Abstract
Background
Expansion of genomic resources for the Pacific white shrimp (Litopenaeus vannamei), such as the construction of dense genetic linkage maps, is crucial for the application of genomic tools in order to improve economically relevant traits. Sexual dimorphism exists in Pacific white shrimp, and the mapping of the sex-determination region in this species may help in future reproductive applications. We have constructed male, female, and sex-averaged high-density genetic maps using a 50 K single-nucleotide polymorphism (SNP) array, followed by a genome-wide association study (GWAS) to identify genomic regions associated with sex in white shrimp.
Results
The genetic map yielded 15,256 SNPs assigned to 44 linkage groups (LG). The lengths of the male, female, and sex-averaged maps were 5,741.36, 5,461.20 and 5,525.26 cM, respectively. LG18 was found to be the largest for both sexes, whereas LG44 was the shortest for males and LG31 for females. A sex-determining region was found in LG31 with 21 statistically significant SNPs. The most important SNP was previously identified as a sex-linked marker and was able to identify 99% of the males and 88% of the females. Although other significant markers had a lower ability to determine sex, putative genes were intercepted or close to them. The oplophorus-luciferin 2-monooxygenase, serine/arginine repetitive matrix protein and spermine oxidase genes were identified as candidates with possible participation in important processes of sexual differentiation in shrimp.
Conclusions
Our results provide novel genomic resources for shrimp, including a high-density linkage map and new insights into the sex-determining region in L. vannamei, which may be usefulfor future genetics and reproduction applications.
Similar content being viewed by others
Background
According to the FAO (2023), the Pacific white shrimp (Litopenaeus vannamei) holds the distinction of being one of the top aquaculture species in terms of global production. In 2020, approximately 5.81 million tons of this species was cultivated worldwide [1]. White shrimp farming is primarily concentrated in Asia, accounting for 79% of the total production amount. The leading contributor to this continent is China, with a market share of 32%, followed by India (15%) and Indonesia (12%). In the Americas, white shrimp production represents 20% of the global output, with Ecuador having the leading market share, with 13%, followed by Mexico at 3% and Brazil at 1% [1]. The shrimp industry faces several challenges that limit its expansion, such as high incidence of diseases [2], low efficiency of reproduction in captivity [3, 4], adaptation to different rearing environments [5,6,7], and optimization of production strategies to maximize yield e.g., mono-sex culture [8, 9]. The application of breeding programs and, more recently, genomic tools may help to address these challenges by providing solutions for the shrimp industry, such as production of animals more resistant to diseases, environment-specific genetic lines and production of mono-sex broodstock [10].
An important step towards the implementation of genomic tools in Pacific white shrimp farming is improving the genomic resources of this species. For example, the construction of genetic linkage maps (LM) may help to understand the genetic architecture of important traits [11]. Several LMs have been constructed in regard to the Pacific white shrimp using different types and low numbers of molecular markers. For instance, by using microsatellites, Pérez et al. [12] used 394 amplified fragment length polymorphisms (AFLPs) to construct a preliminary genetic map, and Alcivar-Warren et al. [13] used 100 simple sequence repeats (SSRs) to identify sex-related markers in L. vannamei. In addition, an LM using 6,359 specific length amplified fragment sequencing (SLAF) markers was constructed to identify potential quantitative trait loci (QTL) related to body weight [14]. More recently, denser genetic maps have been developed using single nucleotide polymorphism (SNPs with SNP numbers ranging from 418 to 6,146) [7, 15,16,17,18,19]. Most of these previous studies have applied restriction site-associated DNA sequencing (RAD-Seq) and double-digest RAD-Seq. The application of a dense SNP array to genotype animals has not been evaluated in regard to the construction of LM for shrimp.
As in other aquaculture species, the Pacific white shrimp exhibits sexual dimorphism, in which females may have a higher growth rate than males [20]. The genetic mechanism of sex determination in L. vannamei and other penaeids was already studied and seems to present a ZZ/ZW model [19, 21]. This model implies that males are homogametic (ZZ), and females are heterogametic (ZW). Genomic studies have been conducted to detect the genomic regions and markers associated with sex determination in the Pacific white shrimp. A sex-determining SNP was first found to be fully associated with linkage group (LG) 18 using linkage mapping analysis of a single family with 205 full-sibs [19]. In a different LG, three other markers were found to be associated with sex in a Mexican commercial population using the RAD-seq method [21, 22]. More recently, Jones et al. [18] found a highly associated SNP that explained more than 79% of the sexual variation. However, the position of this marker was uncertain it was positioned in conjunction with LG42 and 44. Although, some markers have been proposed to be associated with sex determination in this species, there is no consensus regarding the exact position of these markers, and no sex-related genes have been found close to these regions. The objectives of this study were to construct a new LM using a 50 K SNP array, identify genomic regions possibly associated with sex determination through a genome-wide association study (GWAS), and identify potential genes intercepting these regions in the L. vannamei genome.
Methods
Origin of animals and families
The animals used in this study were provided by the Maricultura del Pacífico hatchery (El Rosário, Sinaloa, Mexico). This breeding population was established in 2013 using animals from different countries (Mexico, Ecuador, Panama, and the USA). This population was selected in order to create two different genetic lines, including white-spot syndrome virus resistance and harvest weight, using pedigree information and estimated breeding values to select animals. A total of eight families (n = 1,290) were produced in order to develop this study (mean weight 45.7 g ± 4.98). Animals were identified using elastomers with different color combinations to determine the family origin and were used to generate the LM population with an average number of offspring per family of 20 and a range between 10 and 30. More details on the rearing protocols and production of families can be obtained from Campos-Montes et al. [23] and Castillo-Juárez et al. [24].
DNA extraction, genotyping and quality control
Pleopods from 1,290 animals were collected and stored in microtubes containing 96% ethanol for 72 h. The samples were sent to the Center of Aquaculture Technologies (CAT) for genomic DNA extraction and genotyping. DNA samples were genotyped using the AquaArray HD (50 K) vannamei® panel developed by Neogen® (Nebraska, USA) resulting in 50,811 SNPs in the raw genotype file.
Quality control (QC) filters were applied to the genotyped SNPs using PLINK 1.9 software [25]. SNPs with a minor allele frequency (MAF) < 0.01 and Hardy-Weinberg equilibrium (HWE) p-value < 1.2E− 6 (after Bonferroni correction) were excluded from subsequent analysis. QC filtering also excluded the maximum per-sample (> 0.2) and per-SNP (> 0.1) missing rates. The assignment of the individuals to their respective families was confirmed using CERVUS software [26] and with PLINK to remove SNPs with Mendelian errors (--me 0.1). Samples and markers with high Mendelian errors were removed before map construction.
Linkage map construction
The modules of the Lep-Map3 software [27] were used in order to construct the LM of L. vannamei. ParentCall2 was used to remove the non-informative SNPs (i.e., SNPs that were homozygous for both parents). Filtering2 was used to remove the SNPs with significant segregation distortion (data tolerance = 0.001). The SNPs were assigned to 44 linkage groups (LGs) (corresponding to the 44 chromosome pairs in L. vannamei) using SeparateChromosomes2. In the LG assignment, an optimized LOD score of 20 was achieved using a distortion of LOD = 1. The module JoinSingles2 was used to assign additional SNPs to the existing LGs, decreasing the LOD scores from 19 to 14 (optimized LOD score = 15). The resulting 44 LGs were ordered using OrderMarkers2 in order to generate male-, female-, and sex-averaged LMs. LMs were visualized using the LINKAGEMAPVIEW package available in R software [28].
GWAS for sex determination and gene search
A genome-wide association study (GWAS) for sex determination was performed using the LM coordinates obtained in the previous step for position markers. This population (GWAS population; Table S1) was comprised by 1,049 animals previously subjected to white-spot syndrome virus challenge. Body weight and sex information were available for these animals (44.8% male, and 55.2% female). The DNA extraction and genotyping protocols were the same as those described previously. SNPs that were not assigned to any of the 44 LGs were removed, and the remaining SNPs were repositioned according to the LG and distance (cM) obtained in the LM. Additional QC was applied for genotypes using the following criteria: MAF < 0.05, HWE p-value < 1.2 × 10− 9 and maximum rate per-SNP missing < 0.9. The GCTA software [29] was used for the GWAS and a genomic relationship matrix (GRM) was constructed and then included in the model to account for relatedness within the population. The model used was:
where \(y\) is the phenotype (sex, 1 = male and 2 = female), \(SNPs\) is the fixed SNP effect, and \(e\) is the residual. The --mlma-loco (mixed linear model association leaving-one-chromosome-out) function was used because it excludes candidate markers within the same chromosome from the GRM preventing overfitting and increasing statistical power [30]. The P-value of each SNP-association was -log10 transformed and visualized using a Manhattan plot using the qqman R package [31]. Additionally, linkage disequilibrium (LD) analysis was performed using the top10 SNPs with the lowest p-values to evaluate the effect of historical recombination rates in significant genomic regions within the same LG.
The genotypic frequencies of significant SNPs (lowest p-values) were also evaluated and correlated with sex in order to assess the potential use of these SNPs in determining sex. A nominal logistic model, considering sex as a response variable and genotype as an explanatory variable, was used to estimate the sex detection probabilities for the three most relevant SNPs.
SNP probes of 170 bp corresponding to the top ten significant SNPs, were used to identify any putative genes associated with sex determination. Adjacent probes were BLASTed against the L. vannamei reference genome sequence (Zhang et al. [32] - Gen Bank assembly ASM378908v1). Genes found at 50Kb upstream and downstream of the SNP positions were also considered putative genes.
Results
QC of genotypes and high-density linkage map
QC of the genotypes was applied separately for the two populations to generate the LM and perform the GWAS (Table 1).
For the LM population, of the initial 50,811 SNPs, a total of 40,141 SNPs were retained, presenting MAF values higher than 0.01 and on Hardy-Weinberg equilibrium. SNPs exceeding 10% of the missing genotype data were removed, leaving 35,801 SNPs for downstream analyses. Furthermore, 19 individuals exceeded 20% of the missing genotype data and were discarded, resulting in 1,271 individuals for the correct assignment of the true parents. A Mendelian rate test was performed on the individuals to discard alleles that could not be assigned to their putative biological parents. Thus, 2,341 SNPs that were segregated with Mendelian errors in more than 10% of individuals were excluded. Moreover, 523 individuals with more than 10% of SNPs with Mendelian errors were excluded from map construction. After QC filtering step, 33,460 SNPs and 748 individuals from eight families were used to construct the LM.
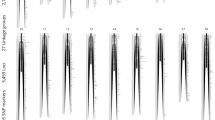
For the linkage analysis, the module ParentCall2 retained a total of 15,289 informative SNPs that were grouped into LGs by the SeparateChromosomes2 module and expressed at male, female, and sex-averaged levels (Table 2). When an LOD score value of 20 was used, the vast majority of SNPs (15,241) were assigned to 44 LGs (corresponding to 44 pairs of chromosomes in L. vannamei), thereafter, a maximum of four markers were assigned to each remaining linkage group. Subsequently, 15 orphan SNPs were assigned to the existing LGs using the JoinSingles2 module, totalling 15,256 SNPs to be ordered. The marker orders with the highest likelihood for each LG were combined to produce the final linkage maps (Fig. 1a and b). The male, female and sex-averaged maps spanned 5,741.36, 5,461.20 and 5,525.26 cM, respectively, with an average distance between markers of 0.377, 0.359 and 0.365 cM, respectively. LG18 was found to be the largest linkage group for both males (206.22 cM) and females (180.48 cM), whereas LG44 was the shortest for males (85.42 cM) and LG31 was the shortest for females (80.56 cM). The size was determined by measuring the genetic distance between the first and last markers within each LG. The LG that presented the highest density of markers (calculated by subtracting the distance between each marker followed by the overall mean of distances) was LG1 for males (0.242 cM) and females (0.213 cM), and LG44 was the least dense for both sexes, with average marker distance of 3.714 and 4.460 cM, respectively. These metrics indicated that the recombination rate within LG1 was higher than that within LG44, as the presence of more markers imply a higher recombination rate in that specific region.
GWAS and gene search for sex determination
The 15,256 SNPs ordered and positioned in the 44 LGs were extracted from the GWAS population with available sex phenotypes. These SNPs were repositioned according to the sex-averaged LM. A new QC of genotypes was applied, removing 222 SNPs with a MAF lower than 5%, 778 with Hardy-Weinberg disequilibrium, and 46 with a high missing rate (Table 1). A total of 14,110 SNPs and 1,049 samples were available for the GWAS.
The GWAS results showed that LG31 was strongly associated with sex determination in this L. vannamei population (Fig. 2). A total of 21 SNPs were found to be significant at the genomic level and the top10 most significant SNPs are listed in Table 3. A complete list of the 21 SNPs is shown in Table S2. The three most significant SNPs showed -log10 p-values of 3.3E− 220, 5.05E− 58 and 2.71E− 52, respectively.
These SNPs were positioned at 86.246, 47.020, and 43.116 cM LG31, respectively, and, a LD analysis was performed on the top10 significant SNPs to evaluate the level of LD in this genomic region (Fig. 3). A low-to-medium LD level (r² < 0.4) was found among the most relevant SNPs, indicating that they were most likely segregated at different rates.
The most significant SNP (Yu_C19299G_B) identified 99% of males (homozygotic) and 88% of females (heterozygotic) (Fig. 4). This marker was already reported as highly associated with sex in a previous study [19]. These results support the proposed ZZ/ZW sex determination system for this species. The other two markers were less reliable in determining sex in the present study, and their potential use in identifing sex could be pruned into errors (Figure S1). Probabilistic analyses revealed a similar trend for sex determination (Table 4).
Comparing the three most relevant SNPs, Yu’s SNP showed the highest r² (0.93), and the other markers were less reliable in determining sex (0.33 and 0.20, respectively).
The gene search performed using the top ten most significant SNPs intercepted some of the genes annotated in the L. vannamei genome. The AX-249,988,993 (LG31, pos: 26.457 cM) and AX-249,687,820 (LG31, pos: 28.246 cM) SNPs intercepted the oplophorus-luciferin 2-monooxygenase and serine/arginine repetitive matrix protein genes, respectively. Although these genes are not directly related to sex determination, some studies shown that they are expressed in the gonads of crustaceans, such as Macrobrachium nipponense [33] and Penaeus monodon [34, 35]. Another interesting gene, spermine oxidase, was found close to the AX-249,610,151 SNP (~ 20Kb downstream). This gene has also been reported to be differentially expressed in other aquaculture species, including L. vannamei [36]. The Yu_C19299G_B SNP was not found close to any gene. However, we used the blastx tool with the same scaffold as the input (JANIEY010001236.1) where this SNP was present. Blasting this scaffold against the Zebra fish (Danio rerio) and Daphnia pulex genomes, the most significant hits (E-value = 2e− 55) showed that this region was rich in retrotransposons, transposase, and integrins (Figure S2) such as, reverse transcriptase (RTs), ribonuclease H (RNase H) of the Ty3/Gypsy family, and integrase (H2C2). These elements may be involved in several sexual determination processes as reported in different species.
Discussion
In the present study, we constructed a high-density linkage map for L. vannamei using populations from a breeding program. To the best of our knowledge, this is the densest linkage map constructed for this species using a 50 K SNP array.
Several linkage maps are available for this species, most of which have a relatively low number of SNPs and use RAD-seq methods for genotyping [16,17,18]. It was proposed that L. vannamei presents 44 pseudochromosome pairs [32]. However, some linkage maps showed 45 [11], 48 [17] and even 49 [37] LGs. The present linkage map resulted in 15,256 SNPs distributed in the 44 LGs which is consistent with the genome assembling [32] and previous linkage maps [15, 16]. The LG44 had a significantly lower number of SNPs (24) than to the other LGs (197–587). Penaeids have unique genomic characteristics, including high levels of heterozygosity, polyploidization in some species, and simple sequence repeats accounting for up to 50% of the genome in some cases [38]. Such characteristics, associated with a large genome size (~ 2.6 Gb), have hampered the construction of a high-quality reference genome and the expansion of genome resources for L. vannamei.
The male and female linkage maps were found to be very similar in terms of total length and overall mean density. Still, it was possible to observe larger recombination gaps, also known as “coldspots”, in the male when comparing against the female map. This was evident in LG31 (the LG found to be associated with sex determination), where a high recombination rate was observed in the female map, whereas for males, the recombination was more concentrated towards the centromere of the LGs. This pattern is common in many fish species. Cooney et al. [39] evaluated the recombination landscape of 61 fish species and found that 42 species had higher recombination rates in females than males. Although there is no clear adaptative explanation for such differences between sexes, some hypotheses were proposed, such as the meiotic drive (i.e., alleles have an advantage during the process of female meiosis changing recombination rates) and protection against aneuploidy (i.e., failure of chromosomes to separate during cell division) [40,41,42]. In crustaceans, a previous study showed similar recombination rates between males and females for Daphnia pulex [43], which conflicts with our results in shrimp. Thus, there is no “rule of thumb” in the recombination dynamics between sexes. Recombination studies are scarce in crustaceans, especially shrimps, and further investigations are necessary to better understand the recombination landscape of this species.
There is an evident interest in understanding the genetic mechanisms of sexual determination in L. vannamei, mainly because of sexual dimorphism in this species. Females have a higher growth rate, and a mono-sex culture may increase the economic return of shrimp farming [20]. The identification of specific genetic regions and genes involved in sex determination may help to manipulate the proportion of females to optimize mono-sex cultures. Associations between markers and sex determination have been reported for L. vannamei. Yu et al. [19] identified a genomic region on LG18 related to sexual determination, and a single marker was found to be completely associated with this trait, being a potential SNP for sex characterization in this species. This SNP (Yu_C19299G_B) was present in the 50 K SNP array and it was the most significant SNP identified in this study. Perez-Enriquez et al. [22] also identified three markers associated with this trait using the RAD-seq method to develop a set of SNPs to characterize Mexican shrimp broodstock populations. The three SNPs were evaluated for sex identification of a different population using the post-PCR high-resolution melting method, and a high accuracy of sex determination was reported. Another major effect marker was identified by Jones et al. [18], who reported a strong association with sex; a high percentage of males were homozygotic and females were heterozygous for this marker in a study of almost 2,000 animals. Recently, a new hypothesis was established regarding the sex determination mechanism of L. vannamei. The sex-determination regions of males and females of this species were analyzed, and the results indicated that these regions could be influenced by variations in both DNA sequence and genome structure [44]. Using GWAS and the new high-density LM, we identified 21 statistically significant SNPs potentially associated with sex determination in this population at LG31. Yu’s marker was effective in determining of sex in our dataset, confirming the ZZ/ZW determination system and its potential use in other L. vannamei populations. These results may help in the fine mapping of the sex determination region, as this important marker was not previously positioned in a high-density genetic map. The other two markers with a higher statistical association with sex determination showed a lower ability to determine sex. A possible explanation for this divergence is the high recombination rate observed in the LM. Most likely, this region experiences strong cross-over events, making it difficult to be detected across multiple families [19]. This can also be visualized in the LD pattern of the most associated SNPs, where low LD was reported among the SNPs in LG31.
In regard to the organisms used in the GWAS, 0.9% (eight females and two males) disagreed with the sex determined in the field (phenotype), with the sex assigned using Yu´s marker. It is possible to infer that these were assignment errors, because when considering AX-249,531,418, sex determination coincided with Yu´s marker. The absence of females with the GC/TT (Yu_C19299G_B / AX-249,531,418) genotype is noteworthy, and could be associated with the presence in this genomic region of genes with an epistatic relationship with lethal effects in females. The main effects of sex-lethal mutations are on the reproductive aspects [45], which were not evaluated in the present study. Because a sex-lethal gene (Sxl) has been previously described in this species [46], further investigations of the deleterious effects of these SNPs are necessary.
At least three important genes have been described in the process of sexual determination in L. vannamei: sex-lethal (Sxl) [46], Pvfem-1 [47] and 25749_180 loci [21]. Another factor often analyzed for sex determination in shrimp is the level of gene expression according to the developmental phase. For example, Unigene0020898 and Unigene0020336 were found to be differentially expressed at the Zoea I stage, indicating that both genes are likely located in the sex-determining region of L. vannamei [48]. Fem-1 genes were also reported to be important during the Zoea III stage and are possible associated with sex differentiation in females [49]. We did not find any genes close to the top10 significative SNPs identified in the GWAS. However, oplophorus-luciferin 2-monooxygenase and serine/arginine repetitive matrix protein (SRRM1) genes were intercepted by significant SNPs. The first gene was downregulated at both the PL15 and PL25 stages in the gonadal tissues of M. nipponense [33]. Although its specific function is still unclear, this gene was found to be related to metabolic and extra cellular matrix processes in other crustacean species [50, 51]. The second gene was downregulated in the testis and ovary tissues of P. monodon using transcriptomic analysis [35]. Serine-arginine proteins have been reported to be differentially expressed during the testis maturation process of the same species [34]. The spermine oxidase gene is closely related to a significant SNP. This gene encodes the precursor of spermidine, a well-known polyamine involved in regulating fertility in mammals [52]. The spermine oxidase is significantly up-regulated in narrow-clawed crayfish (Pontastacus leptodactylus) suggesting its potential role in gonad maturation [53]. It is also differentially expressed in the gonada tissues of L. vannamei individuals [36].
The sex-linked SNP identified by Yu et al. [19] and mapped in this study were not associated with any gene. However, a deeper scan of the scaffold in which this SNP was located revealed conserved domains that were possibly associated with sex determination. We found, for instance, a retrotransposon called reverse transcriptase which is a transposable element. Transposable elements are dynamic DNA sequences that play crucial roles in genomic evolution and species diversification [54]. Given their typical expression in the gonads, transposable elements may be particularly prone to participate in sexual development [55]. Another interesting conserved domain found was a Ribonuclease H (RNase H) of the Ty3/Gypsy family. This transposable element may be associated with sex determination and evolution via two approaches: sequence conservation and co-localization with genomic function [56]. The first aspect is related to sex chromosome degeneration. Ty3/Gypsy suggests a potential contribution to the passive process of sex chromosome degeneration, as it was found to accumulate on the W/Y chromosome of other species, possibly starting from nearby insertions in the proximity of crucial heterogametic sex-linked genes [57]. The second aspect is related to co-localization with regions of known genomic functions on sex chromosomes. In fish, SNP association analyses have shown that sex-specific or sex-linked loci are more strongly associated with Ty3/Gypsy than with other transposable elements, as reported in the North African catfish (Clarias gariepinus) [58] and bighead catfish (Clarias macrocephalus) [59, 60]. In summary, the identification of the Ty3/Gypsy domain in the region where the Yu_C19299G_B SNP is present may indicate a reasonable allignment with the context of the role of these elements in sex chromosome evolution, and thus it supports the hypothesis that Ty3/Gypsy may contribute to structural changes in sex chromosomes.
Our results helped in the identification of genomic regions associated with sex determination in shrimp using informative high-density LM. Future research focused on the fine mapping of this sex-determining region may help disentangle the mechanisms involved in sexual differentiation and the development of mono-sex populations in L. vannamei.
Conclusions
A high-density genetic linkage map for L. vannamei species containing 15,256 SNPs, was constructed using a 50 K array. The genetic map allowed the identification of a region highly associated with sex in LG31. A previously reported sex-linked SNP was identified in the same LG with a high accuracy at the sex determination loci in the studied population.
Data availability
The datasets analyzed during the current study are available in the figshare repository, https://figshare.com/articles/dataset/LM_GWAS_Shrimp/24793677.
References
FAO. The State of World Fisheries and Aquaculture (SOFIA). Rome, Italy; 2023.
Thitamadee S, Prachumwat A, Srisala J, Jaroenlak P, Salachan PV, Sritunyalucksana K, et al. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture. 2016;452:69–87.
Caballero-Zamora A, Cienfuegos-Rivas EG, Montaldo HH, Campos-Montes GR, Martínez-Ortega A, Castillo-Juárez H. Genetic parameters for spawning and growth traits in the Pacific white shrimp (Penaeus (Litopenaeus) Vannamei). Aquac Res. 2013;46:833–9.
Arcos FG, Racotta IS, Ibarra AM. Genetic parameter estimates for reproductive traits and egg composition in Pacific white shrimp Penaeus (Litopenaeus) Vannamei. Aquaculture. 2004;236:151–65.
Li W, Luan S, Luo K, Sui J, Xu X, Tan J, et al. Genetic parameters and genotype by environment interaction for cold tolerance, body weight and survival of the Pacific white shrimp Penaeus vannamei at different temperatures. Aquaculture. 2015;441:8–15.
Nguyen NH, Ninh NH, Hung NH. Evaluation of two genetic lines of Pacific White leg shrimp Liptopenaeus Vannamei selected in tank and pond environments. Aquaculture. 2019; April:734522.
Huang W, Cheng C, Liu J, Zhang X, Ren C, Jiang X, et al. Fine mapping of the High-pH Tolerance and Growth Trait-related quantitative trait loci (QTLs) and identification of the candidate genes in Pacific White shrimp (Litopenaeus vannamei). Mar Biotechnol. 2020;22:1–18.
Perez-Rostro CI, Ibarra AM. Heritabilities and genetic correlations of size traits at harvest size in sexually dimorphic Pacific white shrimp (Litopenaeus vannamei) grown in two environments. Aquac Res. 2003;34:1079–85.
Moss DR, Moss SM. Effects of gender and size on feed Acquisition in the Pacific White shrimp Litopenaeus vannamei. J World Aquac Soc. 2006;37:161–7.
Zenger KR, Khatkar MS, Jones DB, Khalilisamani N, Jerry DR, Raadsma HW. Genomic selection in aquaculture: application, limitations and opportunities with special reference to marine shrimp and pearl oysters. Front Genet. 2019;10(JAN):693.
Du ZQ, Ciobanu DC, Onteru SK, Gorbach D, Mileham AJ, Jaramillo G, et al. A gene-based SNP linkage map for pacific white shrimp, Litopenaeus vannamei. Anim Genet. 2010;41:286–94.
Pérez F, Erazo C, Zhinaula M, Volckaert F, Calderón J. A sex-specific linkage map of the white shrimp Penaeus (Litopenaeus) Vannamei based on AFLP markers. Aquaculture. 2004;242:105–18.
Ac A-W, Meehan-Meola D, Park SW, Xu Z, Delaney M, Zuniga G. SHRIMPMAP: a low-density, microsatellite-based linkage map of the Pacific whiteleg shrimp, Litopenaeus vannamei: identification of sex-linked markers in linkage group 4. J Shellfish Res. 2007;26:1259–77.
Yu Y, Zhang X, Yuan J, Li F, Chen X, Zhao Y, et al. Genome survey and high-density genetic map construction provide genomic and genetic resources for the Pacific White shrimp Litopenaeus vannamei. Sci Rep. 2015;5:1–14.
Jones DB, Jerry DR, Khatkar MS, Raadsma HW, Steen H, Van Der, Prochaska J, et al. A comparative integrated gene-based linkage and locus ordering by linkage disequilibrium map for the Pacific white shrimp, Litopenaeus vannamei. Sci Rep. 2017;7:1–16.
Chen B, Zhong P, Wu X, Peng K, Sun Y, Chen X, et al. Construction of a genetic linkage map, QTLs mapping for low salinity and growth-related traits and identification of the candidate genes in Pacific white shrimp (Litopenaeus vannamei). Aquac Rep. 2022;22:100978.
Lu H, Chen W, Liu F, Huang M, Peng K, Zhao J, et al. A genetic linkage map of the Pacific white shrimp (Litopenaeus vannamei): QTL mapping for low-temperature tolerance and growth-related traits and identification of the candidate genes. Aquaculture. 2023;562:738834.
Jones DB, Nguyen HT, Khatkar MS, Simma DB, Jerry DR, Raadsma HW, et al. The identification of a major sex QTL in the white-leg shrimp, Litopenaeus vannamei. Aquaculture. 2020;529:735673.
Yu Y, Zhang X, Yuan J, Wang Q, Li S, Huang H, et al. Identification of sex-determining loci in Pacific White shrimp Litopeneaus Vannamei using linkage and Association Analysis. Mar Biotechnol. 2017;19:277–86.
Ventura T, Sagi A. The insulin-like androgenic gland hormone in crustaceans: from a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnol Adv. 2012;30:1543–50.
Perez-Enriquez R, Llera-Herrera R, Galindo Torres P, Avila S. A fast sex detection method for the Whiteleg shrimp Litopenaeus vannamei by post-PCR high resolution melting (HRM). 2020. https://doi.org/10.1016/j.aquaculture.2019.734784.
Perez-Enriquez R, Robledo D, Houston RD, Llera-Herrera R. SNP markers for the genetic characterization of Mexican shrimp broodstocks. Genomics. 2018;110:423–9.
Campos-Montes GR, Montaldo HH, Martínez-Ortega A, Jiménez AM, Castillo-Juárez H. Genetic parameters for growth and survival traits in Pacific white shrimp Penaeus (Litopenaeus) Vannamei from a nucleus population undergoing a two-stage selection program. Aquac Int. 2013;21:299–310.
Castillo-Juárez H, Casares JCQ, Campos-Montes G, Villela CC, Ortega AM, Montaldo HH. Heritability for body weight at harvest size in the Pacific white shrimp, Penaeus (Litopenaeus) Vannamei, from a multi-environment experiment using univariate and multivariate animal models. Aquaculture. 2007;273:42–9.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75.
Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program cervus accommodates genotyping error increases success in paternity assignment. Mol Ecol. 2007;16:1099–106.
Rastas P. Lep-MAP3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics. 2017;33:3726–32.
Ouellette LA, Reid RW, Blanchard SG, Brouwer CR. LinkageMapView—rendering high-resolution linkage and QTL maps. Bioinformatics. 2018;34:306–7.
Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82.
Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet 2014 462. 2014;46:100–6.
Turner SD. Qqman: an R package for visualizing GWAS results using Q-Q and manhattan plots. bioRxiv. 2014;:005165.
Zhang X, Yuan J, Sun Y, Li S, Gao Y, Yu Y, et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat Commun. 2019;10:1–14.
Jin S, Zhang W, Xiong Y, Jiang S, Qiao H, Gong Y, et al. Identification of important genes involved in the sex-differentiation mechanism of Oriental River Prawn, Macrobrachium nipponense, during the gonad differentiation and development period. Front Genet. 2022;13:241.
Klinbunga S, Petkorn S, Phaonakrop N, Janpoom S, Prasertlux S, Rongmung P, et al. Comparative proteomics for identification of reproductionrelated proteins in testes of the giant tiger shrimp penaeus monodon. Genet Aquat Org. 2020;4:69–79.
Li Y, Zhou F, Su N, Jiang S, Yang Q, Huang J, et al. Identification of gonadal associated genes in black tiger shrimp (Penaeus monodon) using transcriptome analysis and high-throughput sequencing. Aquac Res. 2022;53:6595–605.
Chen YL, Wang YM, Xu HJ, Li JW, Luo JY, Wang MR, et al. The characterization and knockdown of a male gonad-specific insulin-like receptor gene in the white shrimp Penaeus vannamei. Aquac Rep. 2022;27:101345.
Andriantahina F, Liu X, Huang H. Genetic map construction and quantitative trait locus (QTL) detection of growth-related traits in Litopenaeus vannamei for selective breeding applications. PLoS ONE. 2013;8:1–11.
Yuan J, Zhang X, Li F, Xiang J. Genome sequencing and Assembly Strategies and a comparative analysis of the genomic characteristics in Penaeid Shrimp Species. Front Genet. 2021;12:512.
Cooney CR, Mank JE, Wright AE. Constraint and divergence in the evolution of male and female recombination rates in fishes. Evol (N Y). 2021;75:2857–66.
Brandvain Y, Coop G. Scrambling Eggs: Meiotic Drive and the evolution of female recombination rates. Genetics. 2012;190:709–23.
Johnston SE, Huisman J, Ellis PA, Pemberton JM. A high-density linkage map reveals sexual dimorphism in recombination landscapes in red deer (Cervus elaphus). G3 genes, genomes. Genet. 2017;7:2859–70.
Ariede RB, Freitas MV, Lira LVG, Agudelo JFG, Borges CHS, Mastrochirico-Filho VA, et al. Linkage map for chromosome-level genome anchoring and genome-wide association study for resistance to Aeromonas hydrophila in Colossoma macropomum. Aquaculture. 2022;560:738462.
Molinier C, Lenormand T, Haag CR. No support for a meiosis suppressor in Daphnia pulex: comparison of linkage maps reveals normal recombination in males of obligate parthenogenetic lineages. bioRxiv. 2021. 2021.12.09.471908.
Wang Y, Yu Y, Li S, Zhang X, Li F. Analysis of a sex-biased sequence in sex determination region and exploitation of a fast sex detection method in Litopenaeus vannamei. Aquaculture. 2021;543:736922.
Cline TW. Two closely linked mutations in DROSOPHILA MELANOGASTER that Are Lethal to opposite sexes and interact with Daughterless. Genetics. 1978;90:683.
López-Cuadros I, García-Gasca A, Gomez-Anduro G, Escobedo-Fregoso C, Llera-Herrera RA, Ibarra AM. Isolation of the sex-determining gene sex-lethal (Sxl) in Penaeus (Litopenaeus) Vannamei (Boone, 1931) and characterization of its embryogenic, gametogenic, and tissue-specific expression. Gene. 2018;668:33–47.
Galindo-Torres P, Ventura-López C, Llera-Herrera R, Ibarra AM. A natural antisense transcript of the fem-1 gene was found expressed in female gonads during the characterization, expression profile, and cellular localization of the fem-1 gene in Pacific white shrimp Penaeus vannamei. Gene. 2019;706:19–31.
Wang Y, Yu Y, Li S, Zhang X, Xiang J, Li F. Sex-specific transcriptome sequencing of Zoea I larvae and identification of sex-linked genes using bulked Segregant Analysis in Pacific White shrimp Litopenaeus vannamei. Mar Biotechnol. 2020;22:423–32.
Yu Y, Wang Y, Lv X, Li S, Li F. Sex biased expression of Fem-1 in larval stages suggests its function in early sex differentiation of Litopeneaus Vannamei. Reprod Breed. 2023;3:153–60.
Li M, Wang J, Huang Q, Li C. Proteomic analysis highlights the immune responses of the hepatopancreas against Hematodinium infection in Portunus Trituberculatus. J Proteom. 2019;197:92–105.
Ren X, Zhang Y, Liu P, Li J. Comparative proteomic investigation of Marsupenaeus japonicus hepatopancreas challenged with Vibrio parahaemolyticus and white spot syndrome virus. Fish Shellfish Immunol. 2019;93:851–62.
Lefèvre PLC, Palin MF, Murphy BD. Polyamines on the Reproductive Landscape. Endocr Rev. 2011;32:694–712.
Nazari S, Khoshkholgh M, Baeza JA. Comparative transcriptome sequencing analysis of the narrow-clawed crayfish Pontastacus leptodactylus (Eschscholtz, 1823) and discovery of candidate sex-related genes. Aquac Rep. 2022;25:101235.
Arkhipova IR. Neutral theory, transposable elements, and eukaryotic genome evolution. Mol Biol Evol. 2018;35:1332–7.
Dechaud C, Volff JN, Schartl M, Naville M. Sex and the TEs: transposable elements in sexual development and function in animals. Mob DNA 2019 101. 2019;10:1–15.
Srikulnath K, Ahmad SF, Singchat W, Panthum T. Do Ty3/Gypsy Transposable Elements Play Preferential Roles in Sex Chromosome Differentiation? Life 2022, Vol 12, Page 522. 2022;12:522.
Makałowski W, Gotea V, Pande A, Makałowska I. Transposable elements: classification, identification, and their use as a tool for comparative genomics. Methods Mol Biol. 2019;1910:177–207.
Nguyen DHM, Panthum T, Ponjarat J, Laopichienpong N, Kraichak E, Singchat W, et al. An investigation of ZZ/ZW and XX/XY sex determination systems in North African Catfish (Clarias gariepinus, Burchell, 1822). Front Genet. 2021;11:562856.
Nguyen DHM, Ponjarat J, Laopichienpong N, Panthum T, Singchat W, Ahmad SF, et al. Genome-wide SNP analysis of Hybrid Clariid Fish reflects the existence of polygenic sex-determination in the lineage. Front Genet. 2022;13:789573.
Nguyen DHM, Ponjarat J, Laopichienpong N, Kraichak E, Panthum T, Singchat W, et al. Genome-wide SNP analysis suggests male heterogamety in bighead catfish (Clarias macrocephalus). Aquaculture. 2021;543:737005.
Funding
The authors are thankful to Maricultura del Pacífico S.A. C.V. for providing data used in this study.
Author information
Authors and Affiliations
Contributions
BFG: Conceptualization, Methodology, GWAS and Gene search analyses, Writing - Original Draft and Writing - Review & Editing. VAMF: Construction of linkage map, Writing - Original Draft and Writing - Review & Editing. JGH: Revision of results, Writing - Original Draft and Writing - Review & Editing. GRCM: Resources, Supervision, Project administration, Writing – review & editing. TMM: Revision of results, Writing - Original Draft and Writing - Review & Editing. PVRM: Revision of results, Writing - Original Draft and Writing - Review & Editing. ACZ: Resources, Supervision, Project administration, Writing – review & editing. DTH: Writing - Review & Editing. JMY: Conceptualization, Methodology, Supervision and Writing - Review & Editing. All authors contributed to the design of the study, discussion of results and review of the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Garcia, B.F., Mastrochirico-Filho, V.A., Gallardo-Hidalgo, J. et al. A high-density linkage map and sex-determination loci in Pacific white shrimp (Litopenaeus vannamei). BMC Genomics 25, 565 (2024). https://doi.org/10.1186/s12864-024-10431-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10431-x