Abstract
Background
Paeonia veitchii Lynch, a well-known herb from the Qinghai-Tibet Plateau south of the Himalayas, can synthesize specific monoterpene glycosides (PMGs) with multiple pharmacological activities, and its rhizome has become an indispensable ingredient in many clinical drugs. However, little is known about the molecular background of P. veitchii, especially the genes involved in the biosynthetic pathway of PMGs.
Results
A corrective full-length transcriptome with 30,827 unigenes was generated by combining next-generation sequencing (NGS) and single-molecule real-time sequencing (SMRT) of six tissues (leaf, stem, petal, ovary, phloem and xylem). The enzymes terpene synthase (TPS), cytochrome P450 (CYP), UDP-glycosyltransferase (UGT), and BAHD acyltransferase, which participate in the biosynthesis of PMGs, were systematically characterized, and their functions related to PMG biosynthesis were analysed. With further insight into TPSs, CYPs, UGTs and BAHDs involved in PMG biosynthesis, the weighted gene coexpression network analysis (WGCNA) method was used to identify the relationships between these genes and PMGs. Finally, 8 TPSs, 22 CYPs, 7 UGTs, and 2 BAHD genes were obtained, and these putative genes were very likely to be involved in the biosynthesis of PMGs. In addition, the expression patterns of the putative genes and the accumulation of PMGs in tissues suggested that all tissues are capable of biosynthesizing PMGs and that aerial plant parts could also be used to extract PMGs.
Conclusion
We generated a large-scale transcriptome database across the major tissues in P. veitchii, providing valuable support for further research investigating P. veitchii and understanding the genetic information of plants from the Qinghai-Tibet Plateau. TPSs, CYPs, UGTs and BAHDs further contribute to a better understanding of the biology and complexity of PMGs in P. veitchii. Our study will help reveal the mechanisms underlying the biosynthesis pathway of these specific monoterpene glycosides and aid in the comprehensive utilization of this multifunctional plant.
Similar content being viewed by others
Introduction
Paeonia veitchii of the Paeoniaceae family is a perennial pharmaceutical and ornamental flowering herb inhabiting specific ecological niches of mostly inaccessible locales of the Qinghai-Tibet Plateau in the southern Himalayas. Because of its beautiful flower, P. veitchii is considered a dazzling star in China. The dried rhizomes of P. veitchii, named Chishao, are widely used in traditional Chinese medicine, and they have analgesic effects, regulate menstruation, tonify the blood and act as anti-inflammatory agents [1,2,3].
Previous phytochemical studies of Paeonia revealed that the active compounds in its roots are primarily monoterpene derivatives with a “cage-like” skeleton, including paeoniflorin, albiflorin, oxypaeoniflorin, benzoylpaeoniflorin, oxybenzoylpaeoniflorin, lactiflorin, galloylpaeoniflorin, and paeonin [4,5,6]. Most of these monoterpene derivatives are monoterpene glycosides (PMGs), especially paeoniflorin [7,8,9]. As the most abundant component, paeoniflorin has diverse biological functions, including anti-inflammatory, antioxidant, antithrombotic, anticonvulsive, analgesic, cardioprotective, neuroprotective, hepatoprotective, antidepressant-like, antitumoral, and immunoregulatory activities, as well as enhancing cognition and attenuating learning impairment [10,11,12,13]. In terms of chemical structures, we speculate that many PMGs originate from paeoniflorin, such as benzoylpaeoniflorin and galloylpaeoniflorin.
Unfortunately, the accumulation of PMGs in Paeonia requires approximately 5–6 years [14]. Given their complex molecular structures, PMGs are difficult to chemically synthesize for commercial use. Therefore, metabolic engineering may be an effective method for obtaining increased yields of these components. However, this strategy largely relies on the characterization of their biosynthetic pathway, which still has not been completely elucidated. With the discovery of new PMGs and advances in pharmacological research on PMGs, elucidating the biosynthetic pathways and regulatory mechanisms of active PMGs has attracted the attention of scientists.
The biosynthesis of PMGs begins with terpene pathways, including the 1-deoxy-D-xylulose-5-phosphate/methyl-erythritol-4-phosphate (DXP/MEP) and mevalonate (MVA) pathways [15, 16]. Both pathways result in the biosynthesis of one molecule of isopentenyl diphosphate (IPP) and one molecule of dimethylallyl diphosphate (DMAPP), which is then catalysed by geranyl diphosphate synthase (GPPS) to form geranyl diphosphate (GPP, C10) [17, 18]. In plants, terpene synthases (TPSs) are responsible for the biosynthesis of numerous terpene skeletons from polyisoprene diphosphate precursors, leading to isoprene (C5), monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20) [19]. An early report used transcriptome data of Paeonia lactiflora (another well-known pharmaceutical and ornamental flowering plant of Paeonia) to successfully identify one pinene synthase (PlPIN, KU187411) involved in the conversion of GPP to α-pinene, which is one of the skeletons of PMGs [14]. In the biosynthesis of terpenes in plants, scaffolds catalysed by enzymes of the TPS family are then oxidized through the action of cytochrome P450s (CYPs). Modifications introduced by CYPs significantly increase the structural diversity of terpenoids and provide anchoring points for further linkage of sugar residues, alkylations, or esterifications by other transferases [20, 21]. UGTs can transfer sugar moieties from activated donor molecules to acceptor aglycones [22]. The enzymes involved in the acylation reaction of plant secondary metabolites belong to large acyltransferase families. Among these acyltransferases, the BAHD acyltransferase family can utilize acyl-activated coenzyme A thioesters and catalyse the formation of diverse acyl ester derivatives, such as benzoyl, acetyl, and hydroxycinnamoyl [23, 24]. Based on the chemical structure of PMGs, it can be speculated that their biosynthesis should involve enzymes belonging to UGTs, BAHDs and CYPs families (Fig. 1). Although a number of studies have reported functional genes in Paeonia, they have mainly concentrated on codon usage patterns [25], thermotolerance-related differentially expressed genes [26], fatty acids [27] and anthocyanin biosynthetic genes [28] based on transcriptome sequencing. However, the genes involved in glycosylation, benzoylation, oxidation and reduction of PMGs are unknown.
To date, the methods for mining genes associated with the biosynthesis of natural products mainly depend on the following three strategies: (1) an enzyme activity-guided approach, such as the identification of four BAHD acyltransferases (EpHTT, EpHQT, EpHCT, EpCAS) that complete chicoric acid biosynthesis in purple coneflower [29]; (2) a molecular probe-guided approach, such as discovering two UGTs (SrUGT76G3 & SrUGT73E1) involved in the steviol glycoside biosynthesis pathway from Stevia rebuadiana by the development of a photoaffinity probe [30]; and (3) omics analysis, such as using transcriptome mining in Podophyllum hexandrum to identify six tissue-specific expression genes (CYP71CU1, OMT1,2-ODD, CYP71BE54, CYP82D61, OMT3) that complete the biosynthetic pathway of podophyllotoxin [31].
With the rapid development of high-throughput sequencing technology and large-scale data analysis methods, omics analysis has become the most popular method. Large amounts of genomic and transcriptional data from medicinal plants have been submitted to and generated from public databases, but the progress of candidate gene characterization in specialized plant metabolites remains slow due to the following factors. The divergent evolution of some superfamilies (CYP, UGT, etc.) generated massive numbers of closely related individual members that can modify diverse types of natural metabolites [32, 33]. Compared to microorganisms, the genes involved in plant metabolite biosynthesis are generally scattered throughout the plant genome, further increasing the difficulty of accurately identifying candidate genes through the physical distance of metabolic pathway-related genes [32, 34]. Moreover, the PMG biosynthesis pathway requires the participation of genes of superfamilies such as CYPs and UGTs, making it even more difficult to resolve the genes in its biosynthesis pathway.
Due to their growth in inaccessible places in the Himalayas, no research has examined P. veitchii at the molecular level. The lack of molecular background information about P. veitchii is a huge knowledge gap for scientists trying to understand plateau plants, especially in the Paeonia genus. Therefore, providing the available full-length sequence for each RNA, especially those corrected by NGS reads, is vital for researchers to study P. veitchii at the molecular level and improve knowledge of the existing PMG biosynthesis pathways. In the current study, SMRT long-read sequencing technology and NGS short-read sequencing technology were combined to sequence six different tissues from P. veitchii. WGCNA combining the contents of PMGs, quantitative real-time polymerase chain reaction (qRT–PCR), and phylogenetic analysis was further used to predict the putative genes involved in the biosynthesis of PMGs, especially TPSs, UGTs, CYPs, and BAHDs.
Results
Content of the main PMGs in tissues
Among the numerous PMGs, the concentrations of paeoniflorin were higher than those of other monoterpene glycosides [7,8,9]. In addition, the standards of oxypaeoniflora and albiflorin (also isolated from Paeonia) were easily obtained. Therefore, analysis of the three glycosides in all samples was performed using the HPLC-UV method. All validation projects met the quantitative requirements (Table S1). Then, the production profiles of the 3 metabolites in different tissues (leaves, stems, petals, ovaries, phloem and xylem) of P. veitchii were determined (Fig. 2). Paeoniflorin accumulated in all tested tissues, especially in the xylem and phloem (accounting for more than 45.0 mg/g), while the accumulation levels in stem was only 2.6 mg/g. Albiflorin was hardly detected in petal (0.0 mg/g) and highly accumulated in phloem (1.9 mg/g), xylem (1.1 mg/g), and ovary (1.5 mg/g). The distribution of oxypaeoniflora was undetectable in the ovary (0.0 mg/g) and significantly higher in the phloem (1.4 mg/g) and leaf (1.0 mg/g) than in the other tissues. The analysis suggested that the content of the analyzed PMGs displays variation in different tissues. This variation of content should be useful in predicting the genes involved in the biosynthesis of PMGs.
Transcriptome sequencing and functional annotation
First, leaves, stems, petals, ovaries, phloem and xylem with different concentrations of the PMGs of P. veitchii were used to generate short-read libraries using an Illumina HiSeq X Ten platform, and each sample was sequenced with three biological replicates. After filtering out the adapter sequences and low-quality and contaminated reads, a total of 128.93 Gb clean reads (from 134.55 Gb raw reads) for 18 sequencing libraries, including three biological replicates of leaf, stem, petal, ovary, phloem and xylem, were generated (Table S2). After de novo assembly, a set of 92,776 unigenes was obtained, with a total length, average length, N50, and GC content of 100,517,455 bp, 1,083 bp, 1,475 bp and 44.64%, respectively. According to the statistics, 55,041 (59.3%) unigenes were ≤ 500 bp in length (Fig. S1).
Due to the technical limitations of de novo transcriptome assembly, the unigenes from Illumina sequencing are often fragmented or misassembled [35]. Therefore, the SMRT sequencing platform, which is considered the most reliable means for accessing full-length cDNA molecules [36], was utilized to generate full-length complementary cDNA reads for each RNA extracted from P. veitchii. Finally, a total of 18,603,493 raw reads (approximately 72.2 billion bases) were obtained from the Pacific Biosciences Sequel II platform. After performing the IsoSeq protocols [37], 39,347 high-quality consensus sequences were obtained. Furthermore, Illumina short reads were used to correct the low-quality consensus sequences. Finally, 30,827 full-length transcript sequences with an average length of 2,346 bp were obtained by removing redundant sequences for the high-quality consensus sequences and the corrected low-quality consensus sequences. In addition, 28,468 (92.3%) unigenes were > 1,000 bp, but only 2,359 (7.6%) unigenes were ≤ 1,000 bp in length (Fig. S2). Comparing the unigenes between the Illumina and PacBio Sequel platforms, it was obvious that the unigenes generated with the PacBio Sequel platform more accurately represented the real cDNAs in P. veitchii. Thus, the unigenes obtained by SMRT sequencing were selected as the reference transcriptome for subsequent analyses. In addition, we evaluated the integrity of the Illumina and PacBio transcriptomes using BUSCO [38], and the results showed that complete and single-copy duplicated transcript sequences accounted for 82.6% and 57.3%, respectively.
After annotation in the seven public databases, 28,441 (92.26%), 24,602 (79.81%), 4,850 (15.73%), 18,826 (61.07%), 28,027 (90.92%), 22,356 (72.52%), and 26,288 (85.28%) unigenes from SMRT sequencing were annotated in NR, Swiss-Prot, KEGG, KOG, eggNOG, GO, and Pfam, respectively (Table S3). Most of the unigenes were successfully annotated in at least one of the seven databases. In GO analysis, unigenes annotated in “metabolic process” and “catalytic activity” accounted for relatively higher proportions than unigenes belonging to other GO subcategories (Fig. S3). In KEGG pathway annotation, a large group of unigenes (177) were annotated as “metabolism of terpenoids and polyketides”, which was consistent with the richness of terpenoids in P. veitchii (Fig. S4) [39,40,41].
Identification and functional analysis of the TPS gene family
Based on the 30,827 full-length transcript sequences from PacBio sequencing, 18 nonredundant TPSs were identified using Pfam annotation and the BLAST algorithm. We designated these 18 gene models as TPS1 through TPS18, among which TPS1, TPS5, TPS16, TPS17, and TPS18 appeared to have fragmented open reading frames (ORFs) because they encode for proteins shorter than 450 amino acid residues or do not contain the highly conserved aspartate-rich motif ‘DDxxD’ in their C-terminal domain. Generally, TPS genes in plants are divided into seven clades, with some plant lineages having a majority of their TPS genes in one or two clades [19]. The phylogenetic tree of TPSs with complete ORFs from P. veitchii and other species indicated that these PvTPSs were classified into the TPS-b and TPS-g subfamilies (Fig. 3). However, most of the PvTPSs were grouped into the TPS-b subfamily, which contains the RR(X8) W motif in the N-terminal domain, a key feature of the vast majority of monoterpene synthases [20, 42,43,44]. According to the results of the quantitative analysis of the main PMGs in P. veitchii, monoterpene derivatives account for a large proportion of metabolites in P. veitchii. Therefore, we speculated that the genes of the TPS-b subfamily are involved in the formation of monoterpene skeletons in P. veitchii.
Phylogenetic tree illustrating most of the PvTPSs as the members of TPS-b subfamily. Grouping into subfamilies is based on previous study [19, 42]. Sequence abbreviations: AaLis, linalool synthase (ADD81294); FaNes1, nerolidol synthase1 (CAD57083); VvGwbOcs, Gewürztraminer (E)-beta-ocimene synthase (ADR74204); VvRiLinNer, Riesling linalool/nerolidol synthase (JQ062931); VvTer, (−)-α-terpineol synthase (AAS79352); SITPS38, a-Bergamotene synthase (AEP82768.1). The full-length amino acid sequence of PvTPSs described in additional file 3. The tree was derived by neighbour-joining distance analysis based on a ClustalW protein sequence alignment in MEGA7. Quality of the tree was analyzed by bootstrap analyses consisted of 1000 replicates
Identification and functional analysis of the CYP gene family
In this study, 126 CYPs were characterized from the full-length transcript sequences from PacBio sequencing. According to the cross-kingdom nomenclature system of CYPs [45, 46] with reference to the classification of CYPs in Arabidopsis, these CYPs were assigned to 23 families with the numbers 51 (3), 71 (32), 72 (10), 73 (2), 74 (2), 75 (6), 76 (12), 77 (3), 78 (1), 81 (3), 82 (5), 86 (1), 89 (1), 90 (8), 97 (4), 98 (1), 704 (1), 706 (2), 712 (1), 714 (5), 716 (5), 736 (5), and 749 (12), and one annotated as ent-kaurene oxidase (PvKO). The numbers in brackets are the number of genes in each family. Further analysis showed that the amino sequences of 30 CYPs lacked the heme-binding motif (FxxGxxxCxG) and electron-transfer channel motif (PERF), which are well-conserved motifs in P450s [47]. To predict the enzyme function of this family, the nonredundant CYPs with complete ORFs were further separated into clans defined by the phylogenetic tree (Fig. 4). All functionally characterized plant P450s with activity as monoterpene oxidases belong to the CYP71 clan [46, 48]. In our phylogenetic tree, a total of 58 different PvCYPs belonged to the CYP71 clan. Previous studies have found that among CYP71clan, the vast majority of functionally characterized members identified as involved in monoterpene metabolism in plants belong to the CYP71 and CYP76 subfamilies [46, 49, 50], which made these members interesting candidates for a possible role in PMG biosynthesis.
Phylogenetic tree illustrating the PvCYPs were separated into different clans. The full-length amino acid sequence of PvCYPs described in additional file 4. Clan separation referenced to the data in website: http://www.p450.kvl.dk/cyp_allsubfam_NJ_102103.pdf. Phylogenetic relationship was derived by neighbour-joining distance analysis based on a ClustalW protein sequence alignment in MEGA7. Quality of the tree was analyzed by bootstrap analyses consisted of 1000 replicates
Identification and functional analysis of the UGT gene family
In this work, 53 UGTs were characterized from the full-length transcript sequences from PacBio sequencing. According to the GT nomenclature committee [51] and according to the classification of UGTs in Arabidopsis, these UGTs were classified into 16 subfamilies with the numbers 71 (7), 72 (2), 73 (1), 74 (10), 75 (2), 76 (2), 80 (4), 84 (3), 85 (8), 88 (2), 89 (1), 91 (3), 92 (1), 94 (2), 96 (2), and 709 (3). Among these UGTs, the amino sequence of 8 UGTs was found to be without the PSPG box (plant secondary product glycosyltransferase), which contains a conserved 44 amino acid motif (WAP... DQ) [52]. In addition, a fragmented ORF was found in 9 UGTs. To date, monoterpenoid UGT genes have rarely been reported, whereas diterpenoid and triterpenoid-specific UGTs have been well described. Recently, VvGT14 and VvGT15 from Vitis vinifera and AdGT4 from Actinidia delicosa were reported to be monoterpenol-specific UGTs [53, 54]. Hence, these three monoterpenol-specific UGTs were aligned with the forty-five nonredundant UGTs with complete ORFs, and then a composite phylogenetic tree was constructed (Fig. 5). Interestingly, AdGT4 and VvGT14 were clustered in the UGT85 and UGT709 subfamilies with a very high bootstrap value (over 70%). VvGT15 was clustered in the UGT76 subfamily and had a very high bootstrap value (100%). These results implied that the genes in the UGT85, UGT709, and UGT76 subfamilies may be involved in the glucosylation of monoterpenes in P. veitchii. This work prompted us to further characterize UGT genes associated with the formation of glycosylated PMGs.
Phylogenetic tree of functionally characterized UGT members plus the PvUGT proteins. Glucosyltransferases from Actinidia deliciosa (AdGT4, KF954944), Vitis vinifera (VvGT14, XM_002285734.2), and Vitis vinifera (VvGT15, XM_002281477.2) with known function toward monoterpenol are also shown. The tree was derived by neighbour-joining distance analysis of the full-length amino acid sequence described in additional file 5. Distance bootstrap analyses consisted of 1000 replicates, where values over 50% are indicated above the nodes using symbol
Identification and functional analysis of the BAHD acyltransferase family
According to the annotation information and the two well-conserved motifs in BAHD (HXXXD & DFGWG) [55], 14 BAHD genes in P. veitchii from the transcriptome database of PacBio sequencing were identified. We designated these 14 gene models PvBAHD1 through PvBAHD14. Previous studies have shown that BAHD members are all monomeric enzymes with molecular masses ranging from 48 to 55 kDa, and the average number of amino acids is approximately 445 [23]. Among the 14 identified PvBAHDs, the molecular mass and number of amino acids in PvBAHD13 are 31 kD and 275, respectively. Hence, the PvBAHD13 gene may be a pseudogene.
In an attempt to identify the candidate PvBAHD genes involved in the benzoylation of PMGs, phylogenetic analysis using full-length amino acid sequences of 13 complete PvBAHDs with a number of functionally characterized BAHDs from multiple species was conducted to determine the relationships of the PvBAHDs with the five major BAHD clades previously described [23] (Fig. 6). The results showed that five proteins (PvBAHD4/5/6/7/12) cluster with BAHD clade V-i, which is mainly responsible for utilizing benzoyl-CoA as the major acyl donor to synthesize benzenoid ester production, including CbBEBT from Clarkia breweri [55], NtBEBT from Nicotiana tabacum [55], VhBEBT from Verbena x hybrida [56], BPBT from Petunia x hybrida [57], DBNTBT from Taxus canadensis [58], DBBT from Taxus cuspidata [59], and PtACT49 and PtSABT from Populus trichocarpa [60]. Therefore, these five putative genes may be involved in the benzoylation of PMGs. Interestingly, a new clade consisting of 3 BAHD family members (PvBAHD1/2/8) was generated and has high bootstrap support (100%). Considering the particularity of the growth environment of P. veitchii, the genes in this new clade may have special functions, such as helping plants adapt to the harsh environment or participating in the biosynthesis of other special compounds.
Phylogenetic tree of functionally characterized BAHD members plus the 13 PvBAHD1 proteins. The five major clades (I-V) referenced to the work by D’Auria (2006) [55]. The following proteins and their corresponding GenBank Accession numbers were used: AcAT16 (HO772640), AdAT9 (HO772637), AMAT (AAW22989), AsHHT1 (BAC78633), At5MAT (Q9LJB4), AtACT (Q9FNP9), AtHCT (NP_199704), AtHHT1 (ACY78659), AtSDT (NP_179932), AtSHT (AEC06845), BAPT (AAL92459), BPBT (AAU06226), CbBEAT (AAF04787), CbBEBT (AAN09796), CbRAS (CAK55166), CcHCT (DQ104740), CcHCT1 (EF137954), Cer2 (AAB17946), CHAT (AAN09797), CmAAT1 (CAA94432), CmAAT2 (AAL77060), CmAAT3 (AAW51125), CmAAT4 (AAW51126), CsHCT (AEJ88365), CsHQT (DQ915589), DAT (AAC99311), DBAT (AAF27621), DBBT (Q9FPW3), DBNTBT (AAM75818), Dm3MAT1 (AAQ63615), Dm3MAT2 (AAQ63616), Dm3MAT3 (BAF50706), Dv3MAT (Q8GSN8), EcHQT (AFF19202), Glossy2 (DAA36076), Gt5AT (Q9ZWR8), HCBT (CAB06430), HcHCT (JQ779021), HMT (BAD89275), HvACT (AAO730710), LaAT (AB581532), LaAT1 (DQ886904), Lp3MAT1 (AAS77404), MpAATI (AAU14879), NtBEBT (AAN09798), NtHCT (CAD478300), NtHQT (CAE46932), NtMAT1 (2XR7_A), OsMaT1 (BAD21952), OsMaT2 (NP001046855), Pf3AT (Q9MBC1), Pf5MaT (Q9LJB4), PtFHT1 (XP_002298644), Pun1 (ADN97116), RhAAT1 (AAW31948), SAAT (AAG13130), SalAT (Q94FT4), Sc3MaT (AAO38058), ScSlAT2 (AFM77971), Ss5MaT1 (Q8W1W9), Ss5MaT2 (AAR26385), StFHT (FJ825138), TAT (AAF34254), TpHCT2 (ACI16631), VAAT (AX025504), Vh3MAT1 (AAS77402), VhBEBT (BAE72881), RsVS (CAD89104). PtSABT (Potri.013G074500) and PtACT49 (Potri.019G043600) obtained from Phytozome v10 (http://www.phytozome.net/). The tree was derived by neighbour-joining distance analysis of the full-length amino acid sequence described in additional file 6. Distance bootstrap analyses consisted of 1000 replicates, where values over 50% are indicated above the nodes
Metabolite and gene coexpression analysis to further predict functional genes
WGCNA has been proven to be an effective analytical tool in systematically describing the correlation relationship between clusters of highly correlated genes or modules and external conditions or sample traits [61]. Consequently, this method was selected to further analyse the potential genes involved in the biosynthesis of PMGs. First, 30,827 transcripts from PacBio sequencing were mapped by the NGS clean reads in each tissue, and then the FPKM value of each transcript in tissues was obtained. Then, these 30,827 transcripts were used as raw input data for WGCNA. Based on the FPKM value, the genes with low fluctuation expression (standard deviation < 1) were filtered, and 6,560 genes remained. As shown in Fig. S5, a soft threshold β=30 was used to build the weighted coexpression network. Then, hierarchical clustering on dissimilar matrices was performed by the function hclust, and Dynamic Tree Cut was used to cut the generated cluster tree. Transcripts with a strong correlation were assigned to the same module with different colours. Finally, 6,560 transcripts were divided into 7 modules, and the number of transcripts in each module was 154-1907. Transcripts that failed to be assigned to any module were placed in the grey module, which has no reference significance. The hierarchical clustering dendrogram of gene networks is visualized in Fig. 7a.
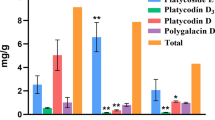
WGCNA analysis to predict that the key genes involved in the biosynthesis of PMGs. A: Clustering dendrograms of 6,560 transcripts (after filtering the low fluctuation expression (standard deviation < 1) transcripts). Each clade corresponding to a module was represented by different color. Modules with certain correlations were merged into the same module. B: Module-PMGs relationships. Each row represents a module of characteristic genes, numbers in bracket are the amount of genes in each module. Each column represents a metabolite. Each cell represents the correlation and the significance (*, **, and *** significant at 10%, 5%, and 1% level, respectively). The list of transcripts in each modules and the expression levels of transcripts in each tissues were provided in the additional file 7 and 8
Then, six generated modules were correlated with the traits (the content of the aforementioned three PMGs in each tissue) to identify potential modules that are significantly related to the PMG content. Based on the absolute value of the Pearson correlation coefficient > 0.3 and p value < 0.05, the modules associated with each PMG were obtained. The module-PMG relationships are colour-coded in Fig. 7b. The larger the absolute value of the Pearson correlation coefficient, the greater the correlation between the genes in the module and the PMG. Depending on the correlation coefficient of the module-PMG, we observed that oxypaeoniflora was positively correlated with the I, III, and IV modules. Albiflorin was positively correlated with the II and VI modules, whereas paeoniflorin clustered well with the VI module (Fig. 7b). The clustering results indicated that the genes in the I, II, III, IV, and VI modules are potentially involved in the biosynthesis of PMGs.
Based on the five predicted modules, 12 TPSs (including 4 pseudogenes), 80 CYPs (including 15 pseudogenes), 8 BAHDs (including 1 pseudogene), and 30 UGTs (including 3 pseudogenes) were obtained (Table S4). The Pearson correlation coefficients between these genes and the PMGs are shown in Fig. 8. Combined with the above analysis of each family: (1) TPSs in the TPS-b subfamily are involved in the formation of the monoterpene skeleton; (2) P450s that can be hydroxylated in the monoterpene skeleton often belong to the CYP71 and CYP76 subfamilies; (3) UGTs in the UGT85 and UGT76 subfamilies are likely to participate in the glucosylation of monoterpenes; and (4) BAHDs clustering with clade V-i may be involved in the benzoylation of metabolites. Genes that have fragmented ORFs were excluded. Finally, we narrowed the TPSs, CYPs, UGTs, and BAHDs to 8, 22, 7, and 2 candidate genes, respectively (Table S5). Notably, PvTPS3, one of the 8 TPSs, was the same as pinene synthase (PlPIN), which proved to be involved in the conversion of GPP to a-pinene, one of the skeletons of PMGs [14]. This further demonstrated the reliability of our analytical result.
The expression levels of these putative genes in leaves, stems, petals, ovaries, phloem and xylem are shown in Fig. 9, and there are significant differences in their expression patterns in different tissues. It is reasonable to believe that these genes are among those most likely to participate in the biosynthesis of monoterpene glycosides, and we will systematically verify them in subsequent research work.
Validation of gene expression profiles by qRT–PCR
To confirm the reliability of the RNA sequencing results, sixteen genes from PvTPSs, PvCYPs, PvBAHDs and PvUGTs were selected for relative quantitative analysis. The primers used in qRT–PCR were listed in Table S6. Regardless of whether actin or GAPDH was used as a housekeeping gene, the relative expression of the sixteen selected genes showed that the tendency of these genes to be expressed was similar between the qRT–PCR and transcriptomic analyses, confirming that the transcriptomic results were reliable (Fig. 10). In addition, the qRT–PCR results of the sixteen genes were significantly different in different tissues, which is consistent with the results of the PMG content analysis.
qRT-PCR validation of RNA sequencing data. Expression profiles of sixteen selected genes were determined by transcriptome and qRT-PCR data. The left vertical axis represents the relative expression based on qRT-PCR. The right vertical axis represents the expression level based on RNA sequencing. The letter denotes statistical significance based on the RNA-seq data (different letters denote P < 0.05). The Le, Ov, Pe, Ph, St, and Xy represented the leaf, ovary, petal, phloem, xylem, and stem, respectively
Discussion
To adapt to the local ecological niche, each plant species may synthesize a different set of secondary metabolites to help facilitate interactions with the local biotic and abiotic environment [18]. Among plant metabolites, terpenoids are the most chemically and structurally diverse family of natural products, encompassing over 80,000 individual compounds, most of which have specialized roles in plant-environment interactions [62,63,64]. Furthermore, a variety of molecules with pharmacological properties, both from medicinal plants and clinical drugs derived from natural products, are terpenoids.
The Qinghai-Tibet Plateau, which is located in the southern Himalayas, is the highest altitude plateau in the world and is called the “Roof of the World”. The environment in the Qinghai-Tibet Plateau has a special plateau mountain climate because of the altitude and terrain [65]. Therefore, terpenoids synthesized by plants in this region should be specialized metabolites that are not wholly shared by all plant lineages. The genetic information of plants in this region, especially the genes involved in the biosynthesis of specialized terpenoids, is also unique. P. veitchii is usually distributed at altitudes ranging from 3,000 to 4,000 metres and is a very representative medicinally perennial plant on the Qinghai-Tibet Plateau, but there are no reports about its transcriptome or other omics. In the present study, we used short-read NGS and long-read SMRT together to sequence P. veitchii and then obtained a high-quality full-length transcriptome (total of unigenes = 30,827, average length = 2,346 bp) and a large-scale NGS assembled transcriptome (total of unigenes = 92,776, average length = 1,083 bp). To the best of our knowledge, this report is the first public study to characterize the structure of transcripts in P. veitchii. These transcriptome data will provide strong support for the further study of plants in the Himalayas, especially on the Qinghai-Tibet Plateau.
Studying the biosynthetic pathway of metabolites originating from medicinal plants is still restricted owing to the complexity of their genomes and the absence of genomic information. As described in the introduction, functional genes involved in the biosynthesis of metabolites in plants are usually diffusely distributed in the chromosomes, and gene redundancy and strict genetic regulation in plants lead to difficulty in resolving metabolic pathways [32, 34, 66]. At present, the traditional methods for analysing the biological pathways of metabolites are limited and often yield a large number of false-positive results, especially for large datasets from heterogeneous sources [32, 61].
Chromatography analysis (HPLC, LC–MS/MS, etc.) of P. veitchii indicated that the majority of its constituents were classified as monoterpenoids, which have many biological activities [67,68,69]. P. lactiflora, another Paeonia plant, contains monoterpenoids similar to those of P. veitchii. Several studies have reported the transcriptome data of P. lactiflora, and some of them were focused on the biosynthesis of monoterpenoids [7, 26, 70, 71]. Due to the existence of hybridization between the cultivars in different regions, the genetic background of P. lactiflora was mixed, and all of these transcriptome measurements of P. lactiflora were dependent on the Illumina platform. Therefore, these studies hardly reflected the complete transcriptome information involved in the biosynthesis of terpenoids. Moreover, studies on the biosynthesis of monoterpenoids have mainly focused on upstream genes in terpene pathways, such as genes in the MEP and MVA pathways [7].
SMRT long-read sequencing technology was proven to be the most reliable means of sequencing full-length cDNA molecules and was widely used to offer access to more complete transcriptome data due to its long reads (average 4-8 kb), higher throughput, faster detection speed and fewer systematic errors caused by in vitro reverse transcription [36]. In this study, the genes TPS, CYP, UGT, and BAHD were systematically characterized through full-length transcriptome sequencing, and their functions related to PMG biosynthesis were successfully analysed. With further insight into TPSs, CYPs, UGTs and BAHDs involved in PMG biosynthesis, we constructed a coexpression network of metabolites with gene expression levels in different tissues to further determine the relationships between these genes and PMGs. Finally, 8 TPSs, 22 CYPs, 7 UGTs, and 2 BAHD putative genes were found to be involved in the biosynthesis of PMGs. Compared with previous reports, our study could provide useful information for the accurate prediction of the biosynthetic pathway of PMGs. It should be noted that the filtered CYP genes in this study were mainly from the CYP71 and CYP76 families. However, a few members of the CYP750, CYP82 and CYP736 families have also been shown to be involved in the metabolism of monoterpenes in plants [46], so genes screened by WGCNA in these families are also worthy of attention. In addition, CYP71D family genes were reported to be involved in the oxidation of cyclic monoterpenes such as limonene, menthol, and carvone [72]. Therefore, PvCYP71D7 and PvCYP71D9 identified in this study are also worthy of attention. Prokaryotic expression systems (e.g., Escherichia coli) have proven to be an efficient way to heterologously express plant TPS [73], UGT [74], and BAHD [60] family genes, while plant CYP family genes (especially for CYP71 subfamily) seem to be preferentially expressed in eukaryotic systems (e.g., yeast) [75]. Interestingly, in some studies, a prokaryotic system was also used to heterologously express plant CYP genes, but N-terminal modification and codon optimization were required [75]. Therefore, the genes filtered in this study can be further analysed for their functional attributes.
According to the phytochemical results, the aerial plant part was also found to be capable of producing PMGs. Consequently, the aerial parts (especially leaves) could be profitable as an alternative resource to increase PMG yields in the future. It would be beneficial to protect this precious plateau plant, whose resources have been threatened due to its excessive overexploitation in the herbal market as well as its short reproductive phase and low germination rate [1,2,3, 14]. In short, this finding significantly improves our understanding of the biosynthetic pathways of monoterpenoids in Paeonia and provides an efficient strategy to elucidate the complex pathway of secondary metabolites in other species.
Methods
Plant material
Four-year-old P. veitchii plants were cultivated in an experimental field on the Qinghai-Tibet Plateau of Southwest Minzu University (Hongyuan, Sichuan Province in China). Thirty independent healthy plants in the later period of blooming were harvested and equally divided into three portions. The leaves, stems, petals, ovaries, phloem and xylem were collected from each portion. Since the xylem and phloem can be easily separated from the root of P. veitchii, we used sharp blade to quickly separate the xylem and phloem from the clean roots. To obtain ideal full-length transcriptome unigenes, samples of the above six tissues were separately collected from the seedling, adult and flowering stages, and three independent healthy plants were selected in each stage. All samples were equally divided into two portions. One portion used for transcriptome sequencing was frozen in liquid nitrogen immediately and stored at -80 °C. Another portion used for PMG analysis was dried at 40 °C. Material collection was conducted in accordance with local legislation, and there was no need for permission from other organizations. We complied with the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Quantitative analysis of the main PMGs
The authentic standards of oxypaeoniflora (purity≥98%, Pufeidebio, China), albiflorin (purity≥98%, Pufeidebio) and paeoniflorin (purity≥98%, Pufeidebio) were accurately weighed and then mixed with methanol. Powdered tissues (0.5 g) were accurately weighed and transferred to a conical bottle (100 mL). Then, 25 mL 80% methanol (1:50 ratio, g/mL) was added. After soaking for 2 hours, the bottle was placed in an ultrasonic machine (KQ-300DE, Kun-shan Ultrasonic Instrument Co., Ltd., China) to extract for 45 min at 60 °C and 300 W. Then, the lost weight was made up with 80% methanol and shaken well. Finally, the mixture was filtered with a 0.45 μm microporous membrane. HPLC analysis was performed using an Agilent HC-C18 column (250×4.6 mm, Agilent) with the following conditions: injection volume, 10 μL; flow rate, 1 mL/min; column temperature, 30 °C; and wavelength, 230 nm. The mobile phases of acetonitrile (A) and water with 0.1% phosphoric acid (B) were used for elution under the following conditions: 0-5 min, 5%-10% A; 5-20 min, 10%-15% A; 20-25 min, 15%-17% A; 25-40 min, 17%-19% A; 40-45 min, 19%-40% A; 45-46 min, 40%-70% A; and 46-60 min, 70% A. The method was validated by the limit of detection (LOD), limit of quantitation (LOQ), calibration curve, mean correlation coefficient, linear range, accuracy, injection precision, stability, and system suitability.
RNA isolation, library preparation and transcriptome sequencing
Total RNA was isolated using the mirVana miRNA Isolation Kit (Cat. AM1561, Invitrogen, Thermo Fisher Scientific Inc., USA) following the manufacturer’s protocol. RNA quality and quantification were verified by an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Then, libraries for NGS were constructed using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Eighteen samples of the six different tissues (each in three replicates) were sequenced using the Illumina HiSeq X Ten platform at OE Biotech Co., Ltd. (Shanghai, China). Equal amounts of different tissues from the seedling, adulting and flowering stages were mixed and used for cDNA synthesis using a SMARTer PCR cDNA Synthesis kit (Clontech, USA). A total of 12 PCR cycles of amplification were performed using PrimeSTAR GXL DNA Polymerase (Clontech). After purification with AMPure PB Beads, the cDNA products were then subjected to the construction of SMRTbell template libraries using the SMRTbell Template Prep Kit 1.0 (PacBio, Menlo Park, USA). Finally, the SMRT cells were sequenced using the Pacific Biosciences Sequel II platform at OE Biotech Co., Ltd.
Gene function annotation and expression analysis
The NGS raw reads in FASTQ format were first processed to obtain clean reads using Trimmomatic. Then, the clean reads were de novo assembled into transcripts by Trinity (version: 2.4) with the paired-end method. The assembled transcripts were clustered using CD-HIT (identify=98%) to generate nonredundant unigenes. The SMRT raw reads were processed using PacBio SMRT Link v6.0.0 analysis package, and the high quality isoform sequences with a predicted accuracy greater than 99% were generated. High-quality isoforms were further clustered by CD-HIT (identify=98%) to obtain nonredundant unigenes. Based on the Illumina clean reads, the per kilobase per million (FPKM) values of unigenes in each tissue were obtained using Bowtie2 and eXpress softwares.
The unigenes were functionally annotated by alignment with the NCBI nonredundant (NR), Swiss-Prot, evolutionary genealogy of genes: Nonsupervised Orthologous Groups (eggNOG) and Clusters of Orthologous Groups for eukaryotic complete genomes (KOG) databases using diamond with a threshold E-value of 10-5. The proteins with the highest hits to the unigenes were used to assign functional annotations. The unigenes were also mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to annotate the potential metabolic pathways. Gene Ontology (GO) classification was performed through the mapping relation between Swiss-Prot and GO terms. The annotation of protein families in the Pfam database was realized by HMMER3 analysis [76].
Gene expression analysis using qRT–PCR
qRT–PCR was used to verify the reliability of the transcriptional data. Extracted RNA from eighteen sequencing samples was reverse-transcribed into cDNA using a HiScript® II Q RT SuperMix for qPCR (+gDNA wiper) Kit (Vazyme, China). All qRT–PCR primers were designed using Primer6.0 (Premier Biosoft, Canada) and synthesized by Beijing Tsingke Biotechnology Co., Ltd. The sequences were shown in Table S6. The specificity of the primers was checked by RT–PCR. A 2 × Taq Pro Universal SYBR qPCR Master Mix kit (Vazyme, China) was used for qRT–PCR in triplicate. The PCR cycles were as follows: 95 °C for 30 s (hold stage); 40 cycles at 95 °C for 5 s and 57 °C for 30 s (PCR stage); 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s (melting curve stage). The 2-ΔΔCt comparative threshold cycle (Ct) method was used to calculate the relative expression levels [77]. Both Actin and GAPDH were used as housekeeping genes.
Phylogenetic analysis
Unigenes belonging to TPSs, CYPs, BAHDs, and UGTs were identified using Pfam annotation and BLAST algorithm. Unigenes with a length under 1000 bp were removed. Guided by the nomenclature system [45, 51], we systematically classified and named the CYPs and UGTs. Functional sequences from other plants were used to construct a phylogenetic tree using MEGA-X (MEGA, http://www.megasoftware.net/). The phylogenetic trees of TPSs, CYPs, BAHDs, and UGTs were constructed using the neighbour-joining clustering method with the amino acid sequences of the ORFs. The topology of phylogeny was evaluated by a bootstrap resampling analysis with 1000 replicates.
Coexpression network analysis
For further identify unigenes potentially involved in the biosynthesis of PMGs, the unsigned, weighted correlation networks were constructed by R package WGCNA [78]. First, unigenes with low fluctuation expression (standard deviation ≤ 1.0) were filtered. The power value of adjacency functions for unsigned networks was selected based on the scale-free topology criterion. Next, WGCNA network construction and module detection were conducted using an unsigned type of topological overlap matrix (TOM). The modules related to each trait were identified according to the absolute value of the correlation coefficient (≥0.3) and p value (<0.05). Network was visualized by R language, Python, and heatmaps [76, 78].
Availability of data and materials
All data sets supporting the conclusions of this study are included within the article (and additional files). The raw data from the Illumina HiSeq X Ten platform have been submitted to the Sequence Read Archive (SRA) of the NCBI under accession numbers SRR19175075, SRR19175069, SRR19175060, SRR19175059, SRR19175058, SRR19175074, SRR19175073, SRR19175072, SRR19175071, SRR19175070, SRR19175068, SRR19175067, SRR19175066, SRR19175064, SRR19175065, SRR19175063, SRR19175062, and SRR19175061. The accession number of the raw data from Pacific Biosciences Sequel II platform was SRR19240505.
Abbreviations
- PMGs:
-
Paeonia monoterpene glycosides
- TPS:
-
Terpene synthase
- CYP:
-
Cytochrome P450
- UGT:
-
UDP-glycosyltransferase
- BAHDs:
-
BAHD acyltransferases
- SMRT:
-
Single-molecule real-time sequencing
- NGS:
-
Next-generation sequencing
- HPLC:
-
High-performance liquid chromatography
- WGCNA:
-
Weighted gene coexpression network analysis
- DXP/MEP:
-
1-deoxy-D-xylulose-5-phosphate/methyl-erythritol-4-phosphate
- MVA:
-
Mevalonate
- IPP:
-
Isopentenyl diphosphate
- DMAPP:
-
Dimethylallyl diphosphate
- GPPS:
-
Geranyl diphosphate synthase
- GPP:
-
Geranyl diphosphate
- PlPIN:
-
Pinene synthase
- qRT–PCR:
-
Quantitative real-time polymerase chain reaction
- ORF:
-
Open reading frame
- LOD:
-
Limit of detection
- LOQ:
-
Limit of quantitation
- CCS:
-
Circular consensus sequences
- FLNC:
-
Full-length nonchimeric
- FPKM:
-
Fragments per kilobase per million
- PSPG:
-
Plant secondary product glycosyltransferase
- ACCT:
-
Acetyl-coenzymeA (CoA) C-acetyltransferase
- HMGS:
-
3-hydroxy-3-methylglutaryl-CoA synthase
- HMGR:
-
3-hydroxy-3-methylglutaryl-CoA reductase
- MVK:
-
Mevalonic acid kinase
- PMK:
-
Phosphomevalonate kinase
- MDC:
-
Mevalonate-5-diphosphate decarboxylase
- IDI2:
-
IPP isomerase
- DXS:
-
Deoxyxyulose-5-phosphate synthase
- DXR:
-
Deoxyxyulose-5-phosphate reductoisomerase
- CMS:
-
4-diphosphocytidyl-2-C-methyl-D-erythritol synthase
- CMK:
-
4-diphosphocytidyl-2-C-methyl-D-erythritol kinase
- MCS:
-
4-diphosphocytidyl-2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
- HDS:
-
1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate synthase
- HDR:
-
1-hydroxy-2-methyl-2(E)-butenyl 4-diphosphate reductase
- NR:
-
NCBI nonredundant
- eggNOG:
-
Nonsupervised orthologous groups
- KOG:
-
Clusters of orthologous groups for eukaryotic complete genomes
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- GO:
-
Gene ontology
References
He CN, Peng Y, Zhang YC, Xu LJ, Xiao PG. Phytochemical and Biological Studies of Paeoniaceae [J]. Chem Biodivers. 2010;7(4):805–38.
Ahmad M, Malik K,Tariq A, Zhang GL,Yaseen G, Rashid N, Sultana S,Zafar M, Ullah K, Khan MPZ. Botany, ethnomedicines, phytochemistry and pharmacology of Himalayan paeony (Paeonia emodi Royle.)[J]. J Ethnopharmacol 2018; 220: 197-219.
Chinese Pharmacopoeia Committee. The pharmacopoeia of the people’s republic of China. Beijing: China Medical Science Press; 2015.
Li R, Zhang JF, Wu YZ, Li YC, Wang LY, Qiu BL, et al. Structures and Biological Evaluation of Monoterpenoid Glycosides from the Roots of Paeonia lactiflora [J]. J Nat Prod. 2018;81(5):1252–9.
Wang HB, Gu WF, Chu WJ, Zhang S, Tang XC, Qin GW. Monoterpene glucosides from Paeonia lactiflora [J]. J Nat Prod. 2009;72(7):1321–4.
Duan WJ, Yang JY, Chen LX, Zhang LJ, Jiang ZH, Cai XD, et al. Monoterpenes from Paeonia albiflora and their inhibitory activity on nitric oxide production by lipopolysaccharide-activated microglia [J]. J Nat Prod. 2009;72(9):1579–84.
Yuan Y, Yu J, Jiang C, Li MH, Lin SF, Wang XM, et al. Functional diversity of genes for the biosynthesis of paeoniflflorin and its derivatives in Paeonia [J]. Int J Mol Sci. 2013;14(9):18502–19.
Wang C, Yuan J, Zhang LL, Wei W. Pharmacokinetic comparisons of Paeoniflflorin and Paeoniflflorin-6’O-benzene sulfonate in rats via difffferent routes of administration [J]. Xenobiotica. 2016;46(12):1142–50.
Wu YC, Yu Y, Wang HP, Jiang YY, Yang ZJ, Zhou J, et al. Preparation of paeoniflorin from the stems and leaves of Paeonia lactiflora Pall. 'Zhongjiang' through green efficient microwave-assisted extraction and subcritical water extraction [J]. Ind Crops Prod. 2021;163:113332.
Ma X, Zhang WW, Jiang YX, Wen JX, Wei SZ, Zhao YL. Paeoniflorin, a Natural Product With Multiple Targets in Liver Diseases-A Mini Review [J]. Front Pharmacol. 2020;11:531.
Shao YX, Xu XX, Li YY, Qi XM, Wang K, Wu YG, et al. Paeoniflorin inhibits high glucose-induced macrophage activation through TLR2-dependent signal pathways [J]. J Ethnopharmacol. 2016;193:377–86.
Chen HW, Dong Y, He XH, Li J, Wang J. Paeoniflorin improves cardiac function and decreases adverse postinfarction left ventricular remodeling in a rat model of acute myocardial infarction [J]. Drug Des Devel Ther. 2018;12:823–36.
Shi DM, Li XF, Li DM, Zhao QJ, Shen YN, Yan HX, et al. Oral administration of paeoniflorin attenuates allergic contact dermatitis by inhibiting dendritic cell migration and Th1 and Th17 differentiation in a mouse model [J]. Int Immunopharmacol. 2015;25(2):432–9.
Ma XH, Guo J, Ma Y, Jin BL, Zhan ZL, Yuan Y, et al. Characterization of a monoterpene synthase from Paeonia lactiflora producing alpha-pinene as its single product [J]. Biotechnol Lett. 2016;38(7):1213–9.
Zhou F, Pichersky E. More is better: the diversity of terpene metabolism in plants [J]. Curr Opin Plant Biol. 2020;55:1–10.
Jain S, Caforio A, Driessen AJ. Biosynthesis of archaeal membrane ether lipids [J]. Front Microbiol. 2014;26(5):641.
Michael G, Irina O, Thuong THN, Rachel DR, Mario GF, Yaron S, et al. Cytosolic monoterpene biosynthesis is supported by plastid-generated geranyl diphosphate substrate in transgenic tomato fruits [J]. Plant J. 2013;75(3):351–63.
Schmidt A, Gershenzon J. Cloning and characterization of isoprenyl diphosphate synthases with farnesyl diphosphate and geranylgeranyl diphosphate synthase activity from Norway spruce (Picea abies) and their relation to induced oleoresin formation [J]. Phytochemistry. 2007;68(21):2649–59.
Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom [J]. Plant J. 2011;66(1):212–29.
Pateraki I, Heskes AM, Hamberger B. Cytochromes P450 for Terpene Functionalisation and Metabolic Engineering [J]. Adv Biochem Eng Biotechnol. 2015;148:107–39.
Bathe U, Tissier A. Cytochrome P450 enzymes: A driving force of plant diterpene diversity [J]. Phytochemistry. 2019;161:149–62.
Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: Structures, Functions, and Mechanisms [J]. Annu Rev Biochem. 2008;77(1):521–55.
D'Auria JC. Acyltransferases in plants: a good time to be BAHD [J]. Curr Opin Plant Biol. 2006;9(3):331–40.
St-Pierre B, Luca VD. Evolution of Acyltransferase Genes: Origin and Diversification of the BAHD Superfamily of Acyltransferases Involved in Secondary Metabolism [J]. Recent Adv Phytochem. 2000;34:285–315.
Wu YQ, Zhao DQ, Tao J. Analysis of Codon Usage Patterns in Herbaceous Peony (Paeonia lactiflora Pall.) Based on Transcriptome Data [J]. Genes. 2015;6:1125–39.
Hao ZJ, Wei MR, Gong SJ, Zhao DQ, Tao J. Transcriptome and digital gene expression analysis of herbaceous peony (Paeonia lactiflora Pall.) to screen thermo-tolerant related differently expressed genes [J]. Genes Genom. 2016;38:1201–15.
Meng JS, Tang YH, Sun J, Zhao DQ, Zhang KL, Tao J. Identification of genes associated with the biosynthesis of unsaturated fatty acid and oil accumulation in herbaceous peony ‘Hangshao’ (Paeonia lactiflora ‘Hangshao’) seeds based on transcriptome analysis [J]. BMC Genomics. 2021;22(1):94.
Zhao DQ, Jiang Y, Ning CL, Meng JS, Lin SS, Ding W, et al. Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactiflora Pall.)[J]. BMC Genomics. 2014;15(1):689.
Fu R, Zhang PY, Jin G, Wang LL, Qi SQ, Cao Y, et al. Versatility in acyltransferase activity completes chicoric acid biosynthesis in purple coneflower [J]. Nat Commun. 2021;12(1):1563.
Li WC, Zhou YQ, You WY, Yang MQ, Ma YR, Wang ML, et al. Development of photoaffinity probe for the discovery of steviol glycosides biosynthesis pathway in Stevia rebuadiana and rapid substrate screening [J]. ACS Chem Biol. 2018;13(8):1944–9.
Warren L, Elizabeth SS. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone [J]. Science. 2015;349(6253):1224–8.
Hua X, Song W, Wang KZ, Yin X, Hao CQ, Duan BZ, et al. Effective prediction of biosynthetic pathway genes involved in bioactive polyphyllins in Paris polyphylla [J]. Commun Biol. 2022;5(1):50.
Rai A, Saito K, Yamazaki M. Integrated omics analysis of specialized metabolism in medicinal plants [J]. Plant J. 2017;90(4):764–87.
Shang Y, Ma YS, Zhou Y, Zhang HM, Duan LX, Chen HM, et al. Plant science. Biosynthesis, regulation, and domestication of bitterness in cucumber [J]. Science. 2014;346(6213):1084–8.
Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis [J]. Genome Biol. 2016;17(1):181.
Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing [J]. Nature. 2014;517:608–11.
Salah EAG, Michael H, Jennifer LJ, Peter N, Nicholas D, Faye S, et al. A survey of the sorghum transcriptome using single-molecule long reads [J]. Nat Commun. 2016;24(7):11706.
Seppey M, Manni M, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness [J]. Methods Mol Biol. 2019;1962:227–45.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes [J]. Nucleic Acids Res. 2000;28(1):27–30.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms [J]. Protein Sci. 2019;28(11):1947–51.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms [J]. Nucleic Acids Res. 2021;49(D1):D545–51.
Zhu BQ, Cai J, Wang ZQ. Identification of a Plastid-Localized Bifunctional Nerolidol/Linalool Synthase in Relation to Linalool Biosynthesis in Young Grape Berries [J]. Int J Mol Sci. 2014;15(12):21992–2010.
Aubourg S, Lecharny A, Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana [J]. Mol Gen Genomics. 2002;267(6):730–45.
Dudareva N, Martin D, Kish CM, Kolosova N, Gorenstein N, Fäldt J, et al. (E)-β-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily [J]. Plant Cell. 2003;15(5):1227–41.
Nelson D. The cytochrome P450 homepage [J]. Hum Genom. 2019;4(1):59–65.
Hansen CC, Nelson DR, Møller BL, Werck-Reichhart D. Plant cytochrome P450 plasticity and evolution [J]. Mol Plant. 2021;14(8):1244–65.
Werck-Reichhart D, Bak S, Paquette S. Cytochromes p450[J]. Arabidopsis Book. 2002;1:e0028.
Hamberger B, Bak S. Plant P450s as versatile drivers for evolution of species-specific chemical diversity [J]. Philos Trans R Soc Lond Ser B Biol Sci. 2013;368(1612):20120426.
Gesell A, Blaukopf M, Madilao L, Yuen MMS, Withers SG, Mattsson J, et al. The Gymnosperm Cytochrome P450 CYP750B1 Catalyzes Stereospecific Monoterpene Hydroxylation of (+)-Sabinene in Thujone Biosynthesis in Western Redcedar [J]. Plant Physiol. 2015;168(1):94–106.
Tina I, Claire P, Benoît B, Nicolas N, Danièle WR. Monoterpenol Oxidative Metabolism: Role in Plant Adaptation and Potential Applications [J]. Front Plant Sci. 2016;7:509.
Mackenzie PI, Owens IS, Burchell B, Bock KW, Bairoch A, Belanger A. The UDP glycosyltransferase gene superfamily: recommended nomenclature update based on evolutionary divergence [J]. Pharmacogenetics. 1997;7(4):255–69.
Hughes J, Hughes MA. Multiple secondary plant product UDP-glucose glucosyltransferase genes expressed in cassava (Manihot esculenta Crantz) cotyledons [J]. Mitochondrial DNA. 1994;5(1):41–9.
Bönisch F, Frotscher J, Stanitzek S, Rühl E, Wüst M, Bitz O, et al. Activity-based profiling of a physiologic aglycone library reveals sugar acceptor promiscuity of family 1 UDP-glucosyltransferases from grape [J]. Plant Physiol. 2014;166(1):23–39.
Yauk YK, Ged C, Wang MY, Matich AJ, Tessarotto L, Cooney JM, et al. Manipulation of flavour and aroma compound sequestration and release using a glycosyltransferase with specificity for terpene alcohols [J]. Plant J. 2014;80(2):317–30.
D’Auria JC, Chen F, Pichersky E. Characterization of an acyltransferase capable of synthesizing benzylbenzoate and other volatile esters in flowers and damaged leaves of Clarkia breweri [J]. Plant Physiol. 2002;130(1):466–76.
Togami J, Tamura M, Ishiguro K, Hirose C, Okuhara H, Ueyama Y, et al. Molecular characterization of the flavonoid biosynthesis of Verbena hybrida and the functional analysis of verbena and Clitoria ternatea F3’5’H genes in transgenic verbena [J]. Plant Biotechnol. 2006;23(1):5–11.
Boatright J, Negre F, Chen XL, Kish CM, Wood B, Peel G, et al. Understanding in vivo benzenoid metabolism in Petunia petal tissue [J]. Plant Physiol. 2004;135(4):1993–2011.
Walker K, Long R, Croteau R. The final acylation step in taxol biosynthesis: cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus [J]. Proc Natl Acad Sci U S A. 2002;99(14):9166–71.
Walker K, Croteau R. Taxol biosynthesis: molecular cloning of a benzoyl-CoA: taxane 2-alpha-O-benzoyltransferase cDNA from Taxus and functional expression in Escherichia coli [J]. Proc Natl Acad Sci U S A. 2000;97(25):13591–6.
Chedgy RJ, Köllner TG, Constabel CP. Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis [J]. Phytochemistry. 2015;113:149–59.
Pei G, Chen L, Zhang W. WGCNA Application to Proteomic and Metabolomic Data Analysis [J]. Methods Enzymol. 2017;585:135–58.
Tholl D. Biosynthesis and biological functions of terpenoids in plants [J]. Biotechnol Isopreno. 2015;148:63–106.
Singh B, Sharma RA. Plant terpenes: defense responses, phylogenetic analysis, regulation and clinical applications [J]. 3 Biotech. 2015;5(2):129–51.
Christianson DW. Structural and Chemical Biology of Terpenoid Cyclases [J]. Chem Rev. 2017;117(17):11570–648.
Pan G, Wang L, Li R, Yuan S, Ji W, Yin F, et al. Tectonic evolution of the Qinghai-Tibet Plateau [J]. J Asian Earth Sci. 2012;53:3–14.
Karaiskos I, Souli M, Galani I, Giamarellou H. Colistin: still a lifesaver for the 21st century?[J]. Expert Opin Drug Metab Toxicol. 2017;13(1):59–71.
Li SL, Song JZ, Choi FFK, Qiao CF, Zhou Y, Han QB, et al. Chemical profiling of Radix paeoniae evaluated by ultra-performance liquid chromatography/photo-diode-array/quadrupole time-of-flight mass spectrometry [J]. J Pharm Biomed Anal. 2009;49(2):253–66.
Wu SH, Wu DG, Chen YW. Chemical constituents and bioactivities of plants from the genus Paeonia [J]. Chem Biodivers. 2010;7(1):90–104.
Parker S, May B, Zhang C, Zhang AL, Lu CJ, Xue CC. A Pharmacological Review of Bioactive Constituents of Paeonia lactiflora Pallas and Paeonia veitchii Lynch [J]. Phytother Res. 2016;30(9):1445–73.
Sheng MH, She JJ, Xu WY, Hong Y, Su Z, Zhang XD. HpeNet: Co-expression Network Database for de novo Transcriptome Assembly of Paeonia lactiflora Pall [J]. Front Genet. 2020;11:570138.
Lu B, An F, Cao L, Gao Q, Wang X, Yang YJ, et al. Comparative transcriptomics characterized the distinct biosynthetic abilities of terpenoid and paeoniflorin biosynthesis in herbaceous peony strains [J]. PeerJ. 2020;8(10):e8895.
Weitzel C, Simonsen HT. Cytochrome P450-enzymes involved in the biosynthesis of mono- and sesquiterpenes [J]. Phytochem Rev. 2015;14:7–24.
Zhou F, Pichersky E. The complete functional characterisation of the terpene synthase family in tomato [J]. New Phytol. 2020;226:1341–60.
Zhang SS, Chen H, Xiao J, Liu Q, Xiao F, Wu W. Mutations in the uridine diphosphate glucosyltransferase 76G1 gene result in different contents of the major steviol glycosides in Stevia rebaudiana [J]. Phytochemistry. 2019;162:141–7.
Schuler MA, Werck-Reichhart D. Functional genomics of P450s[J]. Annu Rev Plant Biol. 2003;54(1):629–67.
Feng M, Chen C, Qu-Bie J, Qu-Bie A, Bao X, Cui Q, et al. Metabolome and transcriptome associated analysis of sesquiterpenoid metabolism in Nardostachys jatamansi [J]. Front Plant Sci. 2022;13:1041321.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method [J]. Nat Protoc. 2008;3(6):1101–8.
Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis [J]. BMC Bioinformatics. 2008;9:559.
Acknowledgements
We thank Shanghai OE Biotech Inc. (Shanghai, China) for high-throughput sequencing service and bioinformatics support.
Funding
This work was supported by the Natural Science Foundation of Sichuan Province (Grant No. 2022NSFSC1434) and the Fundamental Research Funds for the Central Universities (Grant No. ZYN2022072). The supporters had no role in study design, data collection, data analysis, the writing of the manuscript or decision to publish.
Author information
Authors and Affiliations
Contributions
SZ and QBJ performed most of the experiments and data analysis and wrote the manuscript. FM, QBA, HY, and ZZ helped to plant and collect the materials. LY and YX designed and coordinated the studies. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interests for this work.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Validation method parameters for quantification of three monoterpene glycosides. Table S2. Summary of transcriptome data sequenced by Illumina HiSeq X Ten platform and their pretreatment. Table S3. BLAST analysis of SMRT unigenes against seven public databases. Table S4. List of TPSs, CYPs, BAHDs, and UGTs in I, II, III, IV, and VI modules. Table S5. List of TPSs, CYPs, BAHDs, and UGTs, which are likely to participate in biosynthesis of PMGs. Table S6. Primers and annealing length in qRT-PCR.
Additional file 2: Fig. S1.
Assembled sequence length distribution from Illumina HiSeq X Ten platform. Fig. S2. Non-redundant read length distribution from Pacific Biosciences Sequel platform. Fig. S3. SMRT unigenes distribution in GO categories under Biological process, Cellular component and Molecular function. Fig. S4. KEGG functional classification of unigenes from SMRT sequencing. Fig. S5. The network construction parameters of WGCNA.
Additional file 3.
The full-length amino acid sequence of PvTPSs in this study.
Additional file 4.
The full-length amino acid sequence of PvCYPs in this study.
Additional file 5.
The full-length amino acid sequence of PvUGTs in this study.
Additional file 6.
The full-length amino acid sequence of PvBAHDs in this study.
Additional file 7.
List of transcripts in modules after WGCNA analysis.
Additional file 8.
The nucleotide sequence of unigenes from SMRT sequencing.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zhang, S., Qu-Bie, Jz., Feng, Mk. et al. Illuminating the biosynthesis pathway genes involved in bioactive specific monoterpene glycosides in Paeonia veitchii Lynch by a combination of sequencing platforms. BMC Genomics 24, 45 (2023). https://doi.org/10.1186/s12864-023-09138-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09138-2