Abstract
Background
The sustainable development of rice production is facing severe threats by a variety of pathogens, such as necrotrophic Rhizoctonia solani and hemibiotrophic Xanthomonas oryzae pv. oryzae (Xoo). Mining and applying resistance genes to increase the durable resistance of rice is an effective method that can be used to control these diseases.
Results
In this research, we isolated and characterized CYP716A16, which is a positive regulator of rice to R. solani AG1-IA and Xoo, and belongs to the cytochrome P450 (CYP450) protein 716A subfamily. Overexpression (OE) of CYP716A16 resulted in enhanced resistance to R. solani AG1-IA and Xoo, while RNA interference (RNAi) of CYP716A16 resulted in increased susceptibility compared with wild-type (WT) plants. Additionally, jasmonic acid (JA)-dependent defense responses and reactive oxygen species (ROS) were activated in the CYP716A16-OE lines after R. solani AG1-IA inoculation. The comparative transcriptomic and metabolomics analysis of CYP716A16-OE and the WT lines showed that OE of CYP716A16 activated the biosynthesis of flavonoids and increased the amounts of narcissoside, methylophiopogonanone A, oroxin A, and amentoflavone in plants.
Conclusion
Based on these results, we suggest that JA-dependent response, ROS level, multiple resistance-related proteins, and flavonoid contents play an important role in CYP716A16-regulated R. solani AG1-IA and Xoo resistance. Our results broaden our knowledge regarding the function of a P450 protein 716A subfamily in disease resistance and provide new insight into the molecular mechanism of rice immune response.
Similar content being viewed by others
Background
Rice (Oryza sativa L.) is an important crop with an irreplaceable role in ensuring food security [1]. However, high and stable production of rice is severely threatened by a variety of pathogens, such as necrotrophic Rhizoctonia solani, which causes rice sheath blight (RSB) [2, 3], and hemibiotrophic Xanthomonas oryzae pv. oryzae (Xoo), which causes bacterial leaf blight (BLB) [4]. These two diseases are the most prevalent rice diseases worldwide [5, 6]. Mining resistance gene resources, and applying them to increase the resistance of rice is an effective method for controlling disease [7]. Recently, some genes that are resistant to these diseases have been identified, and several are provided potential solutions for applied to resistance breeding in rice [8,9,10]. However, the transformation of a single gene appears to have a limited effect on rice disease resistance, while in contrast, combining multiple resistance genes contributes to broad-spectrum and durable resistance to pathogens [11, 12]. Therefore, detecting new resistance genes in the rice germplasm is important for controlling disease.
In nature, plants are constantly threatened by a variety of pathogens and thus have developed complex defense systems to defend against invasion. Examples of these are the activation of the jasmonic acid (JA), salicylic acid (SA), and ethylene (ET)-related defense pathways, the up-regulation of pathogenesis-related (PR) genes; the production of reactive oxygen species (ROS), and antimicrobial peptides and phytoalexins accumulation, and callose deposition, to defend against pathogen invasion [13, 14]. For example, the NLR gene Xa1 and receptor-like protein kinase Xa21 imparts resistance against the Xoo through recognizing pathogen avirulence effectors [15, 16]. Moreover, some transcription factors, such as OsC3H12 [17], OsWRKY45 [18], and OsTFX1 [19], are also involved in rice resistance to Xoo. Compared with well-documented studies on resistance against Xoo, the interaction mechanism of rice and R. solani AG1-IA is still very limited [20,21,22]. Thus, the regulation of JA-dependent defence signalling and ROS has an important role in the resistance to R. solani AG1-IA [23, 24]. An example is OsRSR1 and OsRLCK5 imparting resistance against the RSB fungus through mediated ROS levels by the glutathione-ascorbic acid antioxidant system [9].
Cytochrome P450 (CYP450) protein is one of the largest gene families in plants, and is involved in the regulation processes of a variety of secondary metabolism processes, such as those involving phytoalexin, terpenoids, and flavonoids [25, 26]. A study has indicated that CYP450 plays an important role in hormone biosynthesis and signal transduction [27]. Some of the CYP450 members are also involved in plant defense against pathogens. The cotton P450 gene GhCYP82D is involved in disease resistance by modulating the biosynthetic pathway of oxylipins and JA anabolism [28]. In Arabidopsis, the CYP450 protein encoding the CYP82C2 gene plays a key role in JA-induced immunity, and overexpression (OE) of CYP82C2 enhances the resistance to necrotrophic fungus Botrytis cinerea though activation of expression of JA-related defense genes [29].
Among the rice genome (http://bioinfo.cau.edu.cn/~jyyu/drcyp), 534 CYP450 proteins encoded genes were annotationed, but the vast majority of these genes have unknown functions [30]. Studies have shown that rice P450 gene CYP71P1 possesses tryptamine 5-hydroxylase enzymatic activity and increases the resistance to M. oryzae through catalyzing the conversion of tryptamine to serotonin [31]. The CYP78A gene BSR2 could positively regulates the resistance to R. solani in rice [32]. With the identification of genes encoding multiple types of CYP450, cloning and research to investigate the functions of each of these genes may increase our understanding of the roles of CYP450 in plant immunity.
Previously, we obtained 653 genes that exhibit significant association with the RSB resistance through genome-wide association study analysis (GWAS) analysis based on 2,888,332 high-confidence single nucleotide polymorphisms (SNPs) [9]. Among these 653 genes, the CYP450 protein encoding gene LOC_Os07g33440 was found to be strongly expressed under R. solani AG1-IA inoculation conditions, with high expression in the resistant cultivar Teqing as compared with the susceptible cultivar Lemont. A BLAST search against the National Center for Biotechnology Information GenBank database (NCBI) GenBank database, rice gene annotations (http://rice.plantbiology.msu.edu), and literature analysis revealed that this gene belongs to the 716A subfamily of CYP450, therefore we named it as CYP716A16. In addition, OE of CYP716A16 in rice was shown to confer enhanced tolerance to R. solani AG-IA and Xoo pathogens compared to the control plants, while RNA interference (RNAi) of CYP716A16 promoted the colonization of these two pathogens in rice. Furthermore, OE of CYP716A16 was induced increased expression of PR and JA-related defense genes. Comparative transcription analysis of wild type (WT) and CYP716A16-OE plants suggested that CYP716A16 regulated the accumulation of flavonoids and activation of the phytoalexin synthesis-related genes. From these results, we conclude that CYP716A16 plays a key role in regulating rice immunity.
Results
CYP716A16 was significantly associated with R. solani AG1-IA and Xoo resistance
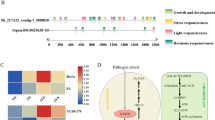
In our previously reported, 653 genes that associate with RSB resistance were obtained through SNP-based and haplotype-based GWAS combined with transcriptome analysis of resistant and susceptible varieties [9]. Among these 653 genes, CYP450 protein encoding gene LOC_Os07g33440 (named CYP716A16) was associated with RSB resistance and located on a 290 kb (19,688-19,978 Kb) haplotype block, based on a significant association with SNP loci Chr. 7_19757768 (−log10P = 12.86) (Fig. 1a). This haplotype block encompassing 58 genes (Fig. 1a), and the expression pattern of these 58 genes in the resistant variety Teqing and susceptible variety Lemont, are shown in Fig. 1b. The expression of CYP716A16 dramatically increased after R. solani AG1-IA infection at different hours post-inoculation (hpi) in Teqing, whereas the expression level of this gene in the Lemont variety was nearly unchanged with inoculation time, as compared with 0 hpi (Fig. 1b). Interestingly, CYP716A16 was also associated with BLB resistance based on GWAS analysis, and up-regulation was induced by Xoo infection in the resistant variety NSIC RC154, although it was not induced in susceptible variety CT 9737-6-1-3P-M [10]. These results suggested that CYP716A16 may be involved in the process of RSB and BLB resistance.
The CYP716A16 was significantly associated with rice sheath blight (RSB) resistance, and its transcription was induced by the infection of Rhizoctonia solani AG1-IA. a Local Manhattan plot of single nucleotide polymorphism (SNP)-based association (top) and LD heatmap (bottom) surrounding the peak on chromosome 7. Red dashed lines indicate the candidate region for associated SNPs; b Expression of 58 candidate genes in resistant and susceptible rice lines at different inoculation times (transcriptome data)
CYP716A16 protein belongs to the cytochrome P450 716A subfamily protein
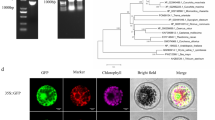
The complete coding sequence (CDS) of CYP716A16 (1479 bp) was obtained from a cDNA library of R. solani AG1-IA-inoculated rice (Teqing variety) by amplifying the cDNA ends. Sequence analysis showed that the CDS of CYP716A16 contains two exons and one intron in the rice genome, and conserved structural analysis indicated that CYP716A16 contains a P450 domain, with the transmembrane domain (TM) was also being predicted (Fig. 2a). The putative homologues were identified by BLASTP searches with rice CYP716A16 as the query, and the proteins were aligned through ClustalW2. A phylogenetic tree was constructed using MEGA 6.0 (Fig. 2b), revealing that the protein shares high homology with the CYP450 716A subfamily proteins of monocotyledonous plants such as Eragrostis curvula, Sorghum bicolor, Zea mays, Panicum hallii, Panicum miliaceum, Digitaria exilis, Setaria viridis, and Setaria italic, and it is distantly related to the CYP450 716A subfamily proteins of dicotyledonous plants such as Gossypium australe, Nicotiana tomentosiformis, Musa acuminate, and Spatholobus suberectus. Based on these results, the CYP716A16 was identified as a CYP450 716A subfamily protein, and it is conserved in land plants.
Overexpression of CYP716A16 enhanced rice resistance to R. solani AG1-IA and Xoo
To investigate the function of CYP716A16 in rice immunity, the OE transformation construct 35S::CYP716A16 was generated and transformed into the Nipponbare background. qRT-PCR results confirmed the high transcription level of CYP716A16 in four homozygous OE lines, CYP716A16-OE2, CYP716A16-OE4, CYP716A16-OE5, and CYP716A16-OE9 [9]. We used stable T1 progenies of four OE lines for disease resistance evaluation. Detached rice leaves from the CYP716A16-OE plants at tillering stage challenged with R. solani AG1-IA showed that growth of R. solani AG1-IA was greatly suppressed in CYP716A16-OE lines relative to WT Nipponbare [9]. Furthermore, to investigate whether CYP716A16 affected rice resistance to BLB, we performed inoculation experiments with the highly pathogenicity Xoo race P6. The results showed that the CYP716A16-OE lines exhibited an enhanced resistance to the Xoo race P6. There were significantly shorter lesions on CYP716A16-OE as compared to the WT (Fig. 3). Importantly, there were no significant differences in agronomic traits between CYP716A16-OE and WT lines (Supplementary Fig. S1). These results suggested that CYP716A16 positively regulates the rice resistance response to RSB and BLB, and plays a positive role in rice basal disease resistance against the fungal and bacterial pathogen.
RNA interference of CYP716A16 decreases rice resistance to R. solani AG1-IA and Xoo
To further determine the function of CYP716A16 in defense against pathogens, we obtained transgenic lines with decreased expression of CYP716A16 in a Teqing (with moderately resistance to R. solani) background. The reduced expression was achieved by RNAi, and the lower transcription level of CYP716A16 in three homozygous RNAi lines, CYP716A16i3, CYP716A16i7, and CYP716A16i10, were verified by qRT-PCR (Fig. 4a). Then, these CYP716A16-RNAi plants were inoculated with R. solani AG1-IA to evaluate the defensive role of CYP716A16. The lesion lengths were measured at 3 d after inoculation, and CYP716A16-RNAi exhibited significantly longer lesions than the WT Teqing (Fig. 4b). In a field measure of resistance against R. solani AG1-IA, we found that the disease degree of the CYP716A16-RNAi transgenic line was significantly higher than that of WT (Fig. 4c). Similarly, we performed inoculation experiments with Xoo race P6. We found that the CYP716A16-RNAi lines exhibited an enhanced susceptibility to the Xoo strain P6 (Fig. 4d). We also found that the agronomic traits were not significantly different between CYP716A16-RNAi and WT plants (Supplementary Fig. S1). These results indicated that the RNAi of CYP716A16 in Teqing plants impaired resistance to R. solani AG1-IA and Xoo.
Interference of CYP716A16 genes enhanced rice susceptibility to R. solani AG1-IA and Xoo. a The qRT-PCR results indicate that CYP716A16 was significantly downregulated in the RNA interference (RNAi) lines. Statistical analysis was determined by one-way ANOVA followed by Tukey’s multiple comparison tests (***P < 0.001); b Detached leaves of CYP716A16-RNAi plants were challenged by R. solani AG1-IA, and the lesion lengths were measured at 3 days post-inoculation (dpi). Three independent experiments were performed (** P < 0.01); c RNAi of CYP716A16 genes enhanced the rice susceptibility to R. solani AG1-IA in the field (*P < 0.05); d The leaves of CYP716A16-RNAi plants were challenged with Xoo and were more susceptible compared to the WT. Three independent experiments were performed (** P < 0.01)
Overexpression of CYP716A16 in rice leads to induced expression of defense-related genes
We examined the PR genes transcription levels in CYP716A16-OE plants and WT during a time course of 0–24 h (0, 12, 24 h) in leaves inoculated with R. solani AG1-IA. In CYP716A16-OE plants, these three genes were constitutively expressed and were further induced to significantly higher levels than those in the WT plants (Fig. 5). We also identified the expression of JA (OsAOC, OsAOS2), ET (OsACS2), and SA (OsNPR1) biosynthesis or signaling-related genes in CYP716A16-OE plants and WT at 0, 12, and 24 hpi [33,34,35,36]. The results showed that pathogen challenge strongly induced the expression of OsAOS2 in the WT and CYP716A16-OE, and the expression levels in the CYP716A16-OE lines were significantly higher than those in the WT at 24 hpi. Interestingly, the expression of OsAOC was only strongly induced in CYP716A16-OE plants at 24 hpi (Supplementary Fig. S2). In contrast, the expression of the ET signaling-related genes OsACS2 was decreased (Supplementary Fig. S2). Additionally, OsNPR1, a marker gene of SA-induced resistance response, showed no significantly difference between CYP716A16-OE plants and the WT (Supplementary Fig. S2).
OE of CYP716A16 activating the jasmonate acid (JA) biosynthetic and pathogenesis-related (PR) genes. The relative expression levels of the tested genes were normalized to UBQ. Three independent experiments were performed for each test of the gene expression levels. Statistical analysis was determined by one-way ANOVA, followed by Tukey’s multiple comparison tests (*P < 0.05; ** P < 0.01; *** P < 0.001)
Furthermore, we did not observe significant differences between ET and JA levels among the WT and CYP716A16-OE plants. Only the level of SA was significantly increased in CYP716A16-OE plants compared with WT (Supplementary Fig. S3). Additionally, JA-isoleucine (JA-Ile) is the active form in JA signal transduction, and thus, we detected JA-Ile in CYP716A16-OE and WT plants, and the amount of JA-Ile in CYP716A16-OE plants was higher than that in the WT (Supplementary Fig. S3). Therefore, we concluded that the defense responses-related to JA were involved in disease resistance regulated by CYP716A16.
CYP716A16 modulates ROS and related antioxidant enzymatic activity
H2O2 and O2− are primary ROS that function as positive signaling molecules and played an important role in disease resistance in plants [37]. To explore whether ROS burst is involved in the defense response mediated by CYP716A16 in rice, levels of H2O2 and O2− were assayed using DAB and NBT staining, respectively. At 24 h after R. solani AG1-IA infection, additional DAB- and NBT- stained spots appeared on the leaves surrounding the lesions on CYP716A16-OE plants as compared to WT plants (Fig. 6a), suggesting that CYP716A16-OE increased the accumulation of H2O2 and O2− in the transgenic rice. Furthermore, superoxide dismutase (SOD), peroxidase (POD), and malondialdehyde (MDA) are three important antioxidant enzymes in the ROS-scavenging system, which is critical for maintaining ROS homeostasis in plants [38]. Thus, we detected the activity levels of SOD, POD, and MDA content in CYP716A16-OE and WT plants. From the results, it was observed that the activation of POD and SOD in CYP716A16-OE plants was significantly elevated compared to the WT at 24 h after R. solani AG1-IA infection, whereas the activation of MDA was reduced (Fig. 6b). These data suggested that CYP716A16 was involved in rice immunity response through mediated ROS burst.
OE of CYP716A16 activating the ROS burst and related antioxidant enzymatic activity. a The levels of H2O2 and O2− in CYP716A16-OE and WT plants were assayed using DAB and NBT staining at 24 hours post-inoculation (hpi); b The amounts of superoxide dismutase (SOD), peroxidase (POD), and malondialdehyde (MDA) in CYP716A16-OE and WT plants were measured at 24 hpi. Three biological repetitions were performed (** P < 0.01; *** P < 0.001)
Biosynthesis of flavonoids was activated in the CYP716A16-OE plant
To explore the roles of CYP716A16 in the defense against pathogens, we performed RNA-Seq using uninfected leaves and leaves of the WT and CYP716A16-OE lines that had been infected with R. solani AG1-IA for 12 h. The sequence number information for the RNA-Seq data is shown in Supplementary Table 1. The transcriptome data indicated that there are 40 genes down-regulated and 235 genes up-regulated in WT-12hpi compared to WT-0hpi (Fig. 7a; Supplementary Table 2). However, the OE of CYP716A16 resulted in increased expression of 892 genes and decreased expression of 97 genes (Fig. 7b; Supplementary Table 2). Additionally, 74 differentially expressed genes (DEGs) were shared by the two lines at 12 hpi, and 915 DEGs were uniquely detected in CYP716A16-OE plants (Fig. 7c). Kyoto Encyclopedia of Genes and Genomes (KEGG) assay revealed that these DEGs were assigned into several functional classes, and among which biosynthesis of flavonoids was significantly enriched (P < 0.05) in CYP716A16-OE lines (Fig. 7e), but no significant enrichment was found in the WT (Fig. 7d). In this class, there are 15 genes putatively related to biosynthesis of flavonoids were uniquely up-regulated in CYP716A16-OE lines (Fig. 7f), including LOC_Os01g53370 (anthocyanidin 5,3-O-glucosyltransferase), LOC_Os02g39850 (transferase family protein), LOC_Os05g45200 (anthocyanidin 5,3-O-glucosyltransferase), LOC_Os03g25150 (transposon protein), LOC_Os04g53810 (leucoanthocyanidin reductase), LOC_Os04g56910 (transferase family protein), LOC_Os05g25640 (cytochrome P450), LOC_Os05g41440 (cytochrome P450), LOC_Os11g07960 (transferase family protein), LOC_Os07g32630 (UDP-glucoronosyl and UDP-glucosyl transferase domain containing protein), LOC_Os10g12050 (expressed protein), LOC_Os10g17260 (cytochrome P450), LOC_Os11g02440 (chalcone--flavonone isomerase), LOC_Os11g32650 (chalcone synthase), and LOC_Os12g02370 (chalcone--flavonone isomerase). Among these 15 genes, the transcript levels of three genes related to phytoalexin biosynthesis (LOC_Os11g02440, LOC_Os11g32650, and LOC_Os12g02370) in CYP716A16-OE lines were validated by qRT-PCR (Fig. 7g). The results indicated that CYP716A16 may activate flavonoid biosynthesis, with the subsequent result of disease resistance.
Analysis of the RNA-Seq data for CYP716A16-OE and WT plants. a The number of differentially expressed genes (DEGs) in WT plants at 12 hpi; b The number of DEGs in CYP716A16-OE plants at 12 hpi; c Venn diagrams showing the overlap of DEGs in CYP716A16-OE and WT plants at 12 hpi; d The KEGG enrichment analysis of the DEGs in WT plants. The sizes and the colours of the dots represent the numbers of DEGs and the p- value, respectively; e The KEGG enrichment analysis of the DEGs in CYP716A16-OE plants. The sizes and the colours of the bars represent the numbers of DEGs and the q- value, respectively; f The expression of 15 DEGs related to the biosynthesis of flavonoids in CYP716A16-OE plants. Three of them are involved in the phytoalexin biosynthesis; g Expression analysis of three phytoalexin biosynthesis-related genes in CYP716A16-OE plants at 12 hpi by qRT–PCR
Furthermore, we determined the different of metabolites in the WT and CYP716A16-OE lines at 12 hpi. In total, 1218 metabolites were detected in rice by liquid chromatography mass spectrometery (LC-MS), including many primary and secondary metabolites, such as amino acids, fatty acyls, phytohormones, sugar alcohols, flavonoids, quinones, and terpenoids (Supplementary Table 3). Compared with the control (0 h), 214 measured metabolites were identified as significantly increased (fold change≥2) in WT plants at 12 h after R. solani AG1-IA inoculation. However, only 51 significantly increased metabolites were observed in CYP716A16-OE plants (Supplementary Table 3). Interestingly, some measured metabolites showed exclusively significant increases in CYP716A16-OE plants that underwent R. solani AG1-IA inoculation, including four flavonoids (narcissoside, methylophiopogonanone A, oroxin A, and amentoflavone) and trans-zeatin (Supplementary Table 3). From these results, we concluded that these metabolites may play important roles in disease resistance mediated by CYP716A16.
Overexpression of CYP716A16 promoted accumulation of phytoalexin
From the transcriptome data, we found several key genes, which involved in phytoalexin biosynthesis that were up-regulated in CYP716A16-OE plants at 24 h after R. solani AG1-IA inoculation, although with no significantly different expression in the WT. This indicated that phytoalexin was involved in the process of disease resistance regulated by CYP716A16. To further confirm this result, we measured the amounts of momilactone A and B, which are important phytoalexins involved in rice defense, in WT and CYP716A16-OE plants. There was a significant increase in the amounts of momilactone B in CYP716A16-OE plants as compared with the WT at 24 hpi (Supplementary Fig. S4).
Discussion
The characterization of resistance genes in rice is basic to the development of rice varieties with disease resistance. Bioinformatics methods, such as GWAS, bulk segregant analysis (BSA), and transcriptomic analyses, have been used to detect the genes that control crop resistance to pathogen infection [1, 39,40,41]. In our previous study, a GWAS was performed using 259 diverse rice germplasm with genotypes based on SNPs and haplotypes with their RSB reactions at three developmental stages, seedlings, tillering, and booting was performed. Furthermore, we performed a combined comparative transcriptomic analysis between Teqing (a line that is resistant line to R. solani AG1-IA) and Lemont (a line that is susceptible to R. solani AG1-IA) after R. solani AG1-IA infection, and we obtained 653 core candidate genes that might regulate rice resistance to RSB [9]. In this study, we selected the CYP450 protein encoding gene CYP716A16 to verify its resistance function, and this further refined our results.
In plants, the CYP450 proteins are divided into 10 separate clans in 61 families [42]. In this study, we isolated and characterized a novel CYP450 gene, CYP716A16, which belongs to the CYP716A subfamily, in rice. CYP716A16 is a widespread family with diversity in structure and function in plants, and members of the CYP716A family are involved in triterpene biosynthesis [43, 44]. For example, licorice CYP450 monooxygenase CYP716A179 plays a key role in the biosynthesis of oleanolic acid and betulinic acid [43]. Although the role of more than a dozen CYP716As in triterpene biosynthesis has been identified, little information is available on the disease resistance functions of the CYP716A subfamily in plants [43]. Here, we show that OE of the CYP716A16 enhanced the resistance of rice to necrotrophic R. solani AG1-IA and hemibiotrophic Xoo. Conversely, the resistance level in CYP716A16-RNAi rice plants was significantly reduced. These results provide new insight into the function of the P450 716A subfamily.
Plant hormones, especially JA, ET, and SA, play important roles in the regulation of plant innate immunity [14]. Additionally, JA-dependent plant immunity plays a key role in resistance to necrotrophic and hemi-necrotrophic fungus [9]. For instance, JA and ET are both involved in the resistance of R. solani AG1-IA in rice, and are mediated by OsWRKY4 and OsWRKY80 [45]. Kouzai et al. [46] found that foliar pretreatment with SA can induce sheath blight resistance in rice and Brachypodium distachyon. In our study, genes related to JA biosynthesis, such as OsAOC and OsAOS2, were upregulated in the CYP716A16-OE plants. Furthermore, the JA response gene PR1b was up-regulated in the CYP716A16-OE plants, indicating that the JA signalling pathway is activated in CYP716A16-OE plants. Therefore, we suggest that activation of a JA-dependent defense in CYP716A16-OE contributes to rice resistance against R. solani AG1-IA and Xoo.
Flavonoids are secondary metabolites that occur widely occur in plants, and can be divided into subgroups including anthocyanidins, flavonols, flavones, flavanols, flavanones, chalcones, dihydrochalcones and dihydroflavonols [47, 48]. Previous studies have shown that flavonoid biosynthesis is an important pathway during the interaction between Medicago truncatula and R. solani [49]. In rice, kaempferol, naringenin, and dihydroquercetin are involved in the defense against the fungal blast pathogen Pyricularis oryzae [50, 51]. We show in this study that the biosynthesis of flavonoids was significantly enriched in CYP716A16-OE plants at 12 hpi, and furthermore, there were increases in the amounts of narcissoside, methylophiopogonanone A, oroxin A, and amentoflavone, which is preliminary proof that the flavonoids are regulated by a CYP716A16. Moreover, some genes involved in phytoalexin were also induced up-regulated in CYP716A16-OE plants. Thus, from these results and the results of transgenic experiments in this study, we showed that CYP716A16 participates in flavonoids biosynthesis and that it plays a positive regulatory role in the resistance to R. solani AG1-IA and Xoo in rice.
Even though there are several RSB resistance genes have been cloned, none of them have been used in rice breeding for RSB resistance. CYP716A16 encodes a CYP450 proteins, and OsBON1 and OsBON3 are copine genes [52]. Similar to OsBON1 and OsBON3, CYP716A16 also confers broad-spectrum disease resistance, constituting a superior disease-resistant characteristic for rice breeding. The CYP716A16 gene, however, carries a further advantage over OsBON1. The RNAi of OsBON1 increases the disease resistance of rice with a decrease in the tillers number [52], and thus, the yields are affected. In contrast, our measurements suggest that CYP716A16 positively regulates resistance to disease without compromising fitness. Therefore, CYP716A16 represents a more optimal alternative genetic resource for rice resistance breeding in rice.
Conclusions
Our findings indicate that the CYP450 protein encoding gene CYP716A16 positively contributes to the immune response in rice, which will expand our understanding of the potential functions of P450 proteins and provide valuable insight into the molecular mechanism of plant immunity. Furthermore, the CYP716A16 displays broad-spectrum disease resistance to both bacterial and fungal pathogens, and this will also provide important gene resources for rice disease resistance breeding in rice.
Materials and methods
Plant materials and growth conditions
The rice cultivars Teqing, Lemont, and Nipponbare were used in this work. All plant materials were planted in the rice transgenic field of the College of Agronomy, Sichuan Agricultural University, Chengdu.
Vector construction and rice transformation
To construct the Nipponbare CYP716A16-OE plants, the CDS of CYP716A16 was amplified from the cultivar Teqing by PCR using the gene-specific primers (Supplementary Table 4). The cDNA product was then inserted into pBWA(V)HS, which harbors a cauliflower mosaic virus (CaMV) 35S promoter, and the constructed vector was introduced into Agrobacterium tumefaciens GV3101. Rice transformation was performed following the methods previously described [53]. Transgenic plants of the T1 generation with positive activity were used in the experiments. For RNAi vector construction, a specific CYP716A16 fragment of approximately 270 bp was selected, and then amplified with the primers listed in Supplementary Table S4. The plasmid was constructed as previously described [54, 55]. The CYP716A16-RNAi cis- and trans-fragments with correct sequences were inserted into the vector pBWA(V)HS. Agrobacterium tumefaciens-mediated transformation of Teqing was used to obtain the CYP716A16-RNAi transgenic plants. Transgenic plants of the T1 generation with positive activity were used in the experiments.
Pathogenic infection
For R. solani AG1-IA, we identified the resistance level of rice plants in indoor and field inoculation. The second youngest leaf from the main tiller was cut around the heading stage and inoculated into a 5 mm potato dextrose agar (PDA) plug containing R. solani AG1-IA mycelia, placed on a moistened filter paper, and maintained in a petri plate. To enhance humidity and increase R. solani AG1-IA infection and development, the moisture of the filter paper was maintained with sterile water, and the plate was covered with protective film. After 72 h, the length of each leaf lesion was measured. In the field, the pathogen was grown on truncated thin matchsticks (0.8–1.0 cm long 2–3 mm wide, and 1 mm thick) on potato dextrose broth medium at 28 °C in the dark for 2-3 d. To perform inoculation, the inoculum was closely affixed to one side of the base of the seedling stem, assuring that hypha was directly touching the plant [39]. Five sheaths per line were inoculated as replications.
For Xoo, we used Xoo virulent strains P6 to artificially inoculate plants. At the rice tillering stage, 15 of the uppermost leaves of each variety were inoculated with the Xoo race P6, using the leaf-clipping method [56]. Lesion lengths were measured on all inoculated leaves at 14 days post-inoculation (dpi), when lesions were easily visible as well as stable.
qRT-PCR analysis
Plant total RNA was extracted using a Plant Total RNA Isolation Kit (Sangon Biotech, Shanghai, China). First strand cDNA was synthesized from total RNA using the Transcriptor First-Strand cDNA Synthesis Kit (Roche, Indianapolis, IN, USA). The cDNA samples were then subjected to qRT-PCR on a Bio-Rad CFX96 Real-Time PCR System (Foster City, CA, USA), according to the manufacturer’s instructions. The PCR reactions were prepared in a 20 μL volume, containing 3 μL cDNA and 1 μL each of the forward and reverse gene specific primers. Each PCR was replicated four times. The ubiquitin (UBQ) gene was used as an internal control for data normalization. Gene expression levels were calculated using the 2-∆∆Ct method. The primers used for qRT-PCR are listed in Supplementary Table 4.
H2O2 measurement and antioxidant enzymatic activity detection
To compare the differences in ROS among WT and OE lines, the quantities of H2O2 and O2− were measured using DAB and NBT staining, respectively, at 72 hpi according to a previously described protocol [57]. The same samples used for H2O2 quantification were used for SOD, POD, and MDA activity analysis. Total SOD, POD, and MDA activity was measured using previously described methods [58, 59].
RNA-Seq and data analyses
Leaves of WT and CYP716A16-OE lines were harvested at 12 hpi. Leaves of each line uninfected at 12 h served as a control. RNA samples were sent to Beijing Novogene Biological Technology Co., Ltd. for cDNA library construction and Illumina sequencing (HiSeq TM 2500, San Diego, NEB, USA). RNA-Seq libraries were constructed using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA), according to the manufacturer’s instructions, and unique index codes were added to each sample. The sequencing was performed using an Illumina Hiseq platform to generate 125 bp paired-end sequences. Sequences with a low quality score and those containing adaptor sequences and stretches of -Ns were removed from the raw data. The reference genome of Nipponbare rice and gene model annotation files (Rice Annotation Project) were directly downloaded from the rice genome website (ftp://ftp.ensemblgenomes. org/ pub/ plants/ release_36/ fasta/ oryza_indica/ dna/). An index of the reference genome was built using Bowtie v2.2.3, and paired-end sequences were aligned to the reference genome using TopHat v2.0.12 [60,61,62]. The number of sequences mapped to each gene was counted using HTSeq v0.6.1 [63], and the number of fragments per kilobase of transcript sequence per million (FPKM) of each gene was calculated based on the length of the gene and number of sequence counts mapped to that gene. The differential expression gene (DEG) analysis (q < 0.05 and |log2 (fold change)| > 1) was conducted through the DEGSeq R package [64]. The Benjamini and Hochberg method was used to adjust P-values [65].
KEGG enrichment analysis of DEGs
The KOBAS (v2.0) software was used for statistical enrichment analysis of DEGs in KEGG pathways [66, 67]. The hypergeometric test was performed using the ‘phyper’ function in R. KEGG terms with P < 0.05 were defined as significantly enriched in DEGs.
Metabolome analyses
The same samples used for RNA-seq were used for metabolome analysis. Samples were analyzed using QTRAP 6500plus LC-MS platforms (AB SCIEX, Boston, MA, USA). Analytical conditions were based on the procedures as described in Wang et al. [68]. Quantification of metabolites was carried out using a multiple reaction monitoring method [69]. Metabolites with significant differences in content were set with thresholds of fold change ≥2 or ≤ 0.5 [70].
Statements
The rice lines used in this study were provided by Sichuan Agricultural University and comply with relevant institutional, national, and international guidelines and legislation. Our study was approved by Sichuan Agricultural University.
Availability of data and materials
All the data generated or analysed during this study were included in this article and its additional data files. The O. sativa transcriptome datasets analyzed during the current study are available in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov) under the accession number: PRJNA777627. The rice samples used in this study were deposited at College of Agronomy, Sichuan Agricultural University, Chengdu, China.
References
Chen W, Gao YQ, Xie WB, Gong L, Lu K, Wang WS, et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet. 2014;46:714–21.
Bernardes-de-Assis J, Storari M, Zala M, Wang W, Jiang D, Shidong L, et al. Genetic structure of populations of the rice-infecting pathogen Rhizoctonia solani AG-1 IA from China. Phytopathology. 2009;99:1090–9.
Wang L, Liu LM, Wang ZG, Huang SW. Genetic structure and aggressiveness of Rhizoctonia solani AG1-IA, the cause of sheath blight of rice in southern China. J Phytopathol. 2013;161:753–62.
Khush GS, Mackill DJ, Sidhu GS. Breeding rice for resistance to bacterial blight. In: Banta SJ, editor. Bacterial blight of rice. Los Banos, Philippines: International Rice Research Institute; 1989. p. 207–17.
Gautam K, Rao PB, Chauhan SVS. Efficacy of some botanicals of the family compositae against Rhizoctonia solani Kuhn. J Mycol Plant Pathol. 2003;33:230–5.
Nino-Liu DO, Ronald PC, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. 2010;7(5):303–24.
Deng Y, Zhai K, Xie Z, Yang D, Zhu X, Liu J, et al. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017;355:962–5.
Xu XM, Xu ZY, Ma WX, Haq F, Li Y, Shah SMA, et al. TALE-triggered and iTALE-suppressed Xa1 resistance to bacterial blight is independent of OsTFIIAγ1 or OsTFIIAγ5 in rice. J Exp Bot. 2021. https://doi.org/10.1093/jxb/erab054.
Wang AJ, Shu XY, Jing X, Jiao CZ, Che, L, Zhang JF, et al. Identification of rice (Oryza sativa L.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnol J 2021; doi: https://doi.org/https://doi.org/10.1111/pbi.13569.
Shu XY, Wang AJ, Jiang B, Jiang YQ, Xiang X, Yi XQ, et al. Genome-wide association study and transcriptome analysis discover new genes for bacterial leaf blight resistance in rice (Oryza sativa L.). BMC Plant Biol. 2021;21:255.
Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, et al. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017;545:491–4.
Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P. Navigating complexity to breed disease-resistant crops. Nat Rev Genet. 2018;19:21–33.
Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9.
van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–62.
Ji CH, Ji ZY, Liu B, Cheng H, Liu H, Liu SZ, et al. Xa1 allelic R genes activate rice blight resistance suppressed by interfering TAL effectors. Plant Communications. 2020;1(4):100087.
Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Gardner THJ, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–6.
Deng HQ, Liu HB, Li XH, Xiao JH, Wang SP. A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiol. 2012;158(2):876–89.
Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, et al. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007;19(6):2064–76.
Liu CT, Schläppi MR, Mao BG, Wang W, Wang AJ, Chu CC. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol J. 2019;17(9):1834–49.
Pinson SRM, Capdevielle FM, Oard JH. Confirming QTLs and finding additional loci conditioning sheath blight resistance in rice using recombinant inbred lines. Crop Sci. 2005;45:503–10.
Zou JH, Pan XB, Chen ZX, Xu JY, Lu JF, Zhai WX, et al. Mapping quantitative trait loci controlling sheath blight resistance in two rice cultivars (Oryza sativa L.). Theor Appl Genet. 2000;101:569–73.
Zuo SM, Yin YJ, Zhang L, Zhang YF, Chen ZX, Pan XB. Breeding value and further mapping of a QTL qSB-11 conferring the rice sheath blight resistance. Chinese J Rice Sci. 2007;21:136–42.
Oreiro EG, Grimares EKS, Grande GA, Quibod LL, Roman Reyna V, Oliva RF. Genome-wide associations and transcriptional profiling reveal ROS regulation as one underlying mechanism of sheath blight resistance in rice. Mol Plant-Microbe Interact. 2019;33:212–22.
Zhang J, Chen L, Fu C, Wang L, Liu H, Cheng Y, et al. Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front Plant Sci. 2017;8:1422.
Fang W, Wang Z, Cui R, Li J, Li Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 2012;70:929–39.
Kai K, Hashidzume H, Yoshimura K, Suzuki H, Ohta D. Metabolomics for the characterization of cytochromes P450-dependent fatty acid hydroxylation reactions in Arabidopsis. Plant Biotechnol-Nar. 2009;26:175–82.
Miyoshi K, Ahn BO, Kawakatsu T, Ito Y, Itoh JI, Nagato Y, et al. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc Natl Acad Sci U S A. 2004;101:875–80.
Sun LQ, Zhu LF, Xu L, Yuan DJ, Min L, Zhang XL. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun. 2014;5:5372.
Liu F, Jiang HL, Ye SQ, Chen WP, Liang WX, Xu YX, et al. The Arabidopsis P450 protein CYP82C2 modulates jasmonate-induced root growth inhibition, defense gene expression and indole glucosinolate biosynthesis. Cell Res. 2010;20(5):539–52.
Cui YJ, Peng YL, Zhang Q, Xia SS, Ruan BP, Xu QK, et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 2020;4(105):942–56.
Fujiwara T, Maisonneuve S, Isshiki M, Mizutani M, Chen L, Wong HL, et al. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem. 2010;285:11308–13.
Maeda S, Dubouzet JG, Kondou Y, Jikumaru Y, Seo S, Oda K, et al. The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and rice. Sci Rep. 2019;9:587.
Guo HM, Li HC, Zhou SR, Xue HW, Miao XX. Cis-12-Oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect. Mol Plant. 2014;7(11):1683–92.
Mei CS, Qi M, Sheng GY, Yang YN. Inducible overexpression of a rice allene oxide synthase gene increases the endogenous jasmonic acid level, PR gene expression, and host resistance to fungal infection. Mol Plant-Microbe Interact. 2006;19(10):1127–37.
Hu J, Zhou JB, Peng XX, Xu HH, Liu CX, Du B, et al. The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol. 2011;156(2):856–72.
Feng JX, Cao L, Li J, Duan CJ, Luo XM, Le N, et al. Involvement of OsNPR1/NH1 in rice basal resistance to blast fungus Magnaporthe oryzae. Eur J Plant Pathol. 2011;131(2):221–35.
Miller G, Suzuki N, Ciftci-Yilmaz S, Mittler R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010;33:453–67.
Lei ST, Zeng B, Xu SJ, Su XL. Membrane responses of Salix variegata and Cinnamomum camphora to complete submergence in the three gorges reservoir region. Acta Ecol Sin. 2012;5(32):227–31.
Chen ZX, Feng ZM, Kang HX, Zhao JH, Chen TX, Li QQ, et al. Identification of new resistance loci against sheath blight disease in rice through genome-wide association study. Rice Sci. 2019;26:21–31.
Zhang F, Wu ZC, Wang MM, Zhang F, Dingkuhn M, Xu JL, et al. Genome-wide association analysis identifies resistance loci for bacterial blight in a diverse collection of indica rice germplasm. PLoS One. 2017;12:e0174598.
Li N, Lin B, Wang H, Li XM, Yang FF, Ding XH, et al. Natural variation in ZmFBL41 confers banded leaf and sheath blight resistance in maize. Nat Genet. 2019;51:1540–8.
Jo HJ, Han JY, Hwang HS, Choi YE. β-Amyrin synthase (EsBAS) and β-amyrin 28-oxidase (CYP716A244) in oleanane-type triterpene saponin biosynthesis in Eleutherococcus senticosus. Phytochemistry. 2017;135:53–63.
Tamura K, Teranishi Y, Ueda S, Suzuki H, Kawano N, Yoshimatsu K, et al. Cytochrome P450 monooxygenase CYP716A141 is a unique b-Amyrin C-16b oxidase involved in triterpenoid saponin biosynthesis in Platycodon grandifloras. Plant Cell Physiol. 2017;58(5):874–84.
Yeon HJ, Su HH, Wan CS, Jung KH, Eui CY. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in panax ginseng. Plant Cell Physiol. 2012;53(9):1535–45.
Wang HH, Meng J, Peng XX, Tang XK, Zhou PL, Xiang JH, et al. Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol Biol. 2015;89:157–71.
Kouzai Y, Kimura M, Watanabe M, Kusunoki K, Osaka D, Suzuki T, et al. Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol. 2018;217(2):771–83.
Liu ZH, Liu YX, Pu ZE, Wang JR, Zheng YL, Li YH, et al. Regulation, evolution, and functionality of flavonoids in cereal crops. Biotechnol Lett. 2013;35:1765–80.
Koes R, Quattrocchio F. The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays. 1994;16(2):123–32.
Liu Y, Hassan S, Kidd BN, Garg G, Mathesius U, Singh KB, et al. Ethylene signaling is important for isoflavonoid-mediated resistance to Rhizoctonia solani in roots of Medicago truncatula. Mol Plant-Microbe Interact. 2017;9(30):691–700.
Padmavati M, Sakthivel N, Thara KV, Reddy AR. Differential sensitivity of rice pathogens to grown inhibition by flavonoids. Phytochemistry. 1997;46:499–502.
Treutter D. Significance of flavonoids in plant resistance: a review. Environ Chem Lett. 2006;4:147–57.
Yin X, Zou BH, Hong XX, Gao MJ, Yang WB, Zhong XB, et al. Rice copine genes OsBON1 and OsBON3 function as suppressors of broad-spectrum disease resistance. Plant Biotechnol J. 2018;16:1476–87.
Li SC, Gao FY, Xie KL, Zeng XH, Cao Y, Zeng J, et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol J. 2016;14:2134–46.
Osato Y, Yokoyama R, Nishitani K. A principal role for AtXTH18 in Arabidopsis thaliana root growth: a functional analysis using RNAi plants. J Plant Res. 2006;119:153–62.
Xu S, Wang LJ, Zhang B, Han B, Xie YJ, Yang J, et al. RNAi knockdown of rice SE5 gene is sensitive to the herbicide methyl viologen by the down-regulation of antioxidant defense. Plant Mol Biol. 2012;80:219–35.
Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. A improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzea. Plant Dis Rep. 1973;57:537–41.
Zhu XL, Yang K, Wei XN, Zhang QF, Rong W, Du LP, et al. The wheat AGC kinase TaAGC1 is a positive contributor to host resistance to the necrotrophic pathogen Rhizoctonia cerealis. J Exp Bot. 2015;66:6591–603.
Goel N, Paul PK. Polyphenol oxidase and lysozyme mediate induction of systemic resistance in tomato, when a bioelicitor is used. J Plant Prot Res. 2015;55:343–50.
Karmakar S, Molla KA, Chanda PK, Sarkar SN, Datta SK, Datta K. Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta. 2016;243:115–30.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat 2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–5.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Anders S, Huber W. Differential expression of RNA-Seq data at the gene level-the DESeq package. Heidelberg: EMBL; 2012.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57(1):289–300.
Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:19.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Wang Z, Cui Y, Vainstein A, Chen S, Ma H. Regulation of fig (Ficus carica L.) fruit color: Metabolomic and transcriptomic analyses of the flavonoid biosynthetic pathway. Front Plant Sci. 2017;8:1990.
Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1–2):155–71.
Yun Y, Liang F, Deng B, Lai G, Vicente Gonçalves CM, Lu H, et al. Informative metabolites identification by variable importance analysis based on random variable combination. Metabolomics. 2015;11(6):1539–51.
Acknowledgements
We thank Boyuan biological science and technology Ltd. (China) for providing Agrobacterium-mediated stable transformation services.
Funding
This work was supported by the Key Project of Sichuan Department of Science and Technology (No. 2019YFN0010 and 2020YFH0117) and the National Natural Science Foundation of China (No. 31400130 and 32001490).
Author information
Authors and Affiliations
Contributions
Aijun Wang and Aiping Zheng: designed the experiments. Li Ma and Xinyue Shu: performed the experiments, formal analysis. Yuqi Jiang and Juan Liang: investigation. AijunWang: writing - original draft. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All methods in this research were carried out in accordance with relevant guidelines and regulations of Sichuan Agricultural University, Chengdu, China.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Supplementary Figure S1
. The data of important agronomic traits of WT, CYP716A16-OE, CYP716A16-RNAi lines. Supplementary Figure S2. The expression pattern of OsAOC, OsACS2, and OsNPR1 in WT and CYP716A16-OE plants after inoculation with R. solani AG1-IA. Supplementary Figure S3. The levels of (JA, JA-Ile, SA, and ET) in WT and CYP716A16-OE plants after inoculation 24 h with R. solani AG1-IA. Supplementary Figure S4. The contents of phytoalexin (MA and MB) in WT and CYP716A16-OE plants after inoculation 24 h with R. solani AG1-IA.
Additional file 2: Supplementary Table S1
. The sequence data for the transcriptome.
Additional file 3: Supplementary Table S2
. Differentially expressed genes between WT and CYP716A16-OE lines.
Additional file 4: Supplementary Table S3
. Differentially expressed metabolites in the leaves of the WT and CYP716A16-OE lines.
Additional file 5: Supplementary Table S4
. The primers sequences used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wang, A., Ma, L., Shu, X. et al. Rice (Oryza sativa L.) cytochrome P450 protein 716A subfamily CYP716A16 regulates disease resistance. BMC Genomics 23, 343 (2022). https://doi.org/10.1186/s12864-022-08568-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08568-8