Abstract
Background
The concept of the functional nutritional value of health-beneficial omega-3 long-chain polyunsaturated fatty acids (n-3 LC-PUFA) is becoming a phenomenon among red meat consumers globally. This study examined the expressions of three lipogenic genes (fatty acid binding protein 4, FABP4, fatty acid synthase, FASN; and stearoyl-CoA desaturase, SCD) in the ribeye (Longissimus thoracis et lumborum) muscle of Tattykeel Australian White (TAW) lambs fed fortified omega-3 diets and correlations with fatty acids. To answer the research question, “are there differences in the expression of lipogenic genes between control, MSM whole grain and omega-3 supplemented lambs?”, we tested the hypothesis that fortification of lamb diets with omega-3 will lead to a down-regulation of lipogenic genes. Seventy-five six-month old TAW lambs were randomly allocated to the (1) omega-3 oil-fortified grain pellets, (2) unfortified grain pellets (control) or (3) unfortified MSM whole grain pellets diet supplements to generate three treatments of 25 lambs each. The feeding trial lasted 47 days.
Results
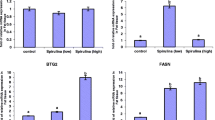
From the Kruskal-Wallis test, the results showed a striking disparity in lipogenic gene expression between the three dietary treatments in which the FABP4 gene was significantly up-regulated by 3-folds in the muscles of lambs fed MSM Milling (MSM) whole grain diet compared to the omega-3 and control diets. A negative correlation was observed between FASN gene expression and intramuscular fat (IMF), eicosapentaenoic acid (EPA), total polyunsaturated fatty acids (PUFA), omega-6 polyunsaturated fatty acids (n-6 PUFA) and monounsaturated fatty acids (MUFA). The FABP4 gene expression was positively correlated (P < 0.05) with EPA and docosahexaenoic acid (DHA).
Conclusion
Taken together, this study’s results suggest that FABP4 and FASN genes perform an important role in the biosynthesis of fatty acids in the ribeye muscle of TAW lambs, and supplementary diet composition is an important factor influencing their expressions.
Similar content being viewed by others
Background
The perception of a healthy nutrient-dense food is increasingly becoming a global and topical discourse amongst meat consumers due to knowledge advancements in medicine, science, and technology that have changed the lifestyles of the populace [1]. The fortification of livestock diets to increase the content of health-claimable fatty acids remains a viable strategy for improving meat quality because fatty acids are important building blocks for cellular structures, tissues, and organs [2]. They are also an integral part of the metabolic processes of synthesising and coordinating the roles of essential biologically active elements [3]. Long chain polyunsaturated fatty acids (LC-PUFA) are required for various biological and physiological processes, but mammals cannot synthesise them [4,5,6] because they lack Δ12 and Δ15 fatty acid desaturase enzymes, thus necessitating the need for dietary supplementation [6]. The α-linolenic (ALA) and linoleic (LA) acids are precursors of n-3 and n-6 LC-PUFA, respectively. ALA is converted to the more potent n-3 LC-PUFA, such as docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) through the de novo synthesis metabolic pathway [7]. These fatty acids contribute to memory improvement, the elevation of visual acuity, the depression of high blood pressure [8], reduction of the risks of inflammatory and degenerative diseases such as cardiovascular diseases, cancer, skin conditions, metabolic syndrome, diabetic neuropathy, allergies, asthma, arthritis and the enhancement of immune function [7, 9, 10]. Similarly, LA is a precursor for the synthesis of arachidonic acid (ARA), an n-6 LC-PUFA that is converted to prostaglandins, leukotrienes, and other associated compounds. However, diets rich in n-6 PUFA are linked to inflammation, blood vessel constriction, and platelet aggregation [11, 12]. Therefore, a high intake of LA relative to ALA, has been reported to interfere with ALA desaturation and elongation pathways [13] as both LA and ALA utilise the same metabolic pathway for synthesising DHA and EPA [14].
An animal’s diet influences its meat fatty acid composition [15], through absorption of the dietary fatty acids [16] and modulation of cellular pathways such as lipogenic gene expression patterns [17]. Gene expression analysis can shed some light on the transcriptional pathways involved in the synthesis of functional gene products. Thus, identifying the framework of gene expression is vital to unravelling the molecular mechanisms controlling complex traits [18]. Only a few studies have evaluated dietary regulation of lipogenic gene expression in the ovine muscles [19,20,21]. In addition, the reported effect of dietary omega-3 rich supplements on gene expression is inconsistent. For instance, Fan et al. [22] reported an increase in the SCD gene expression, while Deng et al. [23] reported a decline in the SCD gene expression in the longissimus thoracis of Hu sheep. Hence, to better comprehend the genetic regulation of fatty acid deposition in Tattykeel Australian White (TAW) lambs, the ribeye muscle from lambs on diverse dietary supplements was utilised to provide a more detailed lipogenic gene expression pattern. To our knowledge, there is no published literature on the impact of dietary supplementation on transcriptional lipogenic gene expression in TAW lambs, hence the objective of this study as the first of its kind, was to fill this gap. We hypothesised that dietary fortification with omega-3 oils influences the transcriptional expression of lipogenic genes in the ribeye muscle in TAW lambs.
Results
The three lipogenic genes showed marked variation in expression levels within the ribeye muscle of lambs in all three dietary supplementation groups (Fig. 1). The Kruskal-Wallis test showed that the SCD gene expression did not differ between treatments (Fig. 1A; P = 0.12). In Fig. 1B, the FABP4 gene was up-regulated 3-folds in lambs fed the MSM whole grain diet compared to the omega-3 fortified diet (P = 0.018). However, the expression of FABP4 between the control and the unfortified MSM whole grain or omega-3 fortified diet fed lambs did not differ (P ≥ 0.11). The expression of the FASN gene tended to be higher in the control lambs than in the MSM whole grain (P = 0.079) and the omega-3 fortified diet (P = 0.059). However, FASN gene expression between lambs fed MSM whole grain or the omega-3 fortified diet did not differ (P = 0.56).
Spearman correlations between the fold changes in SCD, FABP4, and FASN gene expressions and meat quality traits measured are shown in Fig. 2. Some negative associations (P < 0.05) between IMF, total saturated fatty acids (SFA), individual SFA (C16:0, C18:0, C18:2n-6, C18:3n-3 and C20:0), monounsaturated fatty acids (MUFA), PUFA and n-6 PUFA, and fold changes in the expression of the FASN gene were observed. On the other hand, positive correlations (P < 0.05) were detected between the FABP4 gene fold changes and DHA, EPA, EPA + DHA, EPA + DHA + DPA and PUFA/SFA ratio. However, the correlations between the SCD gene expression and meat quality traits did not attain statistical significance (P > 0.05).
The effects of SCD, FABP4, and FASN genes expression on meat quality traits in the ribeye muscle of TAW lambs supplemented with omega-3 fortified diets are depicted in Table 1. The results indicate that the effect of SCD gene expression on all the meat quality traits was negligible. The FABP4 gene significantly influenced LA (C18:2n-6), ALA (C18:3n-3), EPA (C20:5n-3), DHA (C22:6n-3), DPA (C22:5n-3), EPA + DHA + DPA, n-3 PUFA and n-6 PUFA while the FASN gene expression influenced the IMF, C18:0, LA, EPA, C20:0, total PUFA and n-6 PUFA (P < 0.05).
Discussion
To our knowledge, this is the first study, as no previously published literature has described SCD, FABP4, and FASN lipogenic gene expressions in the ribeye muscles of supplemented TAW lambs. The processes of fatty acid metabolism in ruminants are complex. Unsaturated fatty acids (UFA) are converted into SFA due to microbial biohydrogenation in the rumen [23]. The increased levels of these SFA increase the risks of atherosclerosis and coronary heart disease in humans [24]. The modification of fat composition in meat and meat products by the inclusion of omega-3 LC-PUFA is, therefore, an excellent approach to the promotion and improvement of human health [25]. This study utilised a lamb finishing feeding trial with omega-3 fortified, conventional MSM whole grain and control diets to quantify the expression patterns of SCD, FABP4, and FASN genes in the ribeye muscle of TAW lambs.
Stearoyl-CoA desaturase gene
The SCD gene enhances meat quality traits by modifying fat deposition and fatty acid composition [26]. It achieves this by catalysing the synthesis of cis-vaccenic acid from the conjugated linoleic acid (c9, t11 isomer) [27] and converting SFA to MUFA by inserting a double bond between carbon atoms Δ9 and Δ10 of C18:0 fatty acid to synthesise C18:1 c-9 fatty acid [22, 28]. Wang et al. [29] and Cedernaes et al. [30] described the expression of the SCD gene as an indicator of IMF development, hence, it could be an essential regulator of muscle metabolism as it aids lipid biosynthesis and depresses fatty acids degradation [31].
Dietary omega-3 oil fortification in this study did not lead to any significant change in the expression of the SCD gene in the ribeye muscle in agreement with previous studies that reported no change in SCD gene expression in Italian Simmental and Holstein bulls fed linseed [32] and Angus steers fed corn oil [33]. This was in contrast to other studies that reported down-regulation of SCD expression when soybean oil was substituted with 2.7% of linseed oil [34] in cattle and in lambs fed alfalfa [19]. Dietary omega-3 oils supplementation is reported to suppress the SCD gene expression through the retinoic acid receptor-mediated signaling pathway [35], hence the lack of difference observed herein is not clear and warrants further investigation. However, these results corroborate the lack of difference in total MUFA, and oleic acid observed in the muscle of these lambs between treatments [36].
Fatty acid binding protein 4 gene
Lipid synthesis and growth in sheep are influenced by the action of the FABP4 gene [37], and drives the transport, lipogenesis, lipolysis, absorption and storage of long chain fatty acids [38, 39] hence, it is a metabolic indicator of an animal’s capacity to store IMF [40] and contributes to regulating the tenderness of meat in ovine species [41]. It is also associated with the regulation of lipid metabolic syndrome, insulin resistance, diabetes and obesity [42, 43]. Transcription factors, including peroxisomal proliferator-activated receptor (PPAR) α, -β and -γ, are triggered by fatty acids or other hydrophobic ligands and are responsible for stimulating FABP4 gene expression, which occurs mainly in the adipocytes [38, 39]. In the current study, FABP4 gene expression was down-regulated in the ribeye muscle of TAW lambs given omega-3 diet in comparison to the MSM diet.
The expression did not differ between the control and the omega-3 diet in this study. Similarly, Vargas-Bello-Pérez et al. [44] reported no difference in FABP4 expression in subcutaneous adipose tissue of dairy cows fed diet supplemented with fish oil compared to the unsupplemented cows. The lack of difference in FABP4 expression between the control and omega-3 treatments in this study suggests that inclusion of omega-3 oils did not depress fat synthesis [44]. These findings explain the similar total fatty acids content observed in the ribeye muscle of the control and omega-3 treatments presented in our previous study [36]. However, more work is required to elucidate the impact of omega-3 oils fortification in feed on the FABP4 gene expression in sheep.
In the current study, the FABP4 gene was expressed more in the muscle of lambs on MSM than in those consuming the fortified omega-3 supplement, indicating that FABP4 up-regulation is promoted by supplementation with MSM whole grains. Diets regulate the mechanisms governing IMF deposition by triggering transcription factors such as the PPARγ that influence the expression of FABP4 gene that encode proteins involved in fat accumulation and differentiation of adipocytes in muscle tissues [45]. A study carried out by Yang et al. [46] on the influence of diets with varied levels of energy on the efficiency of fat deposition and fatty acid profiles of the yak muscle, revealed dense energy diets boosted the deposition and partial fatty acids content of this muscle primarily by up-regulation of mRNA expression of lipogenic genes including FABP4. However, the MSM whole grains diet in this study had similar energy composition with the omega-3 fortified diet (14.4 versus 15.1 MJ/Kg dry matter) indicating that the difference in FABP4 expression was not influenced by the diet energy composition.
The higher FABP4 gene expression in the MSM whole grain group and the lower expression in the omega-3 fortified group in this study were associated with higher fatty acid deposition levels in the omega-3 than in the MSM supplemented TAW lambs [36]. In addition, FABP4 gene expression was positively correlated with EPA and DHA. Previous studies have also reported the association between FABP4 gene expression and intramuscular fat content [40, 47] and backfat depth [48] in ruminants.
These results indicate that the FABP4 gene has a significant function in the biosynthesis of fatty acids in the ribeye muscle of TAW lambs, and supplementary diet composition is an important factor influencing its expression.
Fatty acid synthase gene
The de novo synthesis of LC fatty acids from acetyl-CoA and malonyl-CoA precursors is carried out by the FASN gene [49, 50]. Hence, the degree of FASN expression performs an important role in fat deposition [51]. Previous reports on FASN gene down-regulation in the Longissimus dorsi muscle of Italian Large White and Duroc pigs [52] and in cattle supplemented with corn oil have been reported [33]. These reports agree with our current results where the expression of FASN gene tended to be lower in the ribeye muscle of lambs consuming omega-3 diet. Previous studies suggest that dietary fat supplementation may reduce FASN gene expression by inhibiting the activity of sterol regulatory element-binding protein [53] and carbohydrate-responsive element-binding protein [54]. The FASN gene expression was negatively correlated with the n-6 PUFA, PUFA, MUFA, SFA, 20:0, 18:3n-3, 18:2n-6, 18:0 and 16:0. Dietary omega-3 supplementation is reported to enhance the level fatty acids composition in the plasma of cattle [55] and the muscle, liver, kidney and heart tissues in sheep [36]. The high fatty acids composition blocks the activation of the carbohydrate-responsive element-binding protein, consequently inhibiting the lipogenic genes expression and de novo lipogenesis [56].
Conclusion
Dietary treatment influenced the composition of fatty acid and lipogenic gene expression in the ribeye muscle of supplemented TAW lambs. The FABP4 gene was expressed 3 folds higher in the muscles of lambs fed MSM whole grain, but not in the omega-3 fortified diet. Furthermore, the FABP4 gene expression was positively correlated with DHA, EPA, and PUFA/SFA ratio. There was a tendency for the FASN gene expression to be lower in the lambs fed omega-3 fortified diet compared to the control diet, and the expression was negatively correlated with most of the fatty acids. Diets had no substantial influence on SCD gene expression in this study and no correlation between expression and the fatty acids was detected. The findings herein buttress the point that dietary omega-3 may improve muscle n-3 PUFA through regulation of the de novo fatty acids synthesis in the ribeye muscle of TAW lambs.
Methods
Management of experimental animals
The reporting in the manuscript follows the recommendations of Kilkenny et al. [57] in the ARRIVE guidelines for animal research. The design of the experiment, animals and location are described previously [36]. In summary, the lamb finishing feeding trial was accomplished from April to June 2019 (Crown Agriculture’s commercial feedlot complex, Borenore, New South Wales, Australia). The feedlot complex was well-ventilated, equipped with automated feeding and watering systems and had a concrete floor spacing of 5 m2 per head. The feeding troughs were equipped with sensors for data capture of every individual lamb’s body weight, rumination time and feed intake which were automatically recorded and cloud-stored. The experimental animals comprised 75 six-months old TAW lambs with a mean liveweight of 30 ± 1.2 kg randomly allocated into three dietary treatments of twenty-five animals per group: (1) omega-3 oil-fortified grain pellets, (2) unfortified grain pellets (control) or (3) unfortified commercial MSM whole grain (MSM Stockfeeds, Manildra, NSW, Australia) pellets. Details of the nutrient compositions of these experimental diets have been published [36]. Furthermore, lambs had unlimited access to hay diet and water. This completely randomised study lasted 47 days including the initial 14-day adaptation period. At the conclusion of the feeding period, lambs were slaughtered humanely at Gundagai Meat Processing Plant according to Meat Standards Australia regulations.
Samples of the ribeye muscles were taken from the 12th and 13th ribs interface 24 h post-mortem and kept in a -80 °C freezer.
RNA isolation, cDNA, and quantitative PCR
Total RNA was extracted from frozen ribeye muscle samples utilising the TRIzol™ Plus RNA Purification Kit (Invitrogen, Thermo Fisher Scientific, Victoria, Australia), and subsequently purified and DNase-treated with ezDNase™ Enzyme (Thermo Fisher Scientific, Victoria, Australia). Total RNA yield and quality were checked with a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Victoria, Australia) and QuantiFluor® RNA System (Promega, WI, USA). Complementary DNA was synthesized from 100 ng RNA using SuperScript™ IV VILO™ Master Mix Reverse Transcription Kit (Thermo Fisher Scientific, Victoria, Australia).
Twenty microliters quantitative polymerase chain reaction (qPCR) reactions were run in duplicate utilising the Fast SYBR Green Chemistry (Thermo Fisher Scientific, Victoria, Australia) with 250 nM primers and 6 µL template on a QuantStudio-3 Real-Time qPCR detection system (Applied Biosystem Inc.). This was carried out under fast-cycling settings of initial 50 °C for 2 min, then 95 °C for 2 min, then 50 cycles at 95 °C for 15s and finally 65 °C for 1 min.
Primer design and housekeeping gene selection
The primers of target and housekeeping genes (Table 2) were designed in the Genious Program version 2.2 (http://www.geneious.com). The suitability of all primers was ascertained by employing a serial dilution of pooled cDNA to generate a standard curve. All primer pairs established acceptable efficiency (90–110%) and R-value (99%). Data normalization for the target FASN, FABP4 and SCD genes utilized two reference genes; the elongation factor 1 A (EF1A, formerly termed EF1α) and Peptidyl-prolyl cis-trans isomerase A (PPIA) using an expression ratio that was constant amongst all samples as the key selection criterion [58,59,60]. The values for the differential gene expression comparison were represented as fold-change values in Fig. 1, calculated using the ∆Ct metric.
Fatty acids analysis
Details of the fatty acid composition of ribeye muscle biopsy samples analysed by means of gas chromatography–mass spectrophotometry procedure was previously described by [36].
Briefly, total lipids in 1 g of un-homogenized muscle tissue samples were extracted overnight. The original phase was a single-phase overnight extraction utilizing CHCl3:MeOH: H2O (1:2:0.8 v/v). The second segment involved phase separation with the addition of CHCl3:Saline-Milli-Q H2O (1:1 v/v) followed by rotary evaporation of the lower chloroform phase at 40 °C to acquire total lipids. The extracted cumulative lipids were separated into lipid classes by thin-layer chromatography (TLC) using 100 mL of the lipid extract reconstituted in hexane. The extract was marked onto silica gel G plates (200 × 200 × 0.25 mm3) using a micropipette. The TLC plate was developed in an acetone/petroleum ether (1:3 v/ v) solvent system in a tank comprising a few crystals of butylated hydroxytoluene (BHT) to hinder oxidation. Triacylglycerols, cholesterol and free fatty acids migrated, while phospholipids remained at the origin of the plate. The phospholipids were scraped off the plate into clean screw-capped test tubes for transmethylation and eventual computation of the lipid conversion factor (LCF) of 0.912 based on fatty acids/g of total lipids (0.083 for phospholipids, 0.829 for triacylglycerols and 0% for cholesterol since cholesterol does not have any fatty acids). An aliquot from each total lipid extract was utilized for transmethylation with MeOH:CHCl3:HCl (10:1:1 v/v) for 2 h at 80 °C. Fatty acid methyl esters (FAME) were extracted thrice using hexane:CHCl3 (4:1 v/v). A known concentration of an internal standard (C19:0) was added in a 1500 µL vial encompassing the extracted FAME. The FAME was analyzed on a 7890B gas chromatograph (Agilent Technologies, Palo Alto, CA, USA) furnished with an EquityTM − 1 fused 15 m silica capillary column with 0.1 mm internal diameter and 0.1 μm film thickness (Supelco, Bellefonte, PA, USA), a flame ionization sensor, a split/ splitless injector and an Agilent Technologies 7683 B Series autosampler. The gas chromatograph settings were splitless mode injection; carrier gas He; original oven temperature 120 °C and then increased to 270 °C at flow rates of 10 °C / min and to 310 °C at 5 °C / min. The Agilent Technologies ChemStation software (Palo Alto, CA, USA) was used to measure fatty acid peaks. The fatty acid identities were established using a Finnigan Thermoquest GCQTM GC/MS fitted with an on-column injector and Thermoquest Xcalibur software (Austin, TX, USA). Fatty acid contents were calculated as follows: FA mg/100 g = (Total lipid) × (LCF [0.912]) × ([%FA]/100) × 1000, where 0.912 was the resultant lipid conversion factor.
Statistical analysis
As gene expression data can exhibit variability and may not always follow a normal distribution, a non-parametric approach provides a suitable alternative for hypothesis testing. Therefore, in this study, data on gene expression, fat melting point (FMP), intramuscular fat (IMF), and fatty acids profiles of TAW MARGRA lamb were analysed using nonparametric statistics in R version 4.0.1. Kruskal-Wallis tests with Bonferroni’s adjusted p-values were used to test for differences in fold changes among dietary treatments. Relationships between variables were explored using Spearman correlation analysis. The effect of gene expression (fold change) on FMP, IMF, and fatty acids was investigated using a quantile regression model. Quantile regression, an extension of linear regression, is often preferred to linear regression because it is robust to outliers [61] and thus superior when the assumptions of linear regression are unmet. We initially fitted animal as a random effect, and the fixed effects of treatment (omega-3 oil-fortified grain pellets, control unfortified grain pellets and unfortified commercial MSM whole grain), lipogenic gene (FASN, FABP4 and SCD) and their interactions in the model. However, the fold changes in gene expression did not follow a normal distribution pattern, hence we used a non-parametric approach in our analysis instead of an individual-level model. Also, the samples emanated from lambs of the same age and breed, hence animal variability would have been at a minimum, if not negligible. Therefore, we used the quantile regression methodology instead of the classical linear regression, because the quantile regression analysis is robust to outliers and enables the estimation of all the conditional quantiles of the response variable instead of the mean as depicted below:
Where \({\text{Q}\text{u}\text{a}\text{n}\text{t}}_{\theta }\left({y}_{i}|{x}_{i}\right)\) denotes the \(\theta -th\) conditional quantile of the response variable \({y}_{i}\), conditional on the set of covariates \({x}_{i}\). \({\beta }_{\theta }\) are the coefficients associated with each covariate for the \(\theta -th\) quantile level.
The comparison between quantile regression and linear regression models is depicted in the Supplementary Fig. S1-S3. Alpha was set to 0.05 for all statistical comparisons.
Data Availability
The datasets generated and/or analysed during the current study are not publicly available due to contractual confidentiality clause obligation, but are available from the corresponding author on reasonable request.
Abbreviations
- ALA:
-
α-linolenic and
- cDNA:
-
Complementary deoxyribonucleic acid
- DHA:
-
Docosahexaenoic acid
- EF1A:
-
Elongation Factor 1-Alpha
- EPA:
-
Eicosapentaenoic acid
- FABP4:
-
Fatty acid binding protein 4
- FASN:
-
Fatty acid synthase
- FMP:
-
Fat melting point
- IMF:
-
Intramuscular fat
- LA:
-
Linoleic acids
- MSM:
-
Unfortified whole grain pellets
- MUFA:
-
Monounsaturated fatty acids
- n-3 LC-PUFA:
-
Omega-3 long-chain polyunsaturated fatty acid
- n-6 PUFA:
-
Omega-6 polyunsaturated fatty acids
- PPAR:
-
Peroxisomal proliferator-activated receptor
- PPIA:
-
Peptidyl-Prolyl Cis-Trans Isomerase A
- qPCR:
-
Quantitative polymerase chain reaction
- RNA:
-
Ribonucleic acid
- SCD:
-
Stearoyl-CoA desaturase
- SFA:
-
Total saturated fatty acids
- TAW:
-
Tattykeel Australian White
- UFA:
-
Unsaturated fatty acids
References
Yeung SSY, Kwan M, Woo J. Healthy diet for healthy aging. Nutrients. 2021;13:4310.
Nudda A, Bee G, Correddu F, Lunesu MF, Cesarani A, Rassu SPG, et al. Linseed supplementation during uterine and early post-natal life markedly affects fatty acid profiles of brain, liver and muscle of lambs. Ital J Anim Sci. 2022;21:361–77.
Sokoła-Wysoczańska E, Wysoczański T, Wagner J, Czyż K, Bodkowski R, Lochyński S, et al. Polyunsaturated fatty acids and their potential therapeutic role in cardiovascular system sisorders—a review. Nutrients. 2018;10:1561.
Choudhary AK, Mishra G. Functional characterization and expression profile of microsomal FAD2 and FAD3 genes involved in linoleic and α-linolenic acid production in Leucas cephalotes. Physiol Mol Biology Plants. 2021;27:1233–44.
Patel A, Karageorgou D, Katapodis P, Sharma A, Rova U, Christakopoulos P, et al. Bioprospecting of thraustochytrids for omega-3 fatty acids: a sustainable approach to reduce dependency on animal sources. Trends Food Sci Technol. 2021;115:433–44.
Wu C, Hong B, Jiang S, Luo X, Lin H, Zhou Y, et al. Recent advances on essential fatty acid biosynthesis and production: clarifying the roles of ∆12/∆15 fatty acid desaturase. Biochem Eng J. 2022;178:108306.
Nigam D, Yadav R, Tiwari U. Omega-3 fatty acids and its role in human health. Functional Food and Human Health. Singapore: Springer Singapore; 2018. pp. 173–98.
Pratiwy FMPDY. The potentiality of microalgae as a source of DHA and EPA for aquaculture feed: a review. Int J Fish Aquat Stud. 2020;8:39–41.
Sala-Vila A, Fleming J, Kris-Etherton P, Ros E. Impact of α-linolenic acid, the vegetable ω-3 fatty acid, on Cardiovascular Disease and cognition. Adv Nutr. 2022;13:1584–602.
Silva RRRLLMMPMSDSS. Conjugated linoleic acid (CLA): a review. Int J Appl Sci Technol. 2014;4:154–70.
Corino C, Vizzarri F, Ratti S, Pellizzer M, Rossi R. Long term dietary supplementation with omega-3 fatty acids in Charolais beef cattle reared in Italian intensive systems: nutritional profile and fatty acids composition of Longissimus lumborum muscle. Animals. 2022;12:1123.
Rogero M, Calder P. Obesity, inflammation, toll-like receptor 4 and fatty acids. Nutrients. 2018;10:432.
Simopoulos A. An increase in the omega-6/omega-3 fatty acid ratio increases the risk for obesity. Nutrients. 2016;8:128.
Marangoni F, Agostoni C, Borghi C, Catapano AL, Cena H, Ghiselli A, et al. Dietary linoleic acid and human health: focus on cardiovascular and cardiometabolic effects. Atherosclerosis. 2020;292:90–8.
Santos-Silva J, Francisco A, Portugal AP, Paulos K, Dentinho MT, Almeida JM, et al. Effects of partial substitution of grain by agroindustrial byproducts and sunflower seed supplementation in beef haylage-based finisher diets on growth, in vitro methane production and carcass and meat quality. Meat Sci. 2022;188:108782.
Nogoy KMC, Sun B, Shin S, Lee Y, Zi Li X, Choi SH, et al. Fatty acid composition of grain- and grass-Fed beef and their nutritional value and health implication. Food Sci Anim Resour. 2022;42:18–33.
Masoudzadeh SH, Mohammadabadi M, Khezri A, Stavetska RV, Oleshko VP, Babenko OI, et al. Effects of diets with different levels of fennel (Foeniculum vulgare) seed powder on DLK1 gene expression in brain, adipose tissue, femur muscle and rumen of Kermani lambs. Small Ruminant Research. 2020;193:106276.
Lee C. Genome-wide expression quantitative trait loci analysis using mixed models. Front Genet. 2018;9.
Dervishi E, Serrano C, Joy M, Serrano M, Rodellar C, Calvo JH. The effect of feeding system in the expression of genes related with fat metabolism in semitendinous muscle in sheep. Meat Sci. 2011;89:91–7.
González-Calvo L, Dervishi E, Joy M, Sarto P, Martin-Hernandez R, Serrano M, et al. Genome-wide expression profiling in muscle and subcutaneous fat of lambs in response to the intake of concentrate supplemented with vitamin E. BMC Genomics. 2017;18:92.
Calvo JH, González-Calvo L, Dervishi E, Blanco M, Iguácel LP, Sarto P, et al. A functional variant in the stearoyl-CoA desaturase (SCD) gene promoter affects gene expression in ovine muscle. Livest Sci. 2019;219:62–70.
Fan Y, Ren C, Meng F, Deng K, Zhang G, Wang F. Effects of algae supplementation in high-energy dietary on fatty acid composition and the expression of genes involved in lipid metabolism in Hu sheep managed under intensive finishing system. Meat Sci. 2019;157:107872.
Deng K, Ma T, Wang Z, TanTai W, Nie H, Guo Y, et al. Effects of perilla frutescens seed supplemented to diet on fatty acid composition and lipogenic gene expression in muscle and liver of Hu lambs. Livest Sci. 2018;211:21–9.
Virtanen JK. Randomized trials of replacing saturated fatty acids with n-6 polyunsaturated fatty acids in coronary Heart Disease prevention: not the gold standard? Prostaglandins Leukot Essent Fatty Acids. 2018;133:8–15.
Nong Q, Wang L, Zhou Y, Sun Y, Chen W, Xie J, et al. Low dietary n-6/n-3 PUFA ratio regulates meat quality, reduces triglyceride content, and improves fatty acid composition of meat in Heigai pigs. Animals. 2020;10:1543.
Li B, Qiao L, An L, Wang W, Liu J, Ren Y, et al. Transcriptome analysis of adipose tissues from two fat-tailed sheep breeds reveals key genes involved in fat deposition. BMC Genomics. 2018;19:338.
Urrutia O, Soret B, Insausti K, Mendizabal JA, Purroy A, Arana A. The effects of linseed or Chia seed dietary supplementation on adipose tissue development, fatty acid composition, and lipogenic gene expression in lambs. Small Ruminant Research. 2015;123:204–11.
Al-Thuwaini TM, Al-Shuhaib MBS. Variants of the SCD gene and their association with fatty acid composition in Awassi sheep. Mol Biol Rep. 2022;49:7807–13.
Wang YH, Bower NI, Reverter A, Tan SH, de Jager N, Wang R, et al. Gene expression patterns during intramuscular fat development in cattle1. J Anim Sci. 2009;87:119–30.
Cedernaes J, Alsiö J, Västermark Ã, Risérus U, Schiöth HB. Adipose tissue stearoyl-CoA desaturase 1 index is increased and linoleic acid is decreased in obesity-prone rats fed a high-fat diet. Lipids Health Dis. 2013;12:2.
Iommelli P, Infascelli F, Musco N, Grossi M, Ferrara M, Sarubbi F, et al. Stearoyl-CoA desaturase activity and gene expression in the adipose tissue of buffalo bulls was unaffected by diets with different fat content and fatty acid profile. Agriculture. 2021;11:1209.
Corazzin M, Bovolenta S, Saccà E, Bianchi G, Piasentier E. Effect of linseed addition on the expression of some lipid metabolism genes in the adipose tissue of young Italian Simmental and Holstein bulls1. J Anim Sci. 2013;91:405–12.
Joseph SJ, Robbins KR, Pavan E, Pratt SL, Duckett SK, Rekaya R. Effect of diet supplementation on the expression of bovine genes associated with fatty acid synthesis and metabolism. Bioinform Biol Insights. 2010;4:BBIS4168.
Jacobs AAA, van Baal J, Smits MA, Taweel HZH, Hendriks WH, van Vuuren AM, et al. Effects of feeding rapeseed oil, soybean oil, or linseed oil on stearoyl-CoA desaturase expression in the mammary gland of dairy cows. J Dairy Sci. 2011;94:874–87.
Weiss-Hersh K, Garcia AL, Marosvölgyi T, Szklenár M, Decsi T, Rühl R. Saturated and monounsaturated fatty acids in membranes are determined by the gene expression of their metabolizing enzymes SCD1 and ELOVL6 regulated by the intake of dietary fat. Eur J Nutr. 2020;59:2759–69.
Pewan SB, Otto JR, Kinobe RT, Adegboye OA, Malau-Aduli AEO. Nutritional enhancement of health beneficial omega-3 long-chain polyunsaturated fatty acids in the muscle, liver, kidney, and heart of Tattykeel Australian White MARGRA lambs fed pellets fortified with omega-3 oil in a feedlot system. Biology (Basel). 2021;10:912.
Yan W, Zhou H, Hu J, Luo Y, Hickford JGH. Variation in the FABP4 gene affects carcass and growth traits in sheep. Meat Sci. 2018;145:334–9.
Gan L, Liu Z, Cao W, Zhang Z, Sun C. FABP4 reversed the regulation of leptin on mitochondrial fatty acid oxidation in mice adipocytes. Sci Rep. 2015;5:13588.
Pećina M, Ivanković A. Candidate genes and fatty acids in beef meat, a review. Ital J Anim Sci. 2021;20:1716–29.
Jurie C, Cassar-Malek I, Bonnet M, Leroux C, Bauchart D, Boulesteix P, et al. Adipocyte fatty acid-binding protein and mitochondrial enzyme activities in muscles as relevant indicators of marbling in cattle1. J Anim Sci. 2007;85:2660–9.
Xu QL, Tang GW, Zhang QL, Huang YK, Liu YX, Quan K, et al. The FABP4 gene polymorphism is associated with meat tenderness in three Chinese native sheep breeds. Czech J Anim Sci. 2011;56:1–6.
Wei S, Zan LS, Wang HB, Cheng G, Du M, Jiang Z, et al. Adenovirus-mediated interference of FABP4 regulates mRNA expression of ADIPOQ, LEP and LEPR in bovine adipocytes. Genet Mol Res. 2013;12:494–505.
Poulos SP, Dodson M, v, Hausman GJ. Cell line models for differentiation: preadipocytes and adipocytes. Exp Biol Med. 2010;235:1185–93.
Vargas-Bello-Pérez E, Bionaz M, Garrido-Sartore M, Cancino-Padilla N, Morales M, Romero J, et al. Effect of Soybean Oil and Fish Oil on lipid-related transcripts in Subcutaneous Adipose tissue of dairy cows. Animals. 2019;10:54.
Lee J-S, Priatno W, Ghassemi Nejad J, Peng D-Q, Park J-S, Moon J-O, et al. Effect of dietary rumen-protected L-tryptophan supplementation on growth performance, blood hematological and biochemical profiles, and gene expression in Korean native steers under cold environment. Animals. 2019;9:1036.
Yang C, Liu J, Wu X, Bao P, Long R, Guo X, et al. The response of gene expression associated with lipid metabolism, fat deposition and fatty acid profile in the longissimus dorsi muscle of Gannan yaks to different energy levels of diets. PLoS ONE. 2017;12:e0187604.
Fernyhough MEHDVJHGHRDM. Dedifferentiation of mature adipocytes to form adipofibroblasts: more than just a possibility. Adipocytes. 2005;1:17–24.
Michal JJ, Zhang ZW, Gaskins CT, Jiang Z. The bovine fatty acid binding protein 4 gene is significantly associated with marbling and subcutaneous fat depth in Wagyu x Limousin F2 crosses. Anim Genet. 2006;37:400–2.
Smith S, Witkowski A, Joshi AK. Structural and functional organization of the animal fatty acid synthase. Prog Lipid Res. 2003;42:289–317.
Berndt J, Kovacs P, Ruschke K, Klöting N, Fasshauer M, Schön MR, et al. Fatty acid synthase gene expression in human adipose tissue: association with obesity and type 2 Diabetes. Diabetologia. 2007;50:1472–80.
Liu G, Ding Y, Chen Y, Yang Y. Effect of energy intake and L-carnitine on fattening performance, carcass traits, meat quality, blood metabolites, and gene expression of lamb. Small Ruminant Research. 2020;183:106025.
Braglia S, Zappaterra M, Zambonelli P, Comella M, Dall’Olio S, Davoli R. Analysis of g.265T>C SNP of fatty acid synthase gene and expression study in skeletal muscle and backfat tissues of Italian Large White and Italian Duroc pigs. Livest Sci. 2014;162:15–22.
Moon YS, Latasa M-J, Griffin MJ, Sul HS. Suppression of fatty acid synthase promoter by polyunsaturated fatty acids. J Lipid Res. 2002;43:691–8.
Dentin R, Benhamed F, Pégorier J-P, Foufelle F, Viollet B, Vaulont S, et al. Polyunsaturated fatty acids suppress glycolytic and lipogenic genes through the inhibition of ChREBP nuclear protein translocation. J Clin Invest. 2005;115:2843–54.
Vargas-Bello-Pérez E, Cancino-Padilla N, Geldsetzer-Mendoza C, Morales MS, Leskinen H, Garnsworthy PC, et al. Effects of dietary polyunsaturated fatty acid sources on expression of lipid-related genes in bovine milk somatic cells. Sci Rep. 2020;10:14850.
Song Z, Xiaoli A, Yang F. Regulation and metabolic significance of De Novo Lipogenesis in adipose tissues. Nutrients. 2018;10:1383.
Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. J Pharmacol Pharmacother. 2010;1:94–9.
Edmunds RC, McIntyre JK, Luckenbach JA, Baldwin DH, Incardona JP. Toward enhanced MIQE compliance: reference residual normalization of qPCR gene expression data. J Biomol Tech. 2014;:jbt.14-2502-003.
Edmunds RC. Evidence for thermal adaptation among geographically, genetically and thermally distinct populations of the Australian barramundi, lates calcarifer (Bloch 1790): a multi-level approach. James Cook University; 2009.
Yang C, Lin Y, Qiu Z, Xiang X, Shao D, Li Y, et al. Reference gene selection for qRT-PCR normalization of gene expression analysis in Melaleuca bracteata F. Muell. Under abiotic stresses and hormonal stimuli. Sci Hortic. 2023;319:112184.
Bianchi A, Salvati N. Asymptotic properties and variance estimators of the M-quantile regression coefficients estimators. Commun Stat Theory Methods. 2015;44:2416–29.
Acknowledgements
The authors gratefully acknowledge James Cook University College of Public Health, Medical and Veterinary Sciences (PhD scholarship for the second-named author—S.B.P.), the Australian Commonwealth Department of Industry, Science and Resources Innovation Connections, the Commonwealth Scientific and Industrial Research Organization SIEF Ross Metcalf STEM Business Industrial Research Fellowship (research funding for the first-named author— J.R.O.), Tattykeel Australian White Pty Ltd. (access to flock, farm resources, research funding), CSIRO Marine and Atmosphere Hobart (fatty acid analysis), and the National Veterinary Research Institute Vom, Nigeria (study leave approval for the second-named author—S.B.P.).
Funding
This research was funded by the Innovation Connections Research Grant from the Australian Commonwealth Government’s Department of Industry grant number ICG001084, Science Industry Endowment Fund Ross Metcalf STEM Business Fellowship co-funded by Tattykeel Australian White Pty Ltd. grant number 00044 (awarded to J.R.O.) and a PhD scholarship funded by the James Cook University Postgraduate Research Scholarship (JCUPRS), Queensland, Australia, (awarded to S.B.P).
Author information
Authors and Affiliations
Contributions
Conceptualization, A.E.O.M.-A.; methodology, A.E.O.M.-A., J.R.O., S.B.P., F.W.M, R.C.E., O.A.A. and R.T.K.; software, A.E.O.M.-A.; validation, A.E.O.M.-A.,S.B.P., F.W.M., J.R.O. and R.C.E.; formal analysis, S.B.P. and O.A.A.; investigation, A.E.O.M.-A., J.R.O., S.B.P., F.W.M, R.C.E., O.A.A. and R.T.K.; resources, A.E.O.M.-A., R.T.K. and O.A.A.; data curation, writing—original draft preparation, S.B.P.; Tables (1-2), Figs. (1-2) and supplementary materials (S1-S3) preparation, J.R.O., S.B.P. and O.A.A.; writing—reviewing and editing, J.R.O., A.E.O.M.-A., R.T.K., F.W.M., R.C.E. and O.A.A.; supervision, A.E.O.M.-A., R.T.K. and O.A.A.; project administration, A.E.O.M.-A.; funding acquisition, A.E.O.M.-A. All authors read, reviewed and approved the publication of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The James Cook University Animal Ethics Committee (Permit No. A0015657) approved the use of animals and all procedures executed in this study in compliance with the Australian Code for Care and Use of Animals for Scientific Purposes (Eighth edition, 2013). The reporting in the manuscript follows the recommendations of Kilkenny et al. [57] in the ARRIVE guidelines for animal research.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Otto, J.R., Pewan, S.B., Edmunds, R.C. et al. Differential expressions of FASN, SCD, and FABP4 genes in the ribeye muscle of omega-3 oil-supplemented Tattykeel Australian White lambs. BMC Genomics 24, 666 (2023). https://doi.org/10.1186/s12864-023-09771-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09771-x