Abstract
Background
The zoonotic agent Toxoplasma gondii is distributed world-wide, and can infect a broad range of hosts including humans. Microneme protein 16 of T. gondii (TgMIC16) is responsible for binding to aldolase, and is associated with rhomboid cleavage and presence of trafficking signals during invasion. However, little is known of the TgMIC16 sequence diversity among T. gondii isolates from different hosts and geographical locations.
Results
In this study, we examined sequence variation in MIC16 gene among T. gondii isolates from different hosts and geographical regions. The entire genomic region of the MIC16 gene was amplified and sequenced, and phylogenetic relationship was reconstructed using Bayesian inference (BI) and maximum parsimony (MP) based on the MIC16 gene sequences. The results of sequence alignments showed two lengths of the sequence of MIC16 gene among all the examined 12 T. gondii strains: 4391 bp for strains TgCatBr5 and MAS, and 4394 bp for strains RH, TgPLH, GT1, PRU, QHO, PTG, PYS, GJS, CTG and TgToucan. Their A+T content ranged from 50.30 to 50.59 %. A total of 107 variable nucleotide positions (0.1–0.9 %) were identified, including 29 variations in 10 exons and 78 variations in 9 introns. Phylogenetic analysis of MIC16 sequences showed that typical genotypes (Type I, II and III) were able to be grouped into their respective genotypes. Moreover, the three major clonal lineages (Type I, II and III) can be differentiated by PCR-RFLP using restriction enzyme Pst I.
Conclusions
Phylogenetic analysis and PCR-RFLP of the MIC16 locus among T. gondii isolates from different hosts and geographical regions allowed the differentiation of three major clonal lineages (Type I, II and III) into their respective genotypes, suggesting that MIC16 gene may provide a novel potential genetic marker for population genetic studies of T. gondii isolates.
Similar content being viewed by others
Background
Toxoplasma gondii is an important zoonotic pathogen which can infect all warm-blooded animals and humans. It is estimated that approximately one-third of human population in the world has been infected with T. gondii [1–3]. Toxoplasmosis causes several clinical syndromes such as chorioretinitis, encephalitis and systemic infections, particularly seriously in pregnant hosts and immuno-compromised individuals such as those with HIV/AIDS [4–6]. Also, toxoplasmosis can lead to considerable economic losses in livestock industry [7–9].
T. gondii isolates from different geographical locations and hosts have been grouped into 15 haplogroups that collectively define six major clades by PCR-restriction fragment length polymorphism (PCR-RFLP) [10, 11]. Despite that genotyping T. gondii strains using polymorphisms has been focused on 12 markers including SAG1, SAG3, BTUB, GRA6, c29-2, L358, PK1, 5′-SAG2, 3′-SAG2, alt. SAG2, C22-8 and Apico, additional gene (s) may contribute to some distinct T. gondii genotypes [12]. The full knowledge of genetic diversity in T. gondii is a key to better understand pathogenicity and epidemiological patterns, and thus to explore a new way for vaccination, treatment and diagnosis of toxoplasmosis.
Microneme proteins (MICs) of T. gondii play key roles in T. gondii survival and the invasion process, and thus affecting host cell signaling, as well as participating in the binding to host cell receptors [13–15]. Among those MICs, the microneme protein 16 of T. gondii (TgMIC16) is responsible for binding to aldolase, is associated with rhomboid cleavage, and thus involved in the trafficking signals during invasion [16]. Although the major functions of TgMIC16 were uncovered, little is known of the sequence variation in MIC16 gene among T. gondii isolates.
Therefore, the objectives of the present study were to examine sequence variation in MIC16 gene among 12 T. gondii strains from different hosts and geographical locations, as well as to assess whether MIC16 gene can provide a potential marker for population genetic studies of T. gondii by phylogenetic analysis and PCR-RFLP.
Methods
Genomic DNA samples of T. gondii
A total of 12 genomic DNA samples of T. gondii were used in this study (Table 1). These T. gondii DNA samples had been genotyped in our previous studies [17–19]. The DNA samples of the reference strains GT1, MAS, TgCatBr5, PTG, CTG and TgToucan were kindly provided by Associate Professor Chunlei Su at the Department of Microbiology, the University of Tennessee, Knoxville, USA.
PCR amplification
The entire genomic sequence of the MIC16 gene was amplified by PCR using a pair of specific primers (forward primer: 5′-ATGGTTGTTTCCTGTCTCTGTAC-3′; reverse primer: 5′-TTAGAGGTAGTTGTCCCGTGTCC-3′) designed based on the MIC16 gene sequence of T. gondii GT1 strain (TGGT1_289630, http://www.toxodb.org/toxo/). The amplification reaction was carried out in a volume of 25 μl containing 10 mM Tris–HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, 250 μM each of dNTP, 0.2 μM of each primer, 100–200 ng of template DNA, and 0.25 U La Taq polymerase (TaKaRa). Amplification of DNA samples from individual isolates was carried out in a thermocycler (Bio-Rad, Hercules, CA, USA) under the following conditions: the initial denaturation at 95 °C for 5 min, followed by 34 cycles consisting of 95 °C for 1 min (denaturation), 64 °C for 45 s (annealing for the two primers), 72 °C for 4 min 30 s (extension for the two primers), and a final extension step was at 72 °C for 10 min. Each (5 μl) amplification was carried out by electrophoresis on a 1 % (w/v) agarose gel with ML5000 DNA ladder marker (TaKaRa), stained with GoldenView™ and photographed using a gel documentation system (UVPGelDoc-It™ Imaging System, Cambridge, UK) [20].
Sequencing of the MIC16 amplicons
To ensure the accuracy of MIC16 sequences from different T. gondii strains, the PCR products of MIC16 gene were purified according to manufacturer’s recommendations (Wizard™ PCR-Preps DNA Purification System, Promega, USA), and then ligated with the pGEM-Teasy (Promega, USA). Thereafter, positive plasmids were transformed into the JM109 competent cells (Promega, USA). Following PCR screening, the positive colonies were sequenced by GenScript (Nanjing) Co., Ltd.
Sequence analysis and phylogenetic reconstruction
The obtained MIC16 gene sequences from different T. gondii strains were aligned using Multiple Sequence Alignment Program, Clustal X 2.11 [21], and sequence variation was determined among the examined T. gondii strains. The phylogenetic reconstructions based on the MIC16 sequences from different T. gondii strains were performed by Bayesian inference (BI) and maximum parsimony (MP). BI analyses were conducted with four independent Markov chains run for 10,000,000 metropolis coupled MCMC generations, sampling a tree every 10,000 generations in MrBayes 3.1.1 [22]. The first 250 trees were omitted as burn-in and the remaining trees were used to calculate Bayesian posterior probabilities (PP). MP analysis was performed using PAUP* 4.0b10 [23]. Bootstrap support for MP tree was calculated from 1000 bootstrap replicates with 10 random additions per replicate [24].
PCR-RFLP
PCR-RFLP was used to further assess whether MIC16 gene sequence is a potential marker for genotyping T. gondii isolates. The amplified MIC16 fragments were digested with restriction enzyme Pst I for 8 h at 37 °C in the appropriate buffer, and then inactivated at 80 °C for 20 min according to the manufacturer’s instructions (NEB). The restriction fragments were analyzed by electrophoresis on 1 % agarose gel stained with GoldenView™ and photographed using a gel documentation system (UVP GelDoc-It™ Imaging System, Cambridge, UK).
Results
Sequence analysis
All the positive transformants of the MIC16 gene were approximately 4000 bp in length on 1 % (w/v) agarose gel (not shown). The MIC16 gene sequences were 4391 bp in length for strains TgCatBr5 and MAS; and they were 4394 bp in length for strains RH, TgPLH, GT1, PRU, QHO, PTG, PYS, GJS, CTG and TgToucan. Their A+T content varied from 50.30 to 50.59 %. Alignment of the obtained MIC16 sequences from 12 T. gondii strains revealed nucleotide polymorphisms at 107 sequence variation positions (0.1–0.9 %), including 78 variations in 9 introns, 29 variations in 10 exons (Additional file 1: Figure S1). In addition, a total of 20 transversions (A<−>T/C<−>G/T<−>G/A<−>C), 76 transitions (C<−>T /A<−>G), 7 deletions and 4 inserts were identified in MIC16 gene (Additional file 1: Figure S1).
Sequences analysis of polymorphisms in the three references genotypes (RH/GTI, PRU/QHO/PTG, CTG) revealed the existence of polymorphic restriction sites. Using PCR-RFLP method, digestion of the amplification products with Pst I allowed the differentiation between genotype I, II, and III (Fig. 1).
Phylogenetic analysis of T. gondii strains based on MIC16 sequences
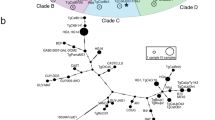
By phylogenetic reconstruction based on MIC16 sequence data of all 12 strains, we have obtained the phylogenetic tree (Fig. 2). Phylogenetic analysis revealed three major clusters, which are corresponding to classical genotypes (I, II, III) respectively, and Type III are clustered more closely with type I than with other strains. Atypical strains TgToucan were phylogenetically linked to Type III, and thus the both strains GYS and PYS were phylogenetically linked to Type II.
PCR-RFLP analysis of MIC16 genomic sequences of Toxoplasma gondii isolates in 1 % agarose gel using restriction endonucleases Pst I. Lane M represents DNA size marker 5000. Lanes 1–7 represent T. gondii Type I (GTI, RH, TgPLH), Type II (PRU, QHO, PTG), Type III (CTG) strains respectively. Refer to Table 1 for isolate information
Discussion
In the present study, we determined the entire genome region of the MIC16 locus for 12 T. gondii strains among different host and geographical origins, and determined genetic diversity between Toxoplasma strains based on MIC16 gene sequences. It has been found that there were a total of 107 variable positions distributed in the genome sequence of the MIC16 locus among 14 isolates, including 29 variations in 10 exons and 78 variations in 9 introns. These results suggested higher sequence variations in MIC16 gene than that in MIC4, MIC13 and eIF4A genes [25–27]. The analyses of MIC16 gene in both nucleotides and amino acids among 12 T. gondii strains showed high ratio of non-synonymous to synonymous polymorphisms, emphasized that MIC16 was undergoing positive selection together with some other loci, such as GRA5, GRA6 and ROP17 [24, 28, 29].
With the purpose of analyzing the evolutionary relationship among the 12 examined T. gondii isolates, the phylogenetic tree was established (Fig. 1) based on the MIC16 sequence using BI and MP methods. Phylogenetic analysis showed that T. gondii strains corresponding to classical genotypes (I, II, III) were grouped into their respective clusters separately, which were consistent with the results from other genetic markers including GRA6 and GRA5 genes [24, 28].
Moreover, the three major clonal lineages (Type I, II and III) can be differentiated by digestion of MIC16 PCR products using endonuclease Pst I (Fig. 2), which further suggested that the MIC16 locus may be a potential new genetic marker for PCR-RFLP genotyping of T. gondii strains. However, some atypical strains including TgCatBr5, TgToucan, PYS, GJS and MAS can not be distinguished from the three major lineage types by PCR-RFLP (not shown), suggesting that PCR-RFLP genotyping with a single locus is not enough for the identification of non-clonal genotypes.
Conclusions
The present study examined sequence variation in MIC16 and demonstrated the existence of sequence variability in MIC16 gene among the examined T. gondii strains from different hosts and geographical localities. Phylogenetic analysis and PCR-RFLP genotyping of the examined T. gondii strains suggest that MIC16 gene may provide a potential marker for population genetic studies of T. gondii isolates, and further research sampling more isolates from different hosts and geographical separations need to be done to verify this possibility.
Abbreviations
AIDS, acquired immune deficiency syndrome; BI, Bayesian inference; HIV, human immunodeficiency virus; MP, maximum parsimony; PCR, polymerase chain reaction; PCR-RFLP, PCR-restriction fragment length polymorphism; TgMIC16, Toxoplasma gondii microneme protein 16
References
Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet. 2004;363:1965–76.
Nardoni S, Angelici MC, Mugnaini L, Mancianti F. Prevalence of Toxoplasma gondii infection in Myocastor coypus in a protected Italian wetland. Parasit Vectors. 2011;4:240.
Schlüter D, Däubener W, Schares G, Groß U, Pleyer U, Lüder C. Animals are key to human toxoplasmosis. Int J Med Miobiol. 2014;304:917–29.
Weiss LM, Dubey JP. Toxoplasmosis: a history of clinical observations. Int J Parasitol. 2009;39:895–901.
Dubey JP. Toxoplasmosis of animals and humans. 2nd ed. Boca Raton: CRC press; 2010. p. 1–313.
Zhou P, Chen Z, Li HL, Zheng H, He S, Lin RQ, et al. Toxoplasma gondii infection in humans in China. Parasit Vectors. 2011;4:165.
Fusco G, Rinaldi L, Guarino A, Proroga YTR, Pesce A, Cringoli G. Toxoplasma gondii in sheep from the Campania region (Italy). Vet Parasitol. 2007;149:271–4.
Innes EA. A brief history and overview of Toxoplasma gondii. Zoonoses Public Health. 2010;57:1–7.
McAuley JB. Congenital toxoplasmosis. J Pediatric Infect Dis Soc. 2014;3:30–5.
Dardé ML, Bouteille B, Pestre-Alexandre M. Isoenzyme analysis of 35 Toxoplasma gondii isolates and the biological and epidemiological implications. J Parasitol. 1992;78:786–94.
Su C, Khan A, Zhou P, Majumdar D, Ajzenberg D, Dardé ML, et al. Globally diverse Toxoplasma gondii isolates comprise six major clades originating from a small number of distinct ancestral lineages. Proc Natl Acad Sci U S A. 2012;109:5844–9.
Dubey JP, Van WK, Verma SK, Choudhary S, Kwok OCH, Khan A, et al. Genotyping Toxoplasma gondii from wildlife in Pennsylvania and identification of natural recombinants virulent to mice. Vet Parasitol. 2014;200:74–84.
Muniz-Feliciano L, Van Grol J, Portillo JA, Liew L, Liu B, Carlin CR, et al. Toxoplasma gondii-induced activation of EGFR prevents autophagy protein-mediated killing of the parasite. PLoS Pathog. 2013;9, e1003809.
Soldati D, Meissner M. Toxoplasma as a novel system for motility. Curr Opin Cell Biol. 2004;16:32–40.
Friedrich N, Santos JM, Liu Y, Palma AS, Leon E, Saouros S, et al. Members of a novel protein family containing microneme adhesive repeat domains act as sialic acid-binding lectins during host cell invasion by apicomplexan parasites. J Biol Chem. 2010;285:2064–76.
Sheiner L, Santos JM, Klages N, Parussini F, Jemmely N, Friedrich N, et al. Toxoplasma gondii transmembrane microneme proteins and their modular design. Mol Microbiol. 2010;77:912–29.
Zhou P, Zhang H, Lin RQ, Zhang DL, Song HQ, Su C, et al. Genetic characterization of Toxoplasma gondii isolates from China. Parasit Int. 2009;58:193–5.
Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology. 2010;137:1–11.
Zhou P, Nie H, Zhang LX, Wang HY, Yin CC, Su C, et al. Genetic characterization of Toxoplasma gondii isolates from pigs in China. J Parasitol. 2010;96:1027–9.
Xu Y, Zhang NZ, Chen J, Liu GH, Xu QM, Zhou DH, et al. Toxoplasma gondii rhoptry protein 38 gene: sequence variation among isolates from different hosts and geographical locations. Genet Mol Res. 2014;13:4839–44.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82.
Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4.
Swofford DL. Paup*: phylogenetic analysis using parsimony, version 4.0b10. 2003.
Chen J, Li ZY, Zhou DH, Liu GH, Zhu XQ. Genetic diversity among Toxoplasma gondii strains from different hosts and geographical regions revealed by sequence analysis of GRA5 gene. Parasit Vectors. 2012;5:279.
Peng GH, Yuan ZG, Zhou DH, He XH, Yan C, Yin CC, et al. Sequence variation in Toxoplasma gondii MIC4 gene and protective effect of an MIC4 DNA vaccine in a murine model against Toxoplasmosis. J Anim Vet Adv. 2010;9:1463–8.
Ren D, Zhou DH, Xu MJ, Zhou Y, Yang JF, Lin ZP, et al. Sequence variation in Toxoplasma gondii MIC13 gene among isolates from different hosts and geographical locations. Afr J Microbiol Res. 2012;6:1333–7.
Chen J, Fang SF, Zhou DH, Li ZY, Liu GH, Zhu XQ. Sequence variation in the Toxoplasma gondii eIF4A gene among strains from different hosts and geographical locations. Genet Mol Res. 2014;13:3356–61.
Fazaeli A, Carter PE, Darde ML, Pennington TH. Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. Int J Parasitol. 2000;30:637–42.
Zhang NZ, Xu Y, Huang SY, Zhou DH, Wang RA, Zhu XQ, et al. Sequence variation in Toxoplasma gondii rop17 gene among strains from different hosts and geographical locations. Sci World J. 2014;2014:349325.
Acknowledgements
None.
Funding
Project support was provided, in part, by the National Natural Science Foundation of China (Grant Nos. 31228022 and 31172316), and The Science Fund for Creative Research Groups of Gansu Province (Grant No. 1210RJIA006).
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional file. GenBank accession numbers for these T. gondii MIC16 sequences are KX147274-KX147285. Study Accession URL of deposition of our phylogenetic data in TreeBase is http://purl.org/phylo/treebase/phylows/study/TB2:S19062.
Authors’ contributions
XQZ and JC conceived and designed the study and revised the manuscript. WGL, XPX and SLL performed the experiments and analyzed the data. QMX helped in the study design and manuscript revision. WGL and XPX wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Author information
Authors and Affiliations
Corresponding authors
Additional file
Additional file 1: Figure S1.
Alignment of MIC16 gene sequences of 12 Toxoplasma gondii strains. Lower case letter “a” indicates the base changes in exons; Lower case letter “b” indicates the base changes in introns. (JPEG 87 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Liu, WG., Xu, XP., Chen, J. et al. MIC16 gene represents a potential novel genetic marker for population genetic studies of Toxoplasma gondii . BMC Microbiol 16, 101 (2016). https://doi.org/10.1186/s12866-016-0726-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-016-0726-3