Abstract
Berberine hydrochloride is the main effective component of Coptis spp. used in Chinese herbal medicine and its underlying molecular mechanisms, responsible for inducing effects in crustacean species, are not fully understood. In this study, the molecular response of the crab Charybdis japonica to berberine hydrochloride exposure was studied using transcriptome sequencing. The survival rate, gene expression and activities of several immune enzymes were measured after berberine hydrochloride treatments, with or without injection of the pathogenic bacterium Aeromonas hydrophila. A total of 962 differentially expressed genes (464 up-regulated and 498 down-regulated) were observed during exposure to 100 mg/L of berberine hydrochloride and in the control group after 48 h. Enrichment analysis revealed that these genes are involved in metabolism, cellular processes, signal transduction and immune functions, indicating that exposure to berberine hydrochloride activated the immune complement system. This bioactive compound simultaneously activated fibrinogen beta (FGB), fibrinogen alpha (FGA), alpha-2-macroglobulin (A2M), kininogen (KNG), fibrinogen gamma chain (FGB), alpha-2-HS-glycoprotein (AHSG), caspase-8 (CASP8), cathepsin L (CTSL), adenylate cyclase 3 (Adcy3) and MMP1. Its action could significantly increase the survival rate of the crabs injected with A. hydrophila and promote the activity of LZM, Caspas8, FGA, ACP and AKP in the hepatopancreas. When A. hydrophila was added, the neutralization of 300 mg/L berberine hydrochloride maximized the activities of Caspas8, LZM, ACP and AKP. Our results provide a new understanding of the potential effects of berberine hydrochloride on the immune system mechanisms in crustaceans.
Similar content being viewed by others
Introduction
Berberine hydrochloride is the hydrochloride form of the natural isoquinoline alkaloid, and is the main active ingredient of several commonly used Chinese herbal medicines, such as those derived from Coptis and Phellodendron plants. It has been shown to have antibacterial [1] and antioxidant properties both in in vitro and in vivo experiments [2], and is considered as a potential biological drug that can replace synthetic antibiotics [3]. However, little information is known about its effects on crustacean species.
Presently, bacterial infections are the leading cause of crab diseases, representing an important limiting factor for the entire crab aquaculture industry [4, 5]. According to reports, several bacterial and viral pathogens regularly infect marine crabs [6], among them, are the gram-negative bacteria Vibrio alginolyticus and Aeromonas hydrophila, that severely affect crab populations causing multiple infections and leading to great economic losses each year [7].
In crabs, the nonspecific immunity is the most important defense against microbial infection. The hepatopancreas, together with body fluids and cell response, controls immune resistance against viruses and bacteria, and it is therefore, the target organ for treatments aimed at improving the response to infections [8,9,10]. In recent years, transcriptome sequencing has become an effective method for evaluating immune responses in aquatic animals, providing valuable data on their key mechanisms and functions [11]. For example, transcriptomics technology has revealed the biological mechanism behind multiple pathogenic microorganisms affecting crustaceans, such as WSSV and Vibrio species. Many immune-related genes and polymorphic microsatellite markers have been identified in crabs [12,13,14], providing important genetic information, essential for further understanding the molecular mechanisms of innate immunity.
The Japanese crab, Charybdis japonica, belongs to the Portunus family and is a common species with considerable commercial value widespread in Southeast Asia [15, 16]. It typically inhabits benthic estuarine environments, characterized by both fine and muddy sand, and can withstand low oxygen concentrations, a wide range of salinity and large temperature fluctuations [17, 18]. In this study, a comparative transcriptome analysis was conducted to identify differentially expressed genes that could potentially emerge in this species under the action of berberine hydrochloride. Subsequently, the variation of gene expression associated with the related immune enzyme activity after Vibrio treatment was measured, and the regulatory mechanisms of immunity were analyzed. Our research aimed to provide a reference for understanding the molecular mechanisms behind the effects of berberine hydrochloride in crustaceans, and to promote the application of this bioactive compound in aquaculture.
Materials and methods
Experimental animals
A total of 580 Charybdis Japonica adult crabs (body weight = 80 ± 2.4 g), collected from the South China Sea, were randomly placed in 29 aquariums containing artificial seawater and equipped with a cyclic exposure system (specific conditions were: salinity of 28 psu, temperature of 25 ± 1 ºC, pH 7.5 ± 0.5, a dissolved oxygen concentration of approximately 5.0 mg L−1 and a 12 h light/dark photoperiod).
Berberine hydrochloride treatment
In this experiment, one berberine hydrochloride (HPLC ≥ 98%, Vicky Biotechnology Co., Ltd.) treatment group and one control group were set every triplicate. Before the treatment, all the crabs were cultured for two weeks to adapt to the laboratory environment and have a 24 h starvation treatment. 100 mg/L of berberine hydrochloride was administered to the treatment group for a 48 h exposure test, crabs were selected randomly from both the experimental control groups, and hepatopancreas samples were removed for RNA isolation.
Bacterial infection test
Samples of Aeromonas hydrophila were kindly provided by Shanghai Ocean University (China). After two weeks, the C. japonica were not fed for 24 h before the experiment. Water quality remained at the same level as acclimation. The 8 treatments were: C. japonica treated with 0, 100, 200, and 300 mg/L of berberine hydrochloride with and without injected with 105 CFU/L of A. hydrophila. According to the preliminary experiment, 300 mg/L is harmless to C. japonica. C. japonica were kept in tanks containing 0, 100, 200 and 300 mg/L berberine hydrochloride in triplicates, and 48 h later, 105 CFU/L of A. hydrophila was injected into the fourth leg in half of the crabs. According to the preliminary experiment, A. hydrophila 105 CFU/L could cause death in C. japonica after 24 h. After the injection of A. hydrophila, the survival rate was calculated (starting at 0 h) for different time periods after 24, 96 and 144 h.
Sample collection
After 24 h injection, 12 crabs were selected randomly, and their hepatopancreas was removed and preserved on ice until further analysis. The hepatopancreas samples of three individuals in each group were mixed in equal amounts. Total RNA was isolated using a TRIzol reagent kit (Invitrogen, USA) following the manufacturer's instructions. An Agilent 2100 Bioanalyzer was used to determine RNA integrity and quality. After extracting total RNA and digesting the DNA with DNase, the eukaryotic mRNA was enriched using magnetic beads with Oligo (dT). A fragmentation reagent was added to break mRNA into short fragments, which were used as templates. Subsequently, one-strand cDNA was synthesized using six-base primers, and double-stranded cDNA was synthesized. After the double-stranded cDNA was purified and repaired, fragment size was selected, and finally PCR amplification was performed. The library was constructed using an Agilent 2100 bioanalyzer (whose quality had been previously evaluated) and an Illumina HiSeqTM 2500 sequencer was used to generate 125 bp or 150 bp paired-end sequence data.
Data pre-processing, quality control and assembly
Quality control was assessed in Trimmomatic [19] and joints were removed; then low-quality bases and N-bases were filtered out, and only high-quality clean reads were retained [20]. The paired-end splicing method in Trinity (version: 2.4) was used to splice clean reads and obtain transcript sequences: the longest one was selected as Unigene based on sequence similarity and length.
Transcriptome quality control, de novo assembly and functional annotation
The original reads were firstly filtered in FASTQ format, and then those containing sequencing adapters were removed, together with nucleotides that were unknown (N than N 10%) or had low quality (quality score ≤ 5), in order to obtain clean reads. At the same time, their Q30 and GC contents were calculated. The remaining high-quality reads were used for downstream analysis. Based on different databases, including non-redundant protein sequences (Nr), non-redundant nucleotides (Nt), Swiss-Prot, Orthologous group (COG), Gene ontology database (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG), a blastx similarity search was performed on all unigenes, with a threshold of e < 1e-5. The GO annotation of Unigene was determined, and finally Unigene was compared to the KEGG [21] database to obtain access information.
Unigene quantification, differential Unigene screening, functional enrichment and cluster analysis
Unigene’s FPKM [22] and count were obtained using bowtie2 [23] and express [23] software. In particular, the latter was used to obtain the number of Unigene reads corresponding to each sample [24]. The “estimateSizeFactors” function of DESeq [25] in the R package was used to standardize the data, and the “nbinomTest” function was used to calculate the p-value and fold change value for ranking differentially expressed genes. The Unigene reads with a p value less than 0.05 and with a multiple of difference greater than 2, were selected and an enrichment analysis of the differentially expressed Unigenes, derived from the GO and KEGG databases [22], was performed to determine the biological functions or pathways that they mainly affect. At the same time, unsupervised hierarchical clustering of Unigenes was performed, and their expression pattern among different samples was displayed using a heat map.
Validation of RNA sequencing and gene expression
To test the reliability of RNA-Seq results, nine candidate genes related to oxidative stress and immune response were selected for qRT-PCR verification. The expression of these genes at different concentrations of berberine hydrochloride, with or without A. hydrophila, was also tested. The design of the primers was based on sequences obtained from the RNAseq and is shown in Table 1. Total RNA was extracted from the hepatopancreas of the control and treatment groups. The reverse transcription of RNA was performed using the PrimeScript RT kit with gDNA eraser (Takara, Nanjing, China) following the kit’s standard protocols. Real-time PCR was conducted in three replicates using reactions containing 10 µL of SYBR mixture, 0.6 µL of each primer, 7.6 µL of ddH2O, and 1.2 µL of cDNA template. The relative gene expression level was evaluated using the 2ΔΔCT method.
Enzyme activity
The hepatopancreas was rinsed with ice-cold physiological saline solution. About 0.1 g of hepatopancreas tissue was homogenized with the solution (hepatopancreas/total extract mass¼10%) and the homogenate was centrifuged at 3000 rpm for 10 min at 4 ℃. The supernatant was used for biochemical assays. The activity of the following enzymes, acid phosphatase (ACP), alkaline phosphatase (AKP), catalase (CAT), lysozyme (LZM), fibrinogen alpha chain (FGA) and Caspase8, was determined using commercial test kits (Nanjing Jiancheng Institute of Biotechnology, Nanjing, China) in accordance with the manufacturer's instructions.
Data analysis
All data were presented as the mean ± SE. To determine the differences between control and treatment groups, one-way analysis of variance (ANOVA) and Tukey’s test were performed. A P < 0.05 was considered significant.
Results
FPKM value distribution of Unigene
As shown in Table 2, the hepatopancreas samples of both the control and experimental groups were subjected to high-throughput transcriptome Illumina sequencing; four replicate samples were sequenced for each group. The average number of original reads after sequencing was 53,737,002, and the average number of clean reads obtained after filtering was 52,435,205. The average effective base percentage was 94.74%, showing 93.82% of bases with a phred value greater than 30 in raw bass.
Unigene expression, volume annotation and statistics
Unigene expression was calculated using the FPKM method, which considers the number of fragments per million of a certain Unigene per kilobase length. The average value of the hepatopancreas median in the control and treatment groups was 1.98. BH1-3 with the smallest average value of FPKM was 13.8121, and the highest CK3 value was 14.93026. The maximum FPKM value and standard deviation for CK1 was 23,433.73 and 166.8806, respectively. The FPKM sum of eight Charybdis japonica samples amounted to 5,781,915.
Results of principal component analysis (PCA analysis) and Unigene statistical diagram of differential expression
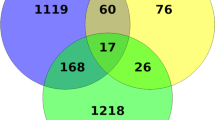
The relationship between the experimental and control group samples was explored through principal component analysis (PCA) (Fig. 1), which showed that BH1-1, BH1-2, BH1-3, and BH1 in the BH1 Charybdis japonica sample presented different dimensions. The sample clustering distance of BH1-4 and that of CK1, CK2, CK3, and CK4 in BCK, were similar in the two-dimensional space. The results showed that the control and experimental groups had multiple up-regulated and down-regulated differences between the samples, corresponding to 464 and 498 Unigenes, respectively; the total number of significantly different expressions of Unigenes was 962 (Fig. 2).
Results of KEGG enrichment analysis
Differentially expressed genes (Fig. 3) in complement and coagulation cascades with innate immune function include fibrinogen beta (FGB), fibrinogen alpha (FGA), alpha-2-macroglobulin (A2M), kininogen (KNG) and the fibrinogen gamma chain (FGB). In particular, adenylate cyclase type 3 (ADCY3), fibrinogen beta (FGB), fibrinogen alpha (FGA) and the fibrinogen gamma chain (FGB) are involved in platelet activation. Lysophospholipase III (LYPLA3) and cathepsin L (CTSL) in lysosomes with innate immune function were significantly changed, and Niemann-Pick C2 protein (NPC2) was down-regulated. NADH-ubiquinone oxidoreductase chain 2 (ND2) in oxidative phosphorylation, alkaline phosphatase activity and molybdenum cofactor biosynthesis protein 1 (Mocs1) in galactose metabolism were all up-regulated. In amino sugar and nucleotide sugar metabolism, UDP-glucose 4-epimerase (Gale), UDP-glucuronate decarboxylase (uxs1), phosphoglucomutase-2 (PGM2) and hydrolase activity CHIT1 were up-regulated. In the same metabolisms, chitinase (Chia) was down-regulated. Fructose and mannose metabolism and Starch and sucrose metabolism genes are also significantly changed.
Differential expression of Unigenes in the KEGG results
The up-regulation and down-regulation of differentially expressed Unigenes are presented in the KEGG level 2 distribution diagram(Fig. 4) One gene in environmental adaptation was on the rise. Nucleotide metabolism had two increasing genes and one decreasing gene. Glycan biosynthesis and metabolism had one up-regulated gene and four down-regulated genes. Lipid metabolism had two up-regulated genes and one down-regulated gene. Because C. japonica can only rely on the innate immune system to respond to viruses or bacteria, the focus of the present study was precisely the immune system, which presented 10 up-regulated genes—FGA, FGB, FGG, KNG, A2M, MMP1, AHSG, caspase-8 (CASP8), cathepsin L (CTSL) and adenylate cyclase 3 (Adcy3)— and five down-regulated genes—egr, adenosine kinase (Adk), chia, angiotensin-converting enzyme (ACE) and surfactant protein-A1 (SFTPA1).
Gene transcription level
Most of the gene expression selected in this study showed an increased trend as berberine hydrochloride concentration increased (Fig. 5). At 200 and 300 mg/L, FGA gene expression significantly increased without A. hydrophila injection (P < 0.05). At 100 mg/L without A. hydrophila injection and higher concentrations (200 and 300 mg/L) with A. hydrophila injection, FGB gene expression significantly increased (P < 0.05). At 200 mg/L with A. hydrophila injection, A2M gene expression level was significantly stimulated (P < 0.05). At 100 and 200 mg/L with A. hydrophila injection, KNG expression level significantly decreased (P < 0.05). The gene expression level of MMP1 significantly increased at all concentrations of berberine hydrochloride without A. hydrophila injection (P < 0.05). CASP8 showed a similar trend to MMP1. ASHG showed no significant changes in all groups.
Survival rate of Charybdis japonica at 24 h, 96 h, and 144 h after being infected with A. hydrophila: CK (control group) AH (A. hydrophila injection of 105 CFU/L), BH1 (exposure to 100 mg/L of berberine hydrochloride), AHBH1(A. hydrophila injection of 105 CFU/L and exposure to 100 mg/L of berberine hydrochloride), BH2 (exposure to 200 mg/L berberine hydrochloride), AHBH2 (A. hydrophila injection of 105 CFU/L and exposure to 200 mg/L of berberine hydrochloride), BH3 (exposure to 300 mg/L of berberine hydrochloride), AHBH3 (A. hydrophila injection of 105 CFU/L and exposure to 300 mg/L of berberine hydrochloride). N per group = 10
Survival rate and activities of selected immune enzymes
Between groups injected with A. hydrophila, the survival rate was highest at the berberine hydrochloride concentration of 300 mg/L (Fig. 6). This group showed a significant increase of the activity of LZM, caspas8, FGA, ACP and AKP in the hepatopancreas (Fig. 7). When A. hydrophila was added, the neutralization of 300 mg/L berberine hydrochloride maximized the activity of Caspas8, LZM, ACP and AKP in the hepatopancreas.
Discussion
This study found that 962 genes were significantly up-regulated or down-regulated under the action of berberine hydrochloride. A number of immune-related genes were also identified, involved in the synthesis of fibrinogen, lysophospholipase, cathepsin L, chitinase, alpha-2-macroglobulin (A2M), kininogen (KNG), cathepsin L (CTSL), caspase-8 (CASP8), alkaline phosphatase (AKP), acid phosphatase (ACP) and lysozyme (LSZ). These genes play an important role in the innate immunity of crustaceans by activating specific antiviral and antibacterial immune responses. Fibrinogen is a protein with coagulative functions and is mainly synthesized by liver cells; it represents the highest coagulation factor in the plasma and is one of the primary components of blood coagulation. It is converted into fibrin through thrombin catalysis processes occurring in the circulatory system, initiating coagulation and physiological hemostasis functions, which are the primary defense reactions during direct exposure of blood to the external environment [26, 27]. Three peptide chains of fibrinogen are respectively encoded by three independent genes: FGA, FGB and FGG [28, 29]. Fibrin is an acute phase response protein, and the concentration of FGA and FGG in the plasma in the event of tissue damage, infection or inflammation can rise significantly [30]. For example, rainbow trout Oncorhynchus mykiss infected with Aeromonas salmonicida presented significant changes in FGA and FGG contents in the plasma [31]. In the present study, under the action of berberine hydrochloride, the expression of FGA, FGB, and FGG in C. japonica increased significantly, which in turn determined an increase in fibrinogen content and provided a strong line of defense against the pathogen.
The antimicrobial peptide Alpha2-macroglobulin (A2M) is involved in the phenoloxidase system, phagocytosis, and the antimicrobial peptide system in crabs [32]. Shrimps express large amounts of the multifunctional A2M protein in plasma and A2M is highly up-regulated after microbial infection [33]. It is known that A2M can inhibit a variety of proteases by inhibiting plasmin and kallikrein, therefore, demonstrating an anti-protease action, and it can also act as a carrier protein [34]. Additionally, it can directly decompose bacteria and fungi, inhibit the replication of bacteria or viruses, or act as an opsonin to increase the amount of phagocytosis, and even neutralize bacteria [35]. It appears that the A2M level detected in C. japonica’s hepatopancreas showed an increasing trend during the response to A. hydrophila infection, which presented in the hemolymph coagulation system and resulted in a strong defense against infection. It seems that the effector cells and inflammatory factors affect the RNA expression of MMPs simultaneously, thereby regulating their reproduction and immunity [36], specifically affecting the expression of MMP1 [37, 38].
Epithelium contributes to mesenchymal transition, and stimulates angiogenesis [39, 40], and induces growth and repair functions [40]. As a result, when the organism responds to pathogens by increasing protease activity and the pathogens in turn release exogenous proteases, A2M attaches to the exogenous proteases, removing them, preventing tissue damage. The present study showed that, under the action of berberine, the hepatopancreas infection caused by A. hydrophila, is counteracted by increased A2M levels that protect the tissues, while MMPs are out of balance with their inhibitors due to the breakdown of the dynamic balance of extracellular mechanisms [41, 42].
A study of the serum protein AHSG reported a correlation between AHSG polymorphisms and AHSG serum concentration levels [43, 44]. The utilization of AHSG by macrophages is essential for regulating the innate immune response in case of tissue damage and infection, and it is noteworthy that berberine hydrochloride promotes the up-regulation of AHSG in C. japonica. AHSG is an endogenous cation, and its increase is necessary for the inactivation of macrophages [45].
Adcy3, which encodes type 3 adenylate cyclase, participates in the cAMP signaling pathway [46]. The significant functions of the gene pathways that are up-regulated by cAMP signaling involve nutrient transport, carbohydrate metabolism, and lysosomal function, as well as those involved in signal transduction, gene transcription, and immune function [47]. cAMP is an important second messenger of intracellular signal transduction operating downstream of key metabolic media (such as ghrelin and the alpha-melanocyte stimulating hormones), and this process controls the development and function of adipose tissue and insulin secretion in the associated beta cells [48]. In fish, the combination of SNP sites in Adcy3 showed a significant effect on the resistance against Vibrio anguillarum [49]. It has been shown that the up-regulation of Adcy3 may activate other immune factors and promote the synthesis of other immune-related proteins.
Kallikrein kinin system (KKS) consists of two main cascades: plasminogen (HMW) and plasma kininogen (KKS). These two proteins are conserved among vertebrates and are involved in inflammatory processes. The concentration of acute C-reactive protein (CRP) in the plasma increases sharply when the body is infected or damaged. As a result of this protein, complement activation and phagocytosis processes are enhanced. Opsonization is subsequently enhanced leading to the elimination of pathogenic microorganisms and prevention of tissue damage, necrosis, and cell death [50]. Previous experiments showed that, during infections, KNG gene expression and C-reactive protein contents increased simultaneously, displaying a new type of interaction between the innate immune CRP system and the inflammatory protein kininogen (KNG) [51]. In this study, a depression of KNG was observed at berberine hydrochloride treated groups injected with A. hydrophila which suggested that some symptoms of the infection probably abated with the treatment of berberine hydrochloride.
Caspases can be divided in three categories based on different functions: initiator apoptotic, effector cell apoptotic and inflammatory [51]. Exogenous cell apoptosis depends on the formation of death-inducing signal complexes, which contain caspase-8 [52]. This enzyme plays a key role in the caspase-dependent apoptotic pathway. Up-regulation of the caspase-8 gene can effectively control exogenous cell apoptosis by reducing inflammation and the action of toxins and viruses [53, 54]. It is known that cadmium induces hepatopancreatic apoptosis through a mitochondrial caspase-dependent pathway, and it has been demonstrated that in this context, the activity of caspase-8 gradually increases in crabs. In this study, it was demonstrated that berberine hydrochloride could significantly increase the caspase-8 level in the hepatopancreas of C. japonica (Fig. 7). When injected with A. hydrophila, the fluctuation range within the group became larger but no significant differences were observed between different groups. It seems that berberine hydrochloride could significantly decrease the adverse effects caused by A. hydrophila by activating the apoptotic process.
Crabs, including C. japonica, do not have an adaptive immune system and depend upon non-specific immune responses. Specific enzymes, such as LZM, ACP and AKP, play an important role in crustacean immunity and are considered reliable indicators for assessing the non-specific immune status of crustaceans. LZM is produced in crustaceans by the activation of complement and phagocytes, through neutrophils and macrophages. After being secreted, it is used to eradicate pathogenic bacteria from the hemolymph and mucus [55]. AKP can enhance the recognition and phagocytosis of pathogens by modifying the structure of the surface of pathogen cells [56]. The lysosomal activity of ACP has similar functions to those of LZM [57]. In this experiment, C. japonica supplemented with berberine hydrochloride, with or without the injection of A. hydrophila, displayed a considerable increase in the activities of LZM, ACP and AKP in the hepatopancreas, suggesting a correlation between berberine hydrochloride and the activity of these specific enzymes. As A. hydrophila was injected and 300 mg/L of berberine hydrochloride was added at the same time, the neutralizing action of the bioactive compound maximized the activity of LZM, ACP and AKP in the hepatopancreas. At 24 h, 96 h and 144 h after injection, the survival rate of the group administered with 300 mg/L of berberine hydrochloride was the highest. It seems that after 24 h, the injection of A. hydrophila had little effect on enzyme activity. The results showed that berberine hydrochloride could significantly increase the immune ability and improve the survival rate of C. japonica when A. hydrophila was injected. At 300 mg/L, it seems to be the best effects on most of the parameters and at 100 mg/L the effects of berberine hydrochloride was obvious and sufficient for survival rate after 144 h.
In conclusion, this is the first report of transcriptome analysis of related immune gene responses under the action of berberine hydrochloride in crustaceans. In particular, LZM, ACP, and AKP were revealed as key enzymes playing a positive role in the immune response caused by berberine hydrochloride. In C. japonica the up-regulation of immune related genes—FGA, AHSG, MPP1, KNG, A2M, FGB and FGG—was simultaneously stimulated by berberine hydrochloride. Bioinformatics analysis showed that these genes may activate the immune complement system in the hemolymph. These findings will contribute to improving the understanding of the molecular mechanisms related to berberine hydrochloride in crustaceans, and to the development and application of treatments using this bioactive compound in aquaculture.
Availability of data and materials
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SUB 11,219,827, Bio project accession: PRJNA819489.
References
Domadia PN, Bhunia A, Sivaraman J, Swarup S, Dasgupta D. Berberine Targets Assembly of Escherichia coli Cell Division Protein FtsZ. Biochemistry. 2008;47(10):3225–34.
Chen PN, Chu SC, Kuo WH, Chou MY, Lin JK, Hsieh YS. Epigallocatechin-3 Gallate Inhibits Invasion, Epithelial-Mesenchymal Transition, and Tumor Growth in Oral Cancer Cells. J Agric Food Chem. 2011;59(8):3836–44.
Zhang D, Li A, Xie J, Ji C, Zhang D, Li A, Xie J, Ji C. In vitro antibacterial effect of berberine hydrochloride and enrofloxacin to fish pathogenic bacteria. Aquac Res. 2010;41(7):1095–100.
Xia XA, Wu QY, Li YY. Isolation and identification of two bacterial pathogens from mixed infection mud crab Scylla serrata and drug therapy. J Trop Oceanogr. 2010;29(5):103–10.
Bonami JR, Zhang S. Viral diseases in commercially exploited crabs: A review. J Invertebr Pathol. 2011;106(1):6–17.
Johnson PT: 1 - Diseases Caused by Viruses, Rickettsiae, Bacteria, and Fungi. In: The Biology of Crustacea. Edited by Provenzano AJ: Academic Press; 1983: 1–78.
Chen DW, Zhang M, Shrestha S. Compositional characteristics and nutritional quality of Chinese mitten crab (Eriocheir sinensis). Food Chem. 2007;103(4):1343–9.
Escobedo-Bonilla CM, Alday-Sanz V, Wille M, Sorgeloos P, Nauwynck HJ. A review on the morphology, molecular characterization, morphogenesis and pathogenesis of white spot syndrome virus. J Fish Dis. 2010;31(1):1–18.
Vaseeharan B, Jayakumar R, Ramasamy P. PCR-based detection of white spot syndrome virus in cultured and captured crustaceans in India. Lett Appl Microbiol. 2010;37(6):443–7.
Hameed ASS, Balasubramanian G, Musthaq SS, Yoganandhan K. Experimental infection of twenty species of Indian marine crabs with white spot syndrome virus (WSSV). Dis Aquat Org. 2003;57(1–2):157.
Yang X, Ikhwanuddin M, Li X, Lin F, Wu Q, Zhang Y, You C, Liu W, Cheng Y, Shi X. Comparative Transcriptome Analysis Provides Insights into Differentially Expressed Genes and Long Non-Coding RNAs between Ovary and Testis of the Mud Crab (Scylla paramamosain). Mar Biotechnol. 2018;20(1):20–34.
Hongyu M, Chunyan M, Shujuan L, Wei J, Xincang L, Yuexing L, Lingbo M, Cynthia G. Transcriptome Analysis of the Mud Crab (Scylla paramamosain) by 454 Deep Sequencing: Assembly, Annotation, and Marker Discovery. PLoS ONE. 2014;9(7): e102668.
Zhao M, Wang W, Chen W, Ma C, Zhang F, Jiang K, Liu J, Diao L, Qian H, Zhao J. Genome survey, high-resolution genetic linkage map construction, growth-related quantitative trait locus (QTL) identification and gene location in Scylla paramamosain. Sci Rep. 2019;9(1):2910.
Jin S, Fu H, Sun S, Jiang S, Xiong Y, Gong Y, Qiao H, Zhang W, Wu Y. iTRAQ-based quantitative proteomic analysis of the androgenic glands of the oriental river prawn, Macrobrachium nipponense, during nonreproductive and reproductive seasons. Comp Biochem Physiol D: Genomics Proteomics. 2018;26:50–7.
Micheletti C, Critto A, Marcomini A. Assessment of ecological risk from bioaccumulation of PCDD/Fs and dioxin-like PCBs in a coastal lagoon. Environ Int. 2007;33(1):45–55.
Gao J, Wang X, Zou Z, Jia X, Wang Y, Zhang Z. Transcriptome analysis of the differences in gene expression between testis and ovary in green mud crab ( Scylla paramamosain ). BMC Genomics. 2014;15(1):585.
Gust N, Inglis GJ. Adaptive Multi-scale Sampling to Determine an Invasive Crab’s Habitat Usage and Range in New Zealand. Biol Invasions. 2006;8(2):339–53.
Hong YuL, Lu Qing P, Debin Z. Effects of Salinity on Biogenic Amines, Hemolymph Osmotic Pressure, and Activity of Gill’s Na/K-ATPase in Charybdis japonica (Crustacea, Decapoda). J World Aquac Soc. 2010;39(6):812–20.
Bolger AM, Marc L, Bjoern U. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;15:2114–20.
Grabherr M, Haas B, Yassour M, Levin JZ, Thompson DA. Amit I: Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. 2011;29:644–52.
Altschul S, Gish W, Miller W, Myers E, Lipman D: Basic local alignment search tool. Journal of Molecular Biology. In: 1990.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480-484.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10(1):71–3.
Anders S. Analysing RNA-Seq data with the DESeq package. Mol Biol. 2010;43:1–17.
Wang Y, Sun Y, Feng J, Li Z, Linghu E. Glycopatterns and Glycoproteins Changes in MCN and SCN: A Prospective Cohort Study. Biomed Res Int. 2019;2019(19):1–11.
Zheng, Cao, Ying, Dong, Jiazi, Zeng, Hongyuan, Zhu, Xin, Xie: Whole-exome sequencing identified novel mutations in FGA and FGG genes in the patients with decreased fibrinogen. Thrombosis research 2019.
Shijie, Duan, Baocheng, Gong, Pengliang, Wang, Hanwei, Huang, Lei, Luo: Novel prognostic biomarkers of gastric cancer based on gene expression microarray: COL12A1, GSTA3, FGA and FGG. Molecular Medicine Reports 2018.
Neerman-Arbez M, de Moerloose P, Honsberger A, Parlier G, Arnuti B, Biron C, Borg JY, Eber S, Meili E, Peter-Salonen K, et al. Molecular analysis of the fibrinogen gene cluster in 16 patients with congenital afibrinogenemia: novel truncating mutations in the FGA and FGG genes. Hum Genet. 2001;108(3):237–40.
Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340(6):448–54.
Ji L, Sun G, Li X, Liu Y. Comparative transcriptome analysis reveals the mechanism of β-glucan in protecting rainbow trout (Oncorhynchus mykiss) from Aeromonas salmonicida infection. Fish Shellfish Immunol. 2020;98:87.
Ning J, Liu Y, Gao F, Song C, Cui Z. Two alpha-2 macroglobulin from Portunus trituberculatus involved in the prophenoloxidase system, phagocytosis and regulation of antimicrobial peptides. Fish Shellfish Immunol. 2019;89:574–85.
Ponprateep S, Vatanavicharn T, Lo CF, Tassanakajon A, Rimphanitchayakit V. Alpha-2-macroglobulin is a modulator of prophenoloxidase system in pacific white shrimp Litopenaeus vannamai. Fish Shellfish Immunol. 2017;62:68–74.
Luan Y, Kong L, Howell DR, Ilalov K, Fajardo M, Bai XH, Cesare PED, Goldring MB, Abramson SB, Liu CJ. Inhibition of ADAMTS-7 and ADAMTS-12 degradation of cartilage oligomeric matrix protein by alpha-2-macroglobulin. Osteoarthritis Cartilage. 2008;16(11):1413–20.
Itami T, Asano M, Tokushige K, Kubono K, Nakagawa A, Takeno N, Nishimura H, Maeda M, Kondo M, Takahashi Y. Enhancement of disease resistance of kuruma shrimp, Penaeus japonicus, after oral administration of peptidoglycan derived from Bifidobacterium thermophilum. Aquaculture. 1998;164(1–4):277–88.
Li R, Meng Q, Huang J, Wang S, Sun J. MMP-14 regulates innate immune responses to Eriocheir sinensis via tissue degradation. Fish Shellfish Immunol. 2020;99:301–9.
Remacle AG, Cieplak P, Nam DH, Shiryaev SA, Ge X, Strongin AY. Selective function-blocking monoclonal human antibody highlights the important role of membrane type-1 matrix metalloproteinase (MT1-MMP) in metastasis. Oncotarget. 2017;8(2):2781–99.
Cepeda MA, Evered CL, Pelling JJH, Damjanovski S. Inhibition of MT1-MMP proteolytic function and ERK1/2 signalling influences cell migration and invasion through changes in MMP-2 and MMP-9 levels. Journal of Cell Communication & Signaling. 2016;11(2):1–13.
Liu Y, Sun X, Feng J, Deng LL, Liu Y, Li B, Zhu M, Lu C, Zhou L. MT2-MMP induces proteolysis and leads to EMT in carcinomas. Oncotarget. 2016;7(30):48193–205.
Itoh Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biology Journal of the International Society for Matrix Biology. 2015;44–46:207–23.
Kanoh Y, Ohtani H, Egawa S, Baba S, Akahoshi T. Changes of proteases and proteinase inhibitors in androgen-dependent advanced prostate cancer patients with alpha2-macroglobulin deficiency. Clin Lab. 2012;58(3–4):217–25.
Md T, Arner EC, Hills R, Easton A, Kortesarfaty J, Fok K, Wittwer AJ, Liu RQ, Malfait AM. Alpha2-macroglobulin is a novel substrate for ADAMTS-4 and ADAMTS-5 and represents an endogenous inhibitor of these enzymes. J Biol Chem. 2004;279(17):17554–61.
Osawa M, Umetsu K, Ohki T, Nagasawa T, Suzuki T, Takeichi S. Molecular evidence for human alpha 2-HS glycoprotein (AHSG) polymorphism. Hum Genet. 1996;99(1):18–21.
Osawa M, Tian W, Horiuchi H, Kaneko M, Umetsu K. Association of α2-HS glycoprotein (AHSG, fetuin-A) polymorphism with AHSG and phosphate serum levels. Hum Genet. 2005;116(3):146–51.
Wang Haichao. Zhang, Minghuang: Fetuin (Alpha2-HS-glycoprotein) opsonizes cationic macrophage-deactivating molecules. Proc Natl Acad Sci U S A. 1998;95(24):14429–34.
Midttun H, Ramsay A, Müller-Harvey I, Williams A. Cocoa procyanidins modulate transcriptional pathways linked to inflammation and metabolism in human dendritic cells. Food Funct. 2018;9(5):2883–90.
Hu J, Wang Y, Le Q, Yu N, Cao X, Kuang S, Zhang M, Gu W, Sun Y, Yang Y. Transcriptome sequencing of olfactory-related genes in olfactory transduction of large yellow croaker (Larimichthy crocea) in response to bile salts. PeerJ. 2019;7:e6627.
Leticia G, Jose RB, Fermín M, Fernando C, Lourdes O, Marta C, Martínez J. Interaction between an ADCY3 Genetic Variant and Two Weight-Lowering Diets Affecting Body Fatness and Body Composition Outcomes Depending on Macronutrient Distribution: A Randomized Trial. Nutrients. 2018;10(6):789.
Zhang K, Han M, Liu Y, Lin X, Liu X, Zhu H, He Y, Zhang Q, Liu J. Whole-genome resequencing from bulked-segregant analysis reveals gene set based association analyses for the Vibrio anguillarum resistance of turbot (Scophthalmus maximus). Fish shellfish immunol. 2019;88:76–83.
Wu Y, Potempa LA, Kebir DE, Filep JG. C-reactive protein and inflammation: conformational changes affect function. Biol Chem. 2015;396(11):1181–97.
Bergsma D, Chen S, Buchweitz J, Gerszten R, Haab BB. Antibody-array interaction mapping, a new method to detect protein complexes applied to the discovery and study of serum amyloid P interactions with kininogen in human plasma. Mol Cell Proteomics. 2010;9(3):446–56.
Tummers B, Green DR. Caspase-8: regulating life and death. Immunol Rev. 2017;277(1):76–89.
Medema JP, Scaffidi C, Kischkel FC, Shevchenko A, Peter ME. FLICE is activated by association with the CD95 Death-inducing signaling complex (DISC). EMBO J. 1997;16(10):2794–804.
LiLi Wei, Li He, Jianping Fu, Yi Liu, Jiming Ruan. Molecular characterization of caspase-8-like and its expression induced by microcystin-LR in grass carp (Ctenopharygodon idella). Fish & Shellfish Immunology. 2019;89:727–35.
Haug T, Kjuul AK. Stensv?G K, Sandsdalen E, Styrvold OB: Antibacterial activity in four marine crustacean decapods. Fish Shellfish Immunol. 2002;12(5):371–85.
Chen QX, Zheng WZ, Lin JY, Shi Y, Xie WZ, Zhou HM. Effect of metal ions on the activity of green crab (Scylla serrata) alkaline phosphatase. Int J Biochem Cell Biol. 2000;32(8):879–85.
Cheng T. Immunodeficiency Diseases in Marine Mollusks: Measurements of Some Variables. J Aquat Anim Health. 1989;1(3):209–16.
Acknowledgements
The authors would like to thank for Mengling Sun her help with sampling.
Funding
This work was supported by the National Key R&D Program of China (2018YFD0901303 and 2020YFD0900305), the Agricultural Project from Jiangsu Province Science and Technology Agency (no.BE2020348) and Jiangsu Agricultural Industry Technology System (JATS[2022]344) in China.
Author information
Authors and Affiliations
Contributions
Mingming Han designed the study, Chenxi Zhu, Zakaria Zuraini, Hui Zhou, and YanXia Guo in the acquisition, analysis, and interpretation of the data, YanXia Guo, Tianheng Gao and Yin Yang Participate in data acquisition and analysis. Qichen Jian drafted the article and all the authors had final approval of the version to be submitted.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Gao, Th., Han, Mm., Zhou, H. et al. Effects of berberine hydrochloride on immune response in the crab Charybdis japonica. BMC Genomics 23, 578 (2022). https://doi.org/10.1186/s12864-022-08798-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08798-w