either end of the transposon. Meanwhile, an adapter of a double-stranded cassette was generated as described previously [11]. Approximately 80 ng purified DNA from the digested DNA was ligated to 1 μg of the adapter using a DNA ligation kit (Takara, Dalian, China) in 10 μl at 16 °C for 12 h. The reaction mixture was diluted with double distilled water to 100 μl as templates in the PCR amplification. Sequencing of DNA flanking the transposon was done by Y linker and P6U primer (Table 1). DNA sequence flanking the transposon insertion site was identified by BLAST-N alignment with the recently sequenced SE P125109 in the NCBI GenBank database (GenBank accession NO. AM933172.1).
Construction of the rfbG gene deletion mutant
The knock-out mutant, SE C50041ΔrfbG was constructed by λ-Red recombination system [17]. Briefly, the rfbG gene was first substituted by a PCR adjusted antibiotic resistance cassette (Cm) using a λ-Red helper plasmid pKD46, which encode a series of phage recombinase. Recombinant clones were selected by plating on LB agar containing Cm. To resolve the antibiotic resistance cassettes (Cm), the temperature sensitive plasmid pCP20 was introduced. Finally, the rfbG gene was completely deleted from the start codon through the stop codon, as confirmed by sequencing. The slide agglutination tests were performed as above to determine whether SE C50041ΔrfbG was rough strain.
Auto-aggregation assay LPS and SDS-PAGE silver staining of the mutants
The auto-aggregation assay was performed based on the method previously described by Zhou et al. [18]. Briefly, Salmonella cultures were statically grown in 5 ml LB medium at 37 °C for 16 h in test tubes. The upper 0.2 ml was carefully removed to measure its optical density (OD600) (recorded as OD600 prevortex). The remaining culture in the test tube was then mixed by vortexing to re-suspend the aggregated cells, and 0.2 ml of the suspension was removed and its OD600 was measured (recorded as OD600 postvortex). The “percent aggregation” was calculated using the formula: 100% *(OD600 postvortex - OD600 prevortex)/OD600 postvortex.
Validation of the LPS phenotype occurred by SDS-PAGE and silver staining [19]. For this purpose LPS was isolated from SE C50041 and SE C50041ΔrfbG using a commercially available LPS extraction kit (Intron biotechnology, Gyeonggi-do, Korea). The obtained LPS was separated by standard SDS-PAGE and was stained using a pierce® silver stain kit (Thermo, Rockford, USA).
Analysis of in vitro growth and biochemical characteristics of the mutants
For in vitro growth analysis of SE C50041, SE C50041ΔspiC, SE C50041ΔspiC - rfbG::Tn5Km2(Cm) and SE C50041ΔrfbG. A single colony of each strain was subcultured in 5 ml LB broth and cultured at 37 °C with shaking at 180 rpm for at least 12 h. Subsequently the absorbance value of each strain was determined by spectrophotometry and cultures were diluted in 20 ml LB broth, then the absorbance value was determined by spectrophotometry to achieve an approx. initial concentration (OD600 = 0.05) as a starting time point (0 h). The cultures were incubated at 37 °C with shaking at 100 rpm and the OD600 was determined at time points of 0.5, 1, 1.5, 2, 2.5, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12 and 13 h. Each strain was tested in triplicate in two independent experiments. Biochemical traits of strains were tested by VITEK® 2 microbial identification system (bioMérieux, Marcy l'Etoile, France), including glucose, maltose, sucrose, mannose, mannitol, lactose, dulcitol, adonitol, sorbitol, malonate, lysine decarboxylase, ornithine decarboxylase, urea, H2S and so on.
Motility Assay
LB plates containing 0.3% (w/v) agar was used to characterize the motility phenotype of SE C50041, SE C50041ΔspiC, SE C50041ΔspiC - rfbG::Tn5Km2(Cm) and SE C50041ΔrfbG. Overnight cultures of each strain were adjusted to the same optical density. Equal volume of suspensions were incubated on spots onto 0.3% LB agar. The plates were incubated at 37 °C for 5 h, and motility was assessed by examining the migration of the bacteria from the center of the inoculation point to the periphery of the plate [20]. The data were representative of three independent experiments, which gave similar results.
Preparation and identification of sera
The SPF chickens were obtained from Poultry Institute of Shandong Academy of Agricultural Science and the chickens were detected for free from any clinical signs of enteric disease and negative for Salmonella. All chickens were given formulated commercial feed and water throughout the experimental period. Experiments were undertaken in accordance with the permission of the Animal Care and Ethics Committee of Yangzhou University.
Three-week-old chickens were inoculated intramuscularly with 100 μl of bacteria suspended in PBS solution and then a boost on day 14 after the first immunization, each bacteria inoculum was 1 × 108 CFU (intramuscularly, n = 5). Chickens were immunized respectively with SE C50041ΔspiC, SE C50041ΔspiC - rfbG::Tn5Km2(Cm) or SE C50041ΔrfbG. A group of chickens was also infected with wild-type strain SE C50041. Five control chickens received 100 μl of PBS via the same route. Sera from each animals (20 μl) were mixed with some SE C50041 and observed for agglutination reaction [4].
The flocktype® Salmonella Ab ELISA kit (QIAGEN, Leipzig, Germany) was also used to determine the presence of serum antibody to the O-antigens 1, 4, 5, 9, and 12 as handbook indicated. An S/P ratio of ≥0.3 was considered positive while <0.2 was considered negative, samples with the S/P ratio ≥0.2 and <0.3 are doubtful.
Virulence assessment
BALB/c mice were obtained from the experimental animal centre of Yangzhou University. The mice were housed in an animal facility under a standard animal study protocol. Experiments were undertaken in accordance with the permission of the Animal Care and Ethics Committee of Yangzhou University.
To investigate the virulence of SE C50041ΔrfbG in BALB/c mice (6 - 8 weeks of age), Two groups of mice (each containing 25 mice) were infected with SE C50041ΔrfbG and SE C50041. Mice in each group were further subdivided into five subgroups, each containing five mice. Each mouse in the C50041ΔrfbG group was injected intraperitoneally with 10-fold dilutions of the strain from 1 × 108 - 1 × 104 CFU in 100 μl PBS. Each mouse in the C50041 groups was injected intraperitoneally with 10-fold dilutions of the strain from 1 × 104 - 1 × 100 CFU in 100 μl PBS. Five control mice received 100 μl of PBS via the same route. Deaths were recorded up to day 14 and the LD50 of each strain was calculated using the Karber and Behrens method [21].
Statistical analysis
All statistical analyses were performed using GraphPad Prism. P values < 0.05 were considered significant when using one-way analysis of variance (ANOVA).
Results
Identification of mutant S. Enteritidis showing smooth-to-rough phenotype
Out of 480 mutants screened, 1 potential mutant was not agglutinated with O9 MAb or O9 factor rabbit antiserum. To confirm that the transposon mutant had a rough phenotype, an acriflavine agglutination test was performed and the result showed that this strain was strongly agglutinated with acriflavine (Fig. 1a).
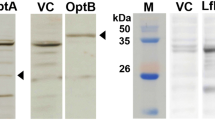
Rough strain characteristics of ΔrfbG mutants. a SDS-PAGE with silver staining of LPS of ΔrfbG mutants (SE C50041ΔrfbG and SE C50041ΔspiC - rfbG::Tn5Km2(Cm)) compared to SE C50041 or SE C50041ΔspiC. b Agglutination was examined with O9 MAb, O9 factor rabbit antiserum and acriflavine. Pictures were taken within 5 min. c Visual aggregation and “percent aggregation” of ΔrfbG mutants (SE C50041ΔrfbG and SE C50041ΔspiC - rfbG::Tn5Km2(Cm)), SE C50041 and SE C50041ΔspiC cultures grown statically for 16 h at 37 °C. 1, SE C50041; 2, SE C50041ΔrfbG; 3, SE C50041ΔspiC; 4, SE C50041ΔspiC - rfbG::Tn5Km2(Cm)
The sequence of the DNA flanking the transposon insertion site from this rough mutant was identified to be rfbG (region from 2176901 to 2177980 in SE P125109), which encodes a putative CDP-glucose 4,6-dehydratase.
Deleted mutant ΔrfbG of S. Enteritidis becoming rough pattern
The deletion mutant ΔrfbG of S. Enteritidis was constructed by λ-Red recombination system. The slide agglutination tests showed that SE C50041ΔrfbG was not agglutinated with O9 MAb or O9 factor rabbit antiserum, but was agglutinated with acriflavine (Fig. 1a). SE C50041ΔrfbG also demonstrated smooth-rough transition.
Auto-aggregation of ΔrfbG mutants and SDS-PAGE silver staining of LPS
The auto-aggregation was tested in Luria-Bertani (LB) broth (Fig. 1b) and the “percent aggregation” was calculated using OD450 measurements from these cultures. Both the SE C50041 and SE C50041ΔspiC showed 5% aggregation, then the SE C50041ΔrfbG and SE C50041ΔspiC - rfbG::Tn5Km2(Cm) demonstrated 48% and 69% aggregation, respectively (Fig. 1b).
LPS patterns obtained by standard Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of SE C50041, SE C50041ΔspiC, SE C50041ΔspiC - rfbG::Tn5Km2(Cm) and SE C50041ΔrfbG are presented in Fig. 1a. It showed a visible loss of O-antigens and Core-LPS antigens for ΔrfbG mutants (SE C50041ΔrfbG and SE C50041ΔspiC - rfbG::Tn5Km2(Cm)) compared to SE C50041 or SE C50041ΔspiC. There was no obvious difference between ΔrfbG mutant and SE C50041ΔspiC - rfbG::Tn5Km2(Cm) (Fig. 1c).
Growth and biochemical characteristics of ΔrfbG mutants
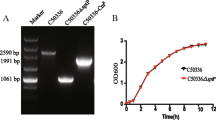
Growth curve analysis revealed no significant differences between the wild-type and each mutant when cultured in LB broth at 37 °C (Fig. 2). Results of biochemical tests including glucose, maltose, sucrose, mannose, mannitol, lactose, dulcitol, adonitol, sorbitol, malonate, lysine decarboxylase, ornithine decarboxylase, urea, H2S and so on were the same between wild-type and each mutant, suggesting mutations in these genes do not alter the biochemical characteristics of S. Enteritidis.
Motility of ΔrfbG mutants
Motility plates (Fig. 3) displayed a marked reduction in migration from the inoculation site to the periphery of the plate for SE C50041ΔrfbG (2.5 mm) and SE C50041ΔspiC - rfbG::Tn5Km2(Cm) (3.0 mm) when compared to SE C50041 (15.5 mm) and SE C50041ΔspiC (14.5 mm). The data were representative of three independent experiments, which gave similar results.
Motility assay of ΔrfbG mutants. Motility assay of the SE C50041, SE C50041ΔspiC, SE C50041ΔspiC - rfbG::Tn5Km2(Cm) and SE C50041ΔrfbG. S. Pullorum S06004, was used as a negative control on swimming agar. After 5 h of incubation at 37 °C on swimming plates, the semi-diameter of each bacterial growth area was measured. These results shown are representative of three independent experiments. The migrations of strains are highlight using red arrows
Sera tests
Sera samples were collected at 14 days after the second immunization with SE C50041ΔspiC, SE C50041ΔspiC - rfbG::Tn5Km2(Cm), SE C50041ΔrfbG, SE C50041 or PBS control. Sera collected from SE C50041ΔspiC - rfbG::Tn5Km2(Cm), SE C50041ΔrfbG and control groups were not agglutinated with SE C50041. In contrast, sera collected from the other two groups showed obvious reaction (Table 2).
Sera samples of chicks immunized with ΔrfbG mutants (SE C50041ΔspiC - rfbG::Tn5Km2(Cm) (n = 5) or SE C50041ΔrfbG (n = 5)) and control animals (n = 5) were considered Salmonella negative when using a commercially available flocktype® Salmonella Ab ELISA kit. Chicks immunized with SE C50041ΔspiC (n = 5) or SE C50041 (n = 5) were considered seropositive for Salmonella in the ELISA test (Table 2). In conclusion, ΔrfbG mutants without O-antigen do not stimulate animals to produce relevant antibody.
Virulence of ΔrfbG mutant
To investigate the role of rfbG on the virulence, mice were injected intraperitoneally with SE C50041ΔrfbG and SE C50041, and half-lethal dose (LD50) values were calculated according to the method of Karber and Behrens. The LD50 of ΔrfbG was 106.69, which was 106.6 -fold higher than that of SE C50041(100.13), implying that the virulence of the SE C50041ΔrfbG was significantly decreased (P < 0.05).
Discussion
For gram-negative bacteria, including Salmonella, LPS are essential components of immunodominant antigens. For Salmonella, some LPS deficient mutants represent a promising research area. These mutants can not only result in attenuation, but also show structural (smooth-rough) transition that can be used as a marker for distinguishing isolates [19]. Therefore, the present study was performed to identify novel factors of S. Enteritidis that are important for smooth-to-rough transition.
STM is a powerful genetic tool that allows identification of genes that are important for different facets of pathogenesis and is well suited for screening rough strain in vitro. In this study, we first attempted to use O9 MAb and O9 factor rabbit antiserum to screen for signature-tagged transposon mutants of S. Enteritidis with slide agglutination tests. We found that rfbG gene was involved in smooth-to-rough transition in S. Enteritidis. Meanwhile, we also identified some other genes that involved in smooth-to-rough transition, e.g., rfc (data not shown), which has been used as a marker for distinguishing in Salmonella enterica vaccine research [22, 23]. In line with this, this new approach of using O9 MAb and O9 factor rabbit antiserum to screen S. Enteritidis signature-tagged transposon mutants to identify novel loci involved in smooth-to-rough transition is useful and reliable.
The rfb gene cluster of S. Typhimurium contains genes that are responsible for all or part of the biosynthetic pathways of dTDP-L-rhamnose, ODP-abequose and GDP-mannose, and are essential for O-antigen biosynthesis. Among them, the rfbG gene, which encodes a CDP-glucose 4,6-dehydratase, is a component of abequose biosynthetic pathway. CDP-glucose 4,6-dehydratase and glucose-1-phosphate cytidylyltransferase (rfbF) are two enzymes required to promote the formation of CDP-4-keto-3,6-dideoxyglucose from CDP-4-keto-6-deoxyglucose (rfbH and rfbI), and abequose synthase (rfbJ) [24]. On the basis of amino-acid sequence homology, S. Typhi CDP-glucose 4,6-dehydratase is known to be a member of the short-chain dehydrogenase/reductase(SDR) superfamily, with the N-terminal domain contains a Rossmann fold and provides the platform for NAD(H) binding. The C-terminal domain is composed mostly of α-helix and houses the bingding pocket for the CDP portion of the CDP-xylose ligand. The xylose moiety extends into the active-site cleft that is located between the two domains [25]. It has also been demonstrated that the rfbG-negative Azotobacter vinelandii grown in liquid medium exhibited agglutination, suggesting that rfbG gene is involved in surface structural transition [26].
It has been described that some LPS deficient mutants (rfaJ, rfaL, rfc, et al.) were used in Salmonella DIVA-vaccine (Differentiation of Infected and Vaccinated Animals) research [19, 27, 28]. In the present study, the virulence change of the rfbG mutant strain were measured in mouse model. The result demonstrated that rfbG mutant strain is safe to mammal. Furthermore, SE Slide Agglutination test and ELISA test showed that rfbG mutants do not stimulate animals to produce agglutinating antibody, which may help to distinguish animals vaccinated with this mutant from those infected by field strains. Overall, rfbG mutant showed a potential “DIVA” capacity. Nevertheless, as a candidate vaccine, rfbG mutant needs further study.
In addition, results obtained from motility assays indicate that the deletion of rfbG gene resulted in variation in swimming motility, suggesting that flagellar assembly and function may be influenced by the altered LPS structures. Deditius et al. described that a rfaG mutant diminished flagellar assembly and significantly reduced transcription of flagellar class II and class III promoters, but not of the class I promoter. Moreover, FlhC protein levels were reduced in the rfaG mutant strain. They concluded that a defect in LPS biosynthesis regulates motility by affecting FlhDC stability or translation of its mRNA on a posttranscriptional level via an unknown mechanism [29]. Moreover, the results obtained from SDS-PAGE silver staining of LPS show that ΔrfbG mutants not only lead to loss of O-antigens but also lead to loss of Core-LPS antigens. It suggested that deficiency in rfbG leads to deep loss of LPS synthesis. The mechanism of rfbG affect LPS biosynthesis need further studies.
Conclusions
We used O9 MAb and O9 factor rabbit antiserum to screen S. Enteritidis signature-tagged transposon mutants to identify novel factors involved in smooth-to-rough transition. The present study demonstrated that rfbG gene inserted/deletion mutant of S. Enteritidis showed almost the same biological characteristics, attenuation, distinguishable reaction (agglutination and ELISA) and other rough strain characteristics. Thus, this approach may be used more broadly in exploring LPS biosynthesis, and as a high-throughput tool for screening rough strains which were used as markers in developing DIVA-vaccine.
Abbreviations
- Amp:
-
Ampicillin
- ANOVA:
-
One-way analysis of variance
- CI:
-
Competitive index
- Cm:
-
Chloromycetin
- DAP:
-
2,6-diaminopimelic acid
- Km:
-
Kanamycin
- LB:
-
Luria-Bertani
- LD50 :
-
Half-lethal dose
- LPS:
-
Lipopolysaccharide
- O9 MAb:
-
Monoclonal antibody against somatic O9 antigen
- PBS:
-
Phosphate-buffered saline
- S. Enteritidis SE:
-
Salmonella enterica serovar Enteritidis
- SDS-PAGE:
-
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- STM:
-
Signature-tagged mutagenesis
References
Patrick ME, Adcock PM, Gomez TM, Altekruse SF, Holland BH, Tauxe RV, Swerdlow DL. Salmonella enteritidis infections, United States, 1985-1999. Emerg Infect Dis. 2004;10(1):1–7.
Keller LH, Benson CE, Krotec K, Eckroade RJ. Salmonella Enteritidis colonization of the reproductive tract and forming and freshly laid eggs of chickens. Infect Immun. 1995;63(7):2443–9.
Jawale CV, Lee JH. Characterization of adaptive immune responses induced by a new genetically inactivated Salmonella Enteritidis vaccine. Comp Immunol Microbiol Infect Dis. 2014;37(3):159–67.
Lalsiamthara J, Gogia N, Goswami TK, Singh RK, Chaudhuri P. Intermediate rough Brucella abortus S19Δper mutant is DIVA enable, safe to pregnant guinea pigs and confers protection to mice. Vaccine. 2015;33(22):2577–83.
Schnaitman CA, Klena JD. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57(3):655–82.
Matiasovic J, Stepanova H, Volf J, Kubala L, Ovesna P, Rychlik I, Faldyna M. Influence of the lipopolysaccharide structure of Salmonella enterica serovar Enteritidis on interactions with pig neutrophils. Vet Microbiol. 2011;150(1-2):167–72.
Bearson BL, Bearson SM, Kich JD. A DIVA vaccine for cross-protection against Salmonella. Vaccine. 2016;34(10):1241–6.
Roman BS, Garrido V, Munoz PM, Arribillaga L, Garcia B, De Andres X, Zabaleta V, Mansilla C, Farran I, Lasa I, De Andres D, Amorena B, Lasarte JJ, Grillo MJ. The extradomain a of fibronectin enhances the efficacy of lipopolysaccharide defective Salmonella bacterins as vaccines in mice. Vet Res. 2012;43:31.
Ku YW, McDonough SP, Palaniappan RU, Chang CF, Chang YF. Novel attenuated Salmonella enterica serovar Choleraesuis strains as live vaccine candidates generated by signature-tagged mutagenesis. Infect Immun. 2005;73(12):8194–203.
Shah DH, Zhou XH, Kim HY, Call DR, Guard J. Transposon mutagenesis of Salmonella enterica serovar Enteritidis identifies genes that contribute to invasiveness in human and chicken cells and survival in egg albumen. Infect Immun. 2012;80(12):4203–15.
Geng S, Jiao X, Barrow P, Pan Z, Chen X. Virulence determinants of Salmonella Gallinarum biovar Pullorum identified by PCR signature-tagged mutagenesis and the spiC mutant as a candidate live attenuated vaccine. Vet Microbiol. 2014;168(2-4):388–94.
Mikael R, Duncan M, Pietro M, John T. Salmonella: Molecular Biology and Pathogenesis. Wymondham: Horizon Bioscience; 2007.
World Organisation for Animal Health. Manual of diagnostic tests and vaccines for terrestrial animals. Chapter 2.9.8: Salmonellosis . 2016. http://www.oie.int/en/international-standard-setting/terrestrial-manual/access-online/.
Jiao X, Zhang R, Gan J, Liu X. Production and characterization of monoclonal antibodies specific for O-antigens of Salmonella serogroups A-F. Chin J Zoonoses. 1989;5:11–4 (in Chinese).
Hu M, Yang Y, Meng C, Pan Z, Jiao X. Responses of macrophages against Salmonella infection compared with phagocytosis. In Vitro Cell Dev Biol Anim. 2013;49(10):778–84.
Zeng L, Li B, Yan S, Pan Z, Geng S, Jiao X. Construction of genetically engineered attenuated vaccines Salmonella enteritidis C50041ΔspiCΔcrp. Chin J Zoonoses. 2015;31(2):97–101 (in Chinese).
Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5.
Zhou X, Liu B, Shi C, Shi X. Mutation of a Salmonella serogroup-C1-specific gene abrogates O7-antigen biosynthesis and triggers NaCl-dependent motility deficiency. PLoS One. 2014;9(9), e106708.
Leyman B, Boyen F, Van Parys A, Verbrugghe E, Haesebrouck F, Pasmans F. Salmonella Typhimurium LPS mutations for use in vaccines allowing differentiation of infected and vaccinated pigs. Vaccine. 2011;29(20):3679–85.
Yim L, Betancor L, Martinez A, Giossa G, Bryant C, Maskell D, Chabalgoity JA. Differential phenotypic diversity among epidemic-spanning Salmonella enterica serovar enteritidis isolates from humans or animals. Appl Environ Microbiol. 2010;76(20):6812–20.
Gilles HJ. Calculation of the index of acute toxicity by the method of linear regression. Comparison with the method of "Karber and Behrens". Eur J Toxicol Environ Hyg. 1974;7(2):77–84.
Collins LV, Attridge S, Hackett J. Mutations at rfc or pmi attenuate Salmonella typhimurium virulence for mice. Infect Immun. 1991;59(3):1079–85.
Kong Q, Liu Q, Jansen AM, Curtiss R. Regulated delayed expression of rfc enhances the immunogenicity and protective efficacy of a heterologous antigen delivered by live attenuated Salmonella enterica vaccines. Vaccine. 2010;28(37):6094–103.
Jiang XM, Neal B, Santiago F, Lee SJ, Romana LK, Reeves PR. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2). Mol Microbiol. 1991;5(3):695–713.
Koropatkin NM, Holden HM. Structure of CDP-D-glucose 4,6-dehydratase from Salmonella typhi complexed with CDP-D-xylose. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):365–73.
Gavini N, Hausman BS, Pulakat L, Schreiner RP, Williamson JA. Identification and mutational analysis of rfbG, the gene encoding CDP-D-glucose-4,6-dehydratase, isolated from free living soil bacterium Azotobacter vinelandii. Biochem Biophys Commun. 1997;240(1):153–61.
Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun. 2011;79(10):4227–39.
Kwon HJ, Cho SH. Pathogenicity of SG 9R, a rough vaccine strain against fowl typhoid. Vaccine. 2011;29(6):1311–8.
Deditius JA, Felgner S, Spöring I, Kühne C, Frahm M, Rohde M, Weiß S, Erhardt M. Characterization of Novel Factors Involved in Swimming and Swarming Motility in Salmonella enterica Serovar Typhimurium. PLoS One. 2015;10(8), e0135351.
Geng SZ, Jiao XA, Pan ZM, Chen XJ, Zhang XM, Chen X. An improved method to knock out the asd Gene of Salmonella enterica serovar Pullorum. J Biomed Biotechnol. 2009;2009:646380.
Acknowledgements
The authors would like to thank Mr Yachen Hu, Ms Xiaochun Wang, Ms. Huijuan Zheng and Mr Zemiao Xia for technical support and Ms Xianyue Zhai and Ms Jiqin Xu for animal husbandry support.
Funding
This work was supported by the National Key Research and Development Program Special Project (2016YFD0501607), the Special Fund for Agroscientific Research in the Public Interest (201403054), the National Natural Science Foundation of China (nos. 31320103907 and 31230070), the Graduate Research and Innovation Projects in Jiangsu Province (KYZZ15_0366), the “Six Talent Peaks Program” of Jiangsu Province (NY-028), the Yangzhou University Science and Technology Innovation Team, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Availability of data and materials
The information supporting the conclusions of this article is included within the article.
Authors’ contributions
ZMP, XFL, SZG and YJ designed of the study; YJ and XLK performed the construction and screening of transposon mutant library screening; YJ, RXG and JLY performed the the tests for molecular and phenotypic characteristics of mutant strains; YJ, RXG, PPT and KYW performed the animal experiment; SZG, QCL, JS and XHZ analyzed the data; JJG contributed reagents/materials; ZMP, XFL, SZG, JJG, XLX and XAJ supervised the study; YJ, ZMP, XFL and XHZ wrote the paper. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
Experiments were undertaken in accordance with the permission of the Animal Care and Ethics Committee of Yangzhou University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Jiao, Y., Guo, R., Tang, P. et al. Signature-tagged mutagenesis screening revealed a novel smooth-to-rough transition determinant of Salmonella enterica serovar Enteritidis. BMC Microbiol 17, 48 (2017). https://doi.org/10.1186/s12866-017-0951-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-017-0951-4