Abstract
Background
Green and blue mold decay, caused by Penicillium digitatum and P. italicum, respectively, are important postharvest diseases of citrus. Biocontrol by microbes is an alternative to synthetic fungicide application. In this study, the antagonistic yeast strain Kloeckera apiculata 34-9 was used to investigate the action mechanisms involved in the biocontrol of postharvest diseases.
Results
An antifungal substance, 2-phenylethanol (PEA), was isolated from K. apiculata and demonstrated to have antimicrobial activity against selected phytopathogenic fungi. Experiments on P. italicum cells identified the mitochondria and the nucleus as particularly sensitive to inhibition. Regulation of P. italicum gene expression was investigated using RNA-Seq. PEA up-regulated genes involved with the peroxisome, regulation of autophagy, phosphatidylinositol signaling system, protein processing in endoplasmic reticulum, fatty acid metabolism, and inhibited ribosome, RNA polymerase, DNA replication, amino acid biosynthesis, aminoacyl-tRNA biosynthesis and cell cycle. Inhibitory responses revealed by RNA-Seq suggest that PEA might compete for attachment on the active site of phenylalanyl-tRNA synthetase (PheRS).
Conclusion
This study provided new insight on the mode of action of biocontrol yeast agents in controlling postharvest pathogenic fungi.
Similar content being viewed by others
Background
Penicillium digitatum and P. italicum, the causal agent of green and blue mold decay, respectively, are important postharvest diseases of citrus and cause heavy losses around the world [1]. Biological control using microbial agents (bacteria, yeast and fungi) is considered to be a viable alternative to the use of synthetic fungicides. Among microbial agents, yeasts have several good properties that make them ideal antagonists, including the ability to survive in adverse environmental conditions, having few nutritional requirements and being amenable to formulation with a long shelf-life [2],[3]. Furthermore, yeast strains can degrade mycotoxins, which are well known for being toxic to humans and animals [4]. Over 30 yeasts have been isolated and investigated for their biocontrol efficacy against postharvest fruit diseases. Some yeast-based products have been registered as commercially available biocontrol agents such as "Aspire" (Candida oleophila 182; Ecogen, Langhorne, PA, USA) [5]. Knowledge about the modes of action of biocontrol agents is essential for developing appropriate commercial formulations and application methods to maximize the potential use of biocontrol agents [6].
Several mechanisms have been proposed to explain the antifungal activity of biocontrol agents. Wound colonization and nutrient competition appear as the primary mechanisms [7]-[11]. Other attributes of yeast that have been associated with their biocontrol activity include the production of antifungal compounds (lytic enzymes, killer toxins, peptides and antibiotic metabolites) [6],[12]-[18]. The most thoroughly studied example is farnesol from Candida albicans[19], which can inhibit various bacteria and fungus [20]. Production of antimicrobial compounds is not restricted to Candida; they can also be found in other yeast genera: Cryptococcus, Saccharomyces, Hanseniaspora, Hansenula, Kluyveromyces, Pichia, Rhodotorula, Tilletiopsis and Meyerozyma[11],[21],[22].
Recently, several studies have focused on antifungal compounds from natural sources as an effective alternative to chemical preservatives, e.g., phenylacetic acid (PAA), phenyllactic acid (PLA) and phenylethanol (PEA). PEA, a colorless liquid with a rose-like odour, occurs widely in nature, including in a variety of essential oils extracted from rose, jasmine, carnation and, hyacinths [23]. Greater attention has been paid to studying the antimicrobial properties of PEA [24]-[27], and in addition screening of yeasts for the production of PEA for natural products in the cosmetic and food industry [28],[29].
In spite of the information available in literature, few extensive isolation studies of antibiotic antifungal compounds from antagonistic yeast are available. The objective of this study is to identify and characterize of antifungal compounds from the antagonistic yeast strain K. apiculata 34-9, and study their fungistatic activities.
Results
Extraction of antifungal substances produced by K. apiculata
In vitro, K. apiculata 34-9 showed antagonistic properties against P. digitatum and P. italicum (Figure 1A) in PDA medium. Meanwhile, antifungal substances were efficiently extracted by ether from both the cell-free culture and cells of K. apiculata (Figure 1B).
Effect of K. apiculata and the (ether) extract on Penicillium molds. A: The agar diffusion assay was performed of K. apiculata against P. italicum (i) and P. digitatum (ii) after 48 h co-cultured at 28°C; B: P. italicum (i) and P. digitatum (ii) incubated for 48 h at 28°C after addition of 10 μL of the extract.
Antifungal activity was not affected by trypase, proteinase K (100 μg/ml, 37°C for 60 min), or high temperature (140°C for 10 min) treatments, but it was sensitive to alkaline pH (data not shown). The activity was stable at pH values between 2.5 and 5.5, but it rapidly decreased between 5.5 and 11.0. A peak of inhibition was observed at the end of log phase (20 h), which is produced by intracellular extraction, and four hours later (24 h) the same peak is produced by extracellular extraction and four hours later (28 h) there was another little increase (Figure 2). The result showed four hours delay between antifungal compound biosynthesis and secretion.
Relationship of population dynamics of K. apiculata to intra/extra-cellular antifungal activity. Effect of the number of K. apiculata on antifungal compounds secretion was investigated by statistic correlation between the cells number and antifungal activity that was extracted from the cell-free culture (extracellular) and cells of K. apiculata (intracellular) respectively. The assays were performed in 50-mL BSM broth at 28°C with 1.0 × 108 cells/mL of K. apiculata initially. Samples were analyzed for the number of K. apiculata, extra- and intracellular activity at intervals of 2 h. Antifungal effects were recorded with inhibition diameter (mm) after 48 h culture at 28°C in PDA medium (2.0 × 105 spore/mL P. italicum spores).
Purification and identification of antifungal substances
The raw extract was first purified by thin-layer chromatography (TLC) eluting with ether and benzene (1:1, v/v). The retention factor (Rf) value of active fractions was 0.74. We collected these active fractions and further separated them by petroleum ether and ethyl acetate (4:1, v/v). Iodine vapor showed that only a white spot (Rf = 0.53) on a light brown background had antifungal activity (Figure 3A). The white spot was collected for the further purification. HPLC revealed that retention time (RT) of this antifungal compound was 5.079 min (Figure 3B).
Purification of the antifungal compound. A: Silica plate TLC purification. The TLC separation was performed with petroleum ether: ethyl acetate (4:1, v/v) and visualized using iodine vapor. Then compounds in the TLC were directly transferred to PDA medium (mixed with 2.0 × 105 spore/mL P. italicum spores) by blotting. The spot of 1, 2 and 3 were antifungal compound (white) in TLC (RF:0.53), its corresponding antifungal spots in PDA. B: semi-preparative HPLC purification. The HPLC were performed with C18 reversed-phase column, 2:3 methanol-H2O (containing 0.1% acetic acid), and 210 nm detection C: LC-MS analysis. The LC-MS was analyzed with C18 reversed-phase column, ion source temperature, gas temperature 200°C, nebulizer 15 psi and HV capillary 3500 V. (i) 220.8 Tandem MS Chromatogram, (ii) 121.9 tandem MS chromatogram, (iii) Total ion chromatogram (TIL, green) and extracted Ion Chromatogram (EIC, red) for 122 and 221.
The collected sample from semi-preparative HPLC was analysed by LC-MS. Through the exploration of conditions, the purified sample indicated prominent m/z ions at 122 and 221, and their extracted ion chromatogram (EIC) were in keeping with the peak of antifungal compound of LC-MS total ion chromatogram (TIL) and DAD spectrogram. The MS/MS revealed that m/z 122 and 221 derived from the same compound (Figure 3C). The sample was then analyzed by GC-EI-MS. Consequently, this antifungal compound was determined to be 2-phenylethanol (PEA; RT = 16.674 min) (Figure 4). In addition, other compounds, including 2-phenylacetic acid (PAA; RT = 17.338 min), phenyllactate (RT = 17.961 min), and phenylpyruvate (RT = 19.243 min), were identified in the raw extract, revealing antifungal substances produced by K. apiculata were closely related to the L-phenylalanine (L-Phe) metabolic pathway.
Identification of the structure of antifungal compound by GC-MS. Purified sample were analyzed by GC-MS with HP-5MS capillary column chromatography after derivatized with N,O-Bis(trimethylsilyl) trifluoroacetamide. Operating conditions: carrier gas (helium) 1 mL/min, split ratio 50:1, injector temperature and interface temperature 280°C, and oven temperature programmed at 40°C (2 min) and ramped to 280°C at 5°C/min, 70 eV. 16.674 min: PEA (Silane, trimethyl(2-phenylethoxy)); 16.960 min: glycerol (Trimethylsilyl ether of glycerol).
L-Phe metabolic pathway of K. apiculata was analyzed using [2-13C] isotope labeling of L-Phe. Figure 5 showed that the largest signal was 13C-labeled PEA, followed by 13C-labeled 2-phenylacetaldehyde (PAD), PAA and phenylpyruvate. The concentration of PEA in culture increased with time (Figure 5C-ii), whereas 13C-labeled PAD decreased with the time (Figure 5C-i). This confirmed that PEA was produced via L-Phe, and that PAD is an intermediate product of PEA [30].
L-Phe metabolic pathway analysis of K. apiculata by [2- 13 C] L-Phe labeling using GC-MS. A: Total ion chromatograph (TIC) of K. apiculata. (B-I) 13.097 min MS and (B-II) PEA standard MS. (C) Total ion chromatograph (TIC) of 2-phenylacetaldehyde (PAD; 10.09 min) and 2-phenylethanol (PEA;13.10 min) for different culture periods in dextrose minimal medium with [2-13C] L-Phe as the sole nitrogen source (0.76% YNB, 2% dextrose, 2% [2-13C] L-Phe). (1), (2), (3), (4) and (5) were 3 h, 6 h, 12 h, 24 h and 48 h analysis results after inoculation, respectively.
Antifungal activity of PEA and PAA
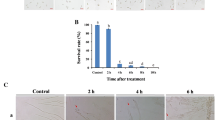
Growth of blue and green molds was inhibited in vitro and in vivo by 10 μL of 1.5 μL/mL PEA. In fruit storage assay, 1.5 μL/mL PEA also effectively protected citrus from infection by susceptible strains of green and blue molds (Figure 6A,B), while not affecting fruit quality factors such as ascorbic acid, soluble solid contents, and titratable acid (data not shown). The incidence of blue and gray molds for PEA treatment was 11.3% after 110 d, which was similar to that achieved with prochloraz (10.7%) (Figure 6C).
We also analyzed the antifungal activity of the L-Phe metabolite, PAA, which is known to possess antifungal properties [31]. We found that PAA inhibited the pathogenic fungus in vitro and in vivo, but it did not effectively control the occurrence of disease in storage (Figure 6C). This result further confirmed PEA as the main antifungal compound of K. apiculata.
Effect of PEA on fungal cells
Transmission electron microscopy (TEM) showed that the action of the raw extract (1000 × dilute for 2 h) on ultra-cellular P. italicum was closely associated with mitochondrial abnormalities (Figure 7), including degraded and disorganized cristae, leakage of the outer membrane, and massive mitochondrial vacuolation. Meanwhile, PEA (1.5 μL/mL for 2 h) induced hyphal cells with abnormal subcellular morphology (Figure 7). The prominent features were massive mitochondrial vacuolation and vacuole mediated organelle degradation.
Effect of the extract and 2-phenylethanol (PEA) on subcellular components of P. italicum . P. italicum was cultured in PDA liquid medium for 8 h, the extract (1000 × dilute) or 1.5 μL/mL PEA were added to medium and further with 2 h culture. (m) mitochondrion, (N) nucleus, (n) nucleolus, (cw) cell wall, (cm) cell membrane, (er) endoplasmic reticulum, white arrow (1) mitochondrialcristae, (2) mitochondria abnormalities and vacuolation, (3) organelles degradation, (4) leakage of the outer membrane of mitochondria.
To test whether PEA's inhibition of fungal cells was associated with a change in the permeability of the cytoplasmic membrane, 4',6-diamidino-2-phenylindole (DAPI) and propidium iodide (PI) staining and membrane electric conductivity were performed (Figure 8). PI is a membrane-impermeable stain in normal healthy cells, but it readily penetrates the membranes of dead cells. As a control, no PI staining in the nucleus was detected in the strain with or without PEA treatment (Figure 8A), consistent with results on conductivity, suggesting that the hyphae were intact (Figure 8B). Similar result was found in hyphae of Aspergillus flavus in the presence of PEA [32].
RNA-Seq profiling of the response of P. italicum to PEA
Regulation of gene expression was investigated using comparative RNA-Seq profiling analysis. Samples of 0 h (CK), 1 h (PEA1) and 3 h (PEA3) treatment were used for the construction of RNA-Seq libraries. After removal of reads with adaptors, unknown bases, low-quality reads (leaving tags of 49 nt long), there were total of 11,605,180, 11,611,106, and 11,672,232 successful sequences (clean reads), produced by PEA1, PEA3, and CK, respectively. The distribution of total clean tags was quite similar at PEA1, PEA3, and CK (Additional file 1).
Clean reads were mapped to reference sequences using SOAPaligner/soap2 [33], and mismatches of no more than 2 bases were allowed in the alignment. A total of mapped genes in PEA1, PEA3, and CK were 6905, 6946, and 7051 of the reference genes (Additional file 2). Analysis of differential expression at PEA1 and PEA3 compared to CK revealed 861 and 1160 up-regulated genes, 1072 and 749 down-regulated genes, respectively. Of these, 654 and 650 genes (68.3% of total) share the same up- and down-regulation expression pattern in PEA1 and PEA3. Gene ontology categories were assigned to the 1304 genes with significantly differential expression using the Blast2GO program (http://www.blast2go.org) to evaluate the potential functions of genes that showed significant transcriptional differences between CK and PEA treatment (Figure 9). Seventeen gene categories were defined. Major categories were metabolism (543), cellular (372), localization (94), and regulation (39). The significant enrichment categories classified on the basis of molecular function were catalytic activity (547), binding (510), structural molecule activity (62) and transporter activity (46). The genes were classified on the basis of cellular compound into cell (409), cell part (409), organelle (157), macromolecular complex (109), organelle part (51), membrane-enclosed lumen (19), envelop (9), virion (1) and virion part (1). Among them, membrane-enclosed lumen, macromolecular complex and organelle were major subcellular organelles that responded to PEA stress.
Functional categorization of the genes with significant transcriptional changes between the PEA treatment and the control type. The total same changed genes (1304) were categorized based on Gene Ontology (GO) annotation. The proportion of each category is displayed based on: Biological process; Molecular function; Cellular component.
The PEA-responsive genes were further assessed using KEGG pathway analysis. A total of 17 different metabolic pathways were found with more than 3 affiliated genes, of which some were consistent with biological processes that were already identified by GO analysis. The most represented pathways are listed in Table 1. The prominent related pathways were ribosome, amino acids biosynthesis, aminoacyl-tRNA biosynthesis, cell cycle, protein processing in endoplasmic reticulum, RNA polymerase, and DNA replication. Data indicated that ribosome, endoplasmic reticulum and nucleus were the major subcellular organelles in response to PEA, which is in accord with TEM data and the physiological indices.
To confirm that the DEGs identified by deep sequencing were indeed differentially expressed, a total of 8 genes were chosen for confirmation in a biologically independent experiment using qRT-PCR, including ribosome, autophagy, proteasome and fatty acid synthesis-related genes, which were detected in the transcriptome and bioinformatic analyses. The relative transcript abundance patterns for the CK and PEA treatment were compared using the transcriptome data. The results of qRT-PCR revealed similar expression patterns as the Illumina sequencing despite some quantitative differences in the expression levels (Figure 10).
Real-time quantitative RT-PCR confirmation of the differentially expressed genes between the PEA treatment and the control. The results were showed by relative transcript times, which acquired by comparison of the transcript abundance of PEA treatment with control. The transcript abundance from RNA-seq data is shown by black columns; Relative transcript levels are calculated by real-time PCR (grey columns) with β-tubulin as the standard (means ± s.d.).
Discussion
Various mechanisms have been proposed to explain the biocontrol of antagonistic yeast to fungal pathogens. Prominent among these is the suggestion that yeast competes with pathogens for nutrients and space [7]. As the safest microbes, yeasts are common on the surfaces of fruits and vegetables, which have used in food preparation for millennia. Previous reports suggest that the antagonistic yeast usually does not depend on the production of antibiotics, but rather on their ability to colonize and grow rapidly in surface wounds [5],[34]. Here, we identified an antifungal compound PEA from K. apiculata 34-9 that produced by Ehrlich pathway from L-Phe, and their potential application in the field of citrus postharvest pathology is unexplored.
As an aromatic alcohol, PEA has been approved for use to modify certain flavor compositions of foods in the USA. The acute toxicity LD50 (1700 mg/kg for rat) of PEA was classified as low toxicity (24,28). It inhibits a range of bacteria, such as Bacillus subtilis, Ralstonia solanacearum and Escherichia coli[23],[24]. In E. coli, PEA appears to inhibit the syntheses of DNA [35], and an increase in membrane fluidity caused by PEA has been demonstrated in Myxococcus xanthus[36]. Also, PEA inhibits synthesis of macromolecules and phospholipids metabolism [37],[38]. Lester [25] showed that PEA exhibited activity in vitro against fungi Neurospora crassa, and inhibition of growth and of the syntheses of RNA, DNA and protein. Furthermore, PEA inhibits RNA, DNA, protein and aminoimidazole ribotide syntheses, cytoplasmic respiratory, and glucose uptake and incorporation in yeast [39]. Zhu et al. [23] inferred that PEA competes for attachment on the active site of the enzyme with L-3,4-dihydroxyphenylalanine due to its -OH group.
Although the mode of action of PEA on bacteria and fungi has been described, our study represents the first comprehensive transcriptome study of the inhibition process. Illumina sequencing was used to monitor the global transcriptional change in the PEA treatment compared with the control, and 1304 differentially expressed genes that were induced or repressed by more than two fold at different treatment times, were identified. RNA-Seq data showed that the action of PEA on P. italicum resembles its effect on N. crassa and yeast; ribosome, mitochondrion, macromolecular complex, endoplasmic reticulum and nucleus were the major subcellular organelles in response to PEA.
A number of new genes possibly related with the inhibition process were found in this study. Functional category analysis revealed that a number of important pathways may work collaboratively in inhibiting the fungal cell growth. The first noticeable pathway is the amino acid and protein biosynthesis pathways which exist in the plastids. 97 out of 99 DGEs of ribosome and aminoacyl-tRNA synthetases (AARSs), and 31 out of 36 DGEs of amino acid biosynthesis were down-regulated (Table 1). Based on these data, it can be speculated that the inhibition of the amino acids and protein biosynthesis resulted in inhibition. The structure of PEA and RNA-Seq data from PEA-inhibited cultures lead us to suppose that the PEA might compete for attachment on the active site of fungi phenylalanyl-tRNA synthetase (PheRS) by the formation of a stable tRNAPhe-PEA [40],[41], thus inhibiting protein synthesis, RNA transcription and energy demanding processes.
The second group of PEA-responsive pathways includes DNA replication, meiosis and cell cycle pathways. Most of these genes also exhibited down-regulation pattern in PEA treatment. The induction of genes associated with cell cycle is consistent with the GO results that indicate that large numbers of genes are located in nucleus; and is in line with our physiological analysis that the nucleus is one of the major subcellular organelles in response to PEA.
The third group of metabolite pathways represents those related with cell death including phagosome, proteasome, peroxisome and regulation of autophagy. Most of these genes were up-regulated indicated that PEA induced fungal cells autophagy or programmed cell death.
Our study also showed that PEA and PAA were identified in the raw extract, and both of them are known to have antimicrobial properties. Both PEA and PAA inhibited citrus green and blue molds in vitro and in vivo. However, PAA was ineffective in fruit storage. Fungus pathogens suppress host cell defense responses by acidification of the fruit with organic acids, such as citric and gluconic [42]-[45]. Maybe, PAA causes low pH in fruit tissue.
Conclusions
We have identified an antifungal compound from biocontrol agent K. apiculata 34-9 and provided a global picture of the gene expression changes in a PEA treatment comparing with control type. The interpretation of the Illumina sequencing data uncovered a large number of genes with previously not known to be involved in the inhibition process. Functional categorization of the differentially expressed genes showed that a number of important pathways, including amino acids and protein biosynthesis, cell cycle and cell death cross communicated and worked collaboratively in inhibiting phytopathogenic fungi. This study provided new insight into the mode of action of biocontrol yeast agents in controlling postharvest pathogenic fungi.
Methods
Antagonist and fungal pathogens
Strain of K. apiculata 34-9 was isolated from the rhizosphere soil [46]. The strain was grown in BSM (20% bean sprout extract, 5% dextrose, 2% agar), YPD (1% yeast extract, 2% peptone, 2% dextrose, 2% agar), and minimal medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate (YNB; Difco), 2% dextrose, 2% L-phenylalanine). The molds of P. digitatum and P. italicum were used as test fungus, which were cultured in PDA (20% potato extract, 2% dextrose, 2% agar).
Collection and extraction of antifungal compounds
K. apiculata was grown in BSM broth at 28°C for 48 hour with shaking at 200 rpm. After incubation, cells were removed by centrifugation at 8000 × g for 10 min and sterile filtered (0.45 μm). The cell-free culture was extracted using a series of organic solvents (1:1; v/v), including cyclohexane, petroleum ether, benzene, chloroform, ether, acetic ether, and n-butyl alcohol. Each extraction was performed twice and extracts were pooled and concentrated using a rotary evaporation (Laborota model 4010, Heidolph, Germany) leaving a yellow oil as the product, which was used directly for in vitro assaying of antifungal activity.
Citrus fruit
Navel orange fruit (Citrus sinensis L. Osbeck) were harvested from the orchard (Yichang, Hubei, China) for in vivo and fruit storage assays. Fruits without physical injuries and infections were selected based on uniformity in size. Prior to use, fruits were disinfected with 2% (v/v) NaOCl solutions for 2 min, rinsed with tap water and air-dried.
Antagonism assays
In vitro, in vivo and fruit storage assaying of antifungal activity were conducted with two biological replicates as described previously [46].
Briefly, in vitro antifungal activity was carried out using a disc-diffusion method. Petri plates were prepared with 15 mL of PDA medium. PEA (1.5 μL/mL, 10 μL), PAA (1 mM, 10 μL) and sterile distilled water (10 μL) were pipetted into the 5 mM (diameter) holes of agar punched in the agar plates with inoculum of 1.0 × 105 conidia/mL of Penicillium suspension.
In vivo, orange fruits were wounded with a bodkin to a 5 mM depth with two wounds per orange. PEA (1.5 μL/mL, 10 μL), 45% prochloraz (PCZ) (1500 × dilute, 10 μL), PAA (1 mM, 10 μL) and sterile distilled water were pipetted onto each wound after inoculation of 10 μL of a 5.0 × 105 conidia/mL of Penicillium suspension. After treatment, the fruits were placed in plastic chambers to maintain a high relative humidity of approximately 95% and maintained at 25°C. The percentage of disease inhibition was determined by measuring the diameter of the lesions produced by the fungus in each fruit wound after five days.
For fruit storage assay, individual fruits were dipped for 5 min in PEA (1.5 μL/mL), 45% prochloraz (PCZ) (1500 × dilute), PAA (1 mM) and sterile distilled water (control). After air dry, fruits were directly stored in a ventilated warehouse (about 5-10°C) for natural storage test. Visual decay was evaluated after 3 months of storage. Each treatment was replicated three times with 100 fruit samples per replicate.
Influence of K. apiculata growth on production of antifungal compounds
Effect of the number of K. apiculata on antifungal compounds secretion was investigated comparing the cells number and antifungal activity that extracted respectively from the cell-free culture (extracellular) and cells of K. apiculata (intracellular). The assays were performed in 50-mL BSM broth at 28°C with 1.0 × 107 cells/mL of K. apiculata initially. Samples were analyzed the number of K. apiculata, extra- and intracellular activity at intervals of 2 hour.
Extracellular extraction of antifungal compound was performed as described above. For intracellular cells, yeast cells were collected by centrifugation at 8000 × g for 10 min and washed twice. The cells were ground into a powder in liquid nitrogen, and then added sterile distilled water to original cultivated volume for extraction.
Purification and identification of antifungal compounds
The (ether) extract was analyzed by thin-layer chromatography (TLC; silica gel G plates) in the following solvent 50% ether, and 50% benzene. The second separation was performed with 75% petroleum ether, and 25% ethyl acetate. The plates were visualized using I2 vapor. The active fractions were eluted with methanol and concentrated using rotary evaporation and pooled. These were then purified by C18 reverse-phase HPLC (Agilent 1200; Santa Clara, CA, USA) employing a C18 column (4.6 mm× 250 mm; 5-μm; Elite, Dalian, China). The column was eluted at 1 mL/min, with an optimized concentration using 40% methanol and 60% H2O (0.1% (v/v) acetic acid). Chromatograms were scanned at 210 nm. The antifungal activity was examined for each substance collected in each fraction.
Separation of the purified compounds for mass spectrometry analysis was achieved with an Agilent 1100 series LC/MSD Trap. The separation was carried out with C18 column (4.6 mm× 150 mm; 5 μm; Agilent). Mass spectrometry operating were ion source temperature and gas temperature 200°C at a drying gas flow 5.00 L/min, nebulizer 15 psi and HV capillary 3500 V. Full scan spectra from m/z 50 to 1000 in both positive and negative ion modes were recorded.
GC-MS analysis of the purified compounds was performed by Agilent 6890A/5975C and equipped with HP-5MS (30 m× 0.25 mm; 0.25 μm; Agilent) capillary column. Helium was used as carrier gas at a flow rate of 1 mL/min and split ratio 50:1. The injector and interface temperature was set to 280°C. The analysis was performed under the following temperature program: 2 min of isothermal heating at 40°C and then ramped to 280°C at 5°C/min. 70 eV of electron energy was used for simple ionization. Mass spectra were scanned from m/z 50 to 1000. In addition, the purified compounds were also analyzed by GC-MS after reacted with N,O-Bis(trimethylsilyl) trifluoroacetamide (BSTFA, Fluka) at 100°C for 15 min.
Microscopic observation of the fungus
The fungus for the transmission electron microscope (TEM) samples were harvested and fixed overnight in 2.5% glutaraldehyde at room temperature. After washing with in a 1 M PBS (pH 7.2), the specimens were postfixed in 1% osmium tetroxide buffer for 2 h, washed again, and then dehydrated by a graded acetone series (30-100%). The samples were embedded in Spurr's low-viscosity resin for 48 h at 45°C. Semithin and ultrathin sections (40-60 nm) were cut with a Leica Ultracut RM2265 (Leica, Vienna, Austria), mounted on regular hexagonal copper grids, stained with lead citrate (10 min), washed three times with ddH2O, stained with uranyl acetate for 30 min, washed again, and examined with a JEOL H-7650 TEM (Hitachi High-Technologies Co., Japan).
K. apiculata L-phenylalanine-related metabolite assay
Experiments were performed in minimal medium containing 2% [2-13C] L-phenylalanine (L-Phe) (99 atom%; Cambridge Isotope Laboratories, Cambridge, MA) as single nitrogen source. Liquid medium inoculated with 1.0 × 107 cells/mL was grown at 28°C for 3 hour, 6 hour, 12 hour, 24 hour and 48 hour, respectively. The concentrations of the various end products of L-Phe metabolites in culture filtrates were determined by using GC-MS, as described above.
6-diamidino-2-phenylindole (DAPI) and propidium iodide (PI) staining
Fungal nuclei to be observed by fluorescence microscopy were stained with DAPI and PI. After 12 hour of growth and 1.5 μL/mL PEA treatment for 2 hour, strains were fixed and stained with DAPI and PI as described previously [18]. Conductivity detection was performed as Wang et al. [47].
Penicillium RNA extraction, Illumina sequencing and data analysis
Untreated (CK), and PEA-treated samples (PEA1 and PEA3 refer to 1.5 μL/mL-treated fungus for 1 hour and 3 hour, respectively), were harvested. Total RNA was extracted from P. italicum by Trizol (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. RNA-Seq profiling was performed by Beijing Genomics Institute (Shenzhen, China). Brifly, mRNA was enriched by using oligo(dT) magnetic beads. The fragmentation buffer was added and mRNA was interrupted to approximately 200 bp. The first and second strand cDNA was synthesized by using reverse transcriptase and random hexamer-primer. Double-strand cDNA was purified with QiaQuick PCR extraction kit and washed with EB buffer for end repair and single nucleotide adenine addition. Finally, sequencing adapters were ligated to the fragments. The required fragments were purified by agarose gel electrophoresis and enriched by PCR amplification. The sequences of the library products were analyzed by using an Illumina HiSeq™ 2000.
Clean reads were generated by removing adapter sequences, unknown bases more than 10% and low quality reads. Each tunnel generated 11.6 million reads with a sequencing length of 49 bp. All clean reads were then aligned to reference sequences of Penicillium chrysogenum Wisconsin 54-1255 by using SOAPaligner/soap2 [33]. Mismatches no more than 2 bases were allowed in the alignment. The expression level of gene (Additional file 2: Table S1) was calculated by using the RPKM (reads per kb per million reads) method [48]. Differentially expressed genes (DEG; Additional file 2: Table S1) in three samples were analyzed as described [49]-[52].
Real-time quantitative RT-PCR (qRT-PCR) verification
Eight genes were chosen for confirmation by qRT-PCR with SYBR Premix Ex Taq™ (Takara, Japan). Primers for the chosen genes were designed with the Primer Express software (Applied Biosystems, USA) and are presented in Additional file 3. qRT-PCR for gene expression analysis was performed on a StepOne Real-time PCR System (Applied Biosystems, USA) using β-tubulin gene as an endogenous control. Briefly, the primers for the target gene and β-tubulin were diluted in the SYBER Mix (Applied Biosystems) and 20 μL of the reaction mix were added to each well. The reactions were performed with an initial incubation at 50°C for 2 min and at 95°C for 1 min followed by 40°Cycles of 95°C for 15 s and 60°C for 1 min. The levels of gene expression were analyzed with StepOne Software v2.0. Zero template controls were included for each primer pair. Each PCR reaction was carried out in triplicate, and the data are presented as the means ± SD.
Additional files
References
Marcet-Houben M, Ballester AR, de la Fuente B, Harries E, Marcos JF, González-Candelas L, Gabaldón T: Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics. 2012, 13: 646-10.1186/1471-2164-13-646.
Sundh I, Melin P: Safety and regulation of yeasts used for biocontrol or biopreservation in the food or feed chain. Antonie Van Leeuwenhoek. 2011, 99: 113-119. 10.1007/s10482-010-9528-z.
Wilson CL, Wisniewski ME: Biological control of postharvest diseases of fruit and vegetables: an emerging technology. Annu Rev Phytopathol. 1989, 27: 425-441. 10.1146/annurev.py.27.090189.002233.
Patharajan S, Reddy KRN, Karthikeyan V, Spadaro D, Gullino ML, Garibaldi A: Potential of yeast antagonists on in vitro biodegradation of ochratoxin A. Food Control. 2011, 22: 290-296. 10.1016/j.foodcont.2010.07.024.
Janisiewicz WJ, Korsten L: Biological control of postharvest diseases of fruits. Annu Rev Phytopathol. 2002, 40: 411-441. 10.1146/annurev.phyto.40.120401.130158.
Zhang D, Spadaro D, Valente S, Garibaldi A, Gullino ML: Cloning, characterization, expression and antifungal activity of an alkaline serine protease of Aureobasidium pullulans PL5 involved in the biological control of postharvest pathogens. Int J Food Microbiol. 2012, 153: 453-464. 10.1016/j.ijfoodmicro.2011.12.016.
Liu P, Luo L, Long CA: Characterization of competition for nutrients in the biocontrol of Penicillium italicum by Kloeckera apiculata. Biol Control. 2013, 67: 157-162. 10.1016/j.biocontrol.2013.07.011.
Liu P, Fang JF, Chen K, Long CA, Cheng YJ: Phenylethanol promotes adhesion and biofilm formation of the antagonistic yeast Kloeckera apiculata for the control of blue mold on citrus. FEMS Yeast Res. 2014, 14: 536-546. 10.1111/1567-1364.12139.
Liu P, Hao HH, Long CA: The effects of Kloeckera apiculata on the cuticular waxes of navel orange fruit. Biol Control. 2014, 73: 16-22. 10.1016/j.biocontrol.2014.03.006.
Vero S, Garmendia G, Gonzalez MB, Bentancur O, Wisniewski M: Evaluation of yeasts obtained from antarctic soil samples as biocontrol agents for the management of postharvest diseases of apple (Malus × domestica). FEMS Yeast Res. 2013, 13: 189-199. 10.1111/1567-1364.12021.
Hershkovitz V, Sela N, Taha-Salaime L, Liu J, Rafael G, Kessler C, Aly R, Levy M, Wisniewski M, Droby S: De-novo assembly and characterization of the transcriptome of Metschnikowia fructicola reveals differences in gene expression following interaction with Penicillium digitatum and grapefruit peel. BMC Genomics. 2013, 14: 168-10.1186/1471-2164-14-168.
Mari M, Martini C, Spadoni A, Rouissi W, Bertolini P: Biocontrol of apple postharvest decay by Aureobasidium pullulans. Postharvest Biol Tec. 2012, 73: 56-62. 10.1016/j.postharvbio.2012.05.014.
Coda R, Rizzello CG, Di Cagno R, Trani A, Cardinali G, Gobbetti M: Antifungal activity of Meyerozyma guilliermondii: identification of active compounds synthesized during dough fermentation and their effect on long-term storage of wheat bread. Food Microbiol. 2013, 33: 243-251. 10.1016/j.fm.2012.09.023.
Friel D, Pessoa NMG, Vandenbo M, Jijakli MH: Separate and combined disruptions of two exo-β-1,3-glucanase genes decrease the efficiency of Pichia anomala (strain K) biocontrol against Botrytis cinerea on apple. Mol Plant Microbe Interact. 2007, 20: 371-379. 10.1094/MPMI-20-4-0371.
López-García B, González-Candelas L, Pérez-Payá E, Marcos JF: Identification and characterization of a hexapeptide with activity against phytopathogenic fungi that cause postharvest decay in fruits. Mol Plant Microbe Interact. 2000, 13: 837-846. 10.1094/MPMI.2000.13.8.837.
Lorenzon EN, Sanches PRS, Nogueira LG, Bauab TM, Cilli EM: Dimerization of aurein 1.2: effects in structure, antimicrobial activity and aggregation of Candida albicans cells. Amino Acids. 2013, 44: 1521-1528. 10.1007/s00726-013-1475-3.
Munoz A, Lopez-Garcia B, Marcos JF: Studies on the mode of action of the antifungal hexapeptide PAF26. Antimicrob Agents Chemother. 2006, 50: 3847-3855. 10.1128/AAC.00650-06.
Santos A, San Mauro M, Bravo E, Marquina D: PMKT2, a new killer toxin from Pichia membranifaciens, and its promising biotechnological properties for control of the spoilage yeast Brettanomyces bruxellensis. Microbiology. 2009, 155: 624-634. 10.1099/mic.0.023663-0.
Semighini CP, Hornby JM, Dumitru R, Nickerson KW, Harris SD: Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol Microbiol. 2006, 59: 753-764. 10.1111/j.1365-2958.2005.04976.x.
Liu P, Luo L, Guo JH, Liu HM, Wang BQ, Deng BX, Long CA, Cheng YJ: Farnesol induces apoptosis and oxidative stress in the fungal pathogen Penicillium expansum. Mycologia. 2010, 102: 311-318. 10.3852/09-176.
Calvente V, Orellano ME, Sansone G, Benuzzi D, Tosetti ML: Effect of nitrogen source and pH on siderophore production by Rhodotorula strains and their application to biocontrol of phytopathogenic moulds. J Ind Microbiol Biotechnol. 2001, 26: 226-229. 10.1038/sj.jim.7000117.
Urquhart EJ, Punja ZK: Hydrolytic enzymes and antifungal compounds produced by Tilletiopsis species, phyllosphere yeasts that are antagonists of powdery mildew fungi. Can J Microbiol. 2002, 48: 219-229. 10.1139/w02-008.
Zhu YJ, Zhou HT, Hu YH, Tang JY, Su MX, Guo YJ, Chen QX, Liu B: Antityrosinase and antimicrobial activities of 2-phenylethanol, 2-phenylacetaldehyde and 2-phenylacetic acid. Food Chem. 2011, 124: 298-302. 10.1016/j.foodchem.2010.06.036.
Fraud S, Rees EL, Mahenthiralingam E, Russell AD, Maillard JY: Aromatic alcohols and their effect on gram-negative bacteria, cocci and mycobacteria. J Antimicrob Chemother. 2003, 51: 1435-1436. 10.1093/jac/dkg246.
Lester G: Inhibition of growth, synthesis, and permeability in Neurospora crassa by phenethyl alcohol. J Bacteriol. 1965, 90: 29-37.
Lucchini JJ, Bonnaveiro N, Cremieux A, Le Goffic F: Mechanism of bactericidal action of phenethyl alcohol in Escherichia coli. Curr Microbiol. 1993, 27: 295-300. 10.1007/BF01575995.
Mo EK, Sung CK: Phenylethyl alcohol (PEA) application slows fungal growth and maintains aroma in strawberry. Postharvest Biol Tec. 2007, 45: 234-239. 10.1016/j.postharvbio.2007.02.005.
Gao F, Daugulis AJ: Bioproduction of the aroma compound 2-phenylethanol in a solid-liquid two-phase partitioning bioreactor system by Kluyveromyces marxianus. Biotechnol Bioeng. 2009, 104: 332-339. 10.1002/bit.22387.
Kim B, Cho BR, Hahn JS: Metabolic engineering of Saccharomyces cerevisiae for the production of 2-phenylethanol via Ehrlich pathway. Biotechnol Bioeng. 2014, 111: 114-124.
Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ: Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proc Natl Acad Sci U S A. 2006, 103: 8287-8292. 10.1073/pnas.0602469103.
Hwang BK, Lim SW, Kim BS, Lee JY, Moon SS: Isolation and in vivo and in vitro antifungal activity of phenylacetic acid and sodium phenylacetate from Streptomyces humidus. Appl Environ Microbiol. 2001, 67: 3739-3745. 10.1128/AEM.67.8.3739-3745.2001.
Hua SS, Beck JJ, Sarreal SB, Gee W: The major volatile compound 2-phenylethanol from the biocontrol yeast, Pichia anomala, inhibits growth and expression of aflatoxin biosynthetic genes of Aspergillus flavus. Mycotoxin Res. 2014, 30: 71-78. 10.1007/s12550-014-0189-z.
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J: SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009, 25: 1966-1967. 10.1093/bioinformatics/btp336.
Droby S, Wisniewski M, Macarisin D, Wilson C: Twenty years of postharvest biocontrol research: Is it time for a new paradigm?. Postharvest Biol Tec. 2009, 52: 137-145. 10.1016/j.postharvbio.2008.11.009.
Treick W, Konetzka WA: Physiological state of Escherichia coli and the inhibition of deoxyribonucleic acid synthesis by phenethyl alcohol. J Bacteriol. 1964, 88: 1580-1584.
Jones MV: Effects of phenethyl alcohol on substrate uptake and cell permeability in Myxococcus xanthus FB. FEMS Microbiol Lett. 1979, 6: 293-296. 10.1111/j.1574-6968.1979.tb03724.x.
Nunn WD, Tropp BE: Effects of phenethyl alcohol on phospholipid metabolism in Escherichia coli. J Bacteriol. 1972, 109: 162-168.
Rosenkranz HS, Carr HS, Rose HM: Phenethyl alcohol I. effect on macromolecular synthesis of Escherichia coli. J Bacteriol. 1965, 89: 1354-1369.
Wilkie D, Maroudas NG: Induction of cytoplasmic respiratory deficiency in yeast by phenethyl alcohol. Genet Res. 1969, 13: 107-111. 10.1017/S0016672300002792.
Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang YK, Hernandez V, Akama T, Baker SJ, Plattner JJ, Shapiro L, Martinis SA, Benkovic SJ, Cusack S, Alley MRK: An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site. Science. 2007, 316: 1759-1761. 10.1126/science.1142189.
Singh UP, Bhat HR, Gahtori P, Singh RK: Hybrid phenylthiazole and 1,3,5-triazine target cytosolic leucyl-tRNA synthetase for antifungal action as revealed by molecular docking studies. In Silico Pharmacol. 2013, 1: 3-10.1186/2193-9616-1-3.
Barad S, Horowitz SB, Moscovitz O, Lichter A, Sherman A, Prusky D: A Penicillium expansum glucose oxidase-encoding gene, GOX2, is essential for gluconic acid production and acidification during colonization of deciduous fruit. Mol Plant Microbe Interact. 2012, 25: 779-788. 10.1094/MPMI-01-12-0002.
Eshel D, Miyara I, Ailing T, Dinoor A, Prusky D: pH regulates endoglucanase expression and virulence of Alternaria alternata in persimmon fruit. Mol Plant Microbe Interact. 2002, 15: 774-779. 10.1094/MPMI.2002.15.8.774.
Miyara I, Shafran H, Davidzon M, Sherman A, Prusky D: pH regulation of ammonia secretion by Colletotrichum gloeosporioides and its effect on appressorium formation and pathogenicity. Mol Plant Microbe Interact. 2010, 23: 304-316. 10.1094/MPMI-23-3-0304.
Prusky D, McEvoy JL, Saftner R, Conway WS, Jones R: The relationship between host acidification and virulence of Penicillium spp. on apple and citrus fruit. Phytopathology. 2004, 94: 44-51. 10.1094/PHYTO.2004.94.1.44.
Long CA, Zheng W, Deng BX: Biological control of Penicillium italicum of citrus and Botrytis cinerea of grape by strain 34-9 of Kloeckera apiculata. Eur Food Res Technol. 2005, 221: 197-201. 10.1007/s00217-005-1199-z.
Wang BQ, Zhang QF, Liu JH, Li GH: Overexpression of PtADC confers enhanced dehydration and drought tolerance in transgenic tobacco and tomato: effect on ROS elimination. Biochem Biophys Res Commun. 2011, 413: 10-16. 10.1016/j.bbrc.2011.08.015.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B: Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008, 5: 621-628. 10.1038/nmeth.1226.
Audic S, Claverie JM: The significance of digital gene expression profiles. Genome Res. 1997, 7: 986-995.
Lysoe E, Seong KY, Kistler HC: The transcriptome of Fusarium graminearum during the infection of wheat. Mol Plant Microbe Interact. 2011, 24: 995-1000. 10.1094/MPMI-02-11-0038.
Peres NT, Sanches PR, Falcão JP, Silveira HC, Paião FG, Maranhão FC, Gras DE, Segato F, Cazzaniga RA, Mazucato M, Cursino-Santos JR, Aquino-Ferreira R, Rossi A, Martinez-Rossi NM: Transcriptional profiling reveals the expression of novel genes in response to various stimuli in the human dermatophyte Trichophyton rubrum. BMC Microbiol. 2010, 10: 39-10.1186/1471-2180-10-39.
Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wirén N, Borriss R: Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol. 2012, 12: 116-10.1186/1471-2180-12-116.
Acknowledgments
We wound like to thank Hongyan Zhang for GC-MS assistance and Prof. Christopher Brian Watkins (Cornell University) for critical reading of the manuscript. The research was financially supported by the 973 programme (2013CB127100), the National Natural Science Foundation of China (31171773, 30972062) and Modern Agriculture (Citrus) Technology System (CARS-27).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
CL, YC, and XD designed research; PL, YL, and KC performed research; XD contributed new reagents/analytic tools; PL and MY analyzed data; and PL and CL wrote the paper. All authors read and approved the final manuscript.
Electronic supplementary material
12866_2014_242_MOESM3_ESM.doc
Additional file 3: Primers used for real-time quantitative RT-PCR for the verification of RNA-Seq profiling data.(DOC 42 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Liu, P., Cheng, Y., Yang, M. et al. Mechanisms of action for 2-phenylethanol isolated from Kloeckera apiculata in control of Penicillium molds of citrus fruits. BMC Microbiol 14, 242 (2014). https://doi.org/10.1186/s12866-014-0242-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-014-0242-2