Abstract
Background
Tick-borne relapsing fever (TBRF) is a globally prevalent, yet under-studied vector-borne disease transmitted by soft and hard bodied ticks. While soft TBRF (sTBRF) spirochetes have been described for over a century, our understanding of the molecular mechanisms facilitating vector and host adaptation is poorly understood. This is due to the complexity of their small (~ 1.5 Mb) but fragmented genomes that typically consist of a linear chromosome and both linear and circular plasmids. A majority of sTBRF spirochete genomes’ plasmid sequences are either missing or are deposited as unassembled sequences. Consequently, our goal was to generate complete, plasmid-resolved genomes for a comparative analysis of sTBRF species of the Western Hemisphere.
Results
Utilizing a Borrelia specific pipeline, genomes of sTBRF spirochetes from the Western Hemisphere were sequenced and assembled using a combination of short- and long-read sequencing technologies. Included in the analysis were the two recently isolated species from Central and South America, Borrelia puertoricensis n. sp. and Borrelia venezuelensis, respectively. Plasmid analyses identified diverse sequences that clustered plasmids into 30 families; however, only three families were conserved and syntenic across all species. We also compared two species, B. venezuelensis and Borrelia turicatae, which were isolated ~ 6,800 km apart and from different tick vector species but were previously reported to be genetically similar.
Conclusions
To truly understand the biological differences observed between species of TBRF spirochetes, complete chromosome and plasmid sequences are needed. This comparative genomic analysis highlights high chromosomal synteny across the species yet diverse plasmid composition. This was particularly true for B. turicatae and B. venezuelensis, which had high average nucleotide identity yet extensive plasmid diversity. These findings are foundational for future endeavors to evaluate the role of plasmids in vector and host adaptation.
Similar content being viewed by others
Introduction
Relapsing fever (RF) is a globally endemic yet neglected vector-borne disease caused by spirochetal bacteria of the Borrelia genus [1]. RF spirochetes are vectored by the human body louse, and both hard (ixodid) and soft (argasid) ticks. Tick-borne RF (TBRF) typically presents in humans with recurring fever and flu-like symptoms; however, neurological complications and death can occur [2]. The disease significantly impacts the impoverished in resource poor countries, while evidence suggests that TBRF spirochetes and their vectors are emerging in densely populated regions in the southwestern United States [3,4,5,6,7,8].
There are seven species of Western Hemisphere soft-tick-borne relapsing fever (WHsTBRF) spirochetes where laboratory isolates have been reported. These include Borrelia anserina, Borrelia coriaceae, Borrelia hermsii, Borrelia parkeri, Borrelia turicatae, Borrelia puertoricensis n. sp., and Borrelia venezuelensis [9,10,11,12,13,14,15,16]. Borrelia coriaceae, B. hermsii, B. turicatae, and B. parkeri are distributed throughout North America [12, 17], while B. anserina has a global distribution [18]. Furthermore, laboratory isolates of B. puertoricensis and B. venezuelensis were only cultured within the last three years and originate from Panama and Brazil, respectively [9, 15]. Interestingly, a phylogenetic analysis of B. venezuelensis using the flaB, rrs, and glpQ loci indicated that this species formed a monophyletic group with B. turicatae with the two species sharing > 99% nucleotide identity [15]. The vast range difference between B. turicatae and B. venezuelensis, their different tick vectors, and the need for completed genomes spurred our interest to perform a genomic comparison between WHsTBRF spirochetes.
Borrelial genomes (Lyme and RF spirochetes) are small (~ 1.5 Mb) but exceptionally complex and distributed across many replicons [19, 20]. Their genomes typically consist of a linear chromosome and linear and circular plasmids. The linear replicons make up most of a genome. The number of plasmids in a given genome can range from five to 23 [21,22,23]. Due to the complex nature of borrelial genomes, particularly the covalently closed linear plasmids and repetitive elements, next-generation short-read DNA sequencing technologies (e.g. Roche 454, Illumina, Ion Torrent) have had difficulties in producing complete genomes [24]. The advent of third generation sequencing technologies (e.g. PacBio and Oxford Nanopore Technologies) opened new avenues for genome sequencing and assembly [25, 26]. However, compared to other bacteria, borrelial genomes require substantially more manual effort to resolve the plasmids. As a result, the plasmids of many publicly available borrelial genomes are highly fragmented across many contigs.
Since the first borrelial genome was completed in 1997 with Borreliella (Borrelia) burgdorferi, there have been 205 Lyme disease (LD) spirochete genomes deposited on GenBank compared to 44 TBRF genomes. Half of the TBRF genomes are of the hard tick-borne species, Borrelia miyamotoi [27]. Furthermore, the chromosomes of B. anserina, B. coriaceae, B. hermsii, B. parkeri, and B. turicatae have been assembled, and analyses between B. hermsii and B. turicatae indicate that they are largely collinear with similar gene content [28]. Thus, we reason that the chromosomes alone do not explain the diversity in vector competence and specificity, host reservoir adaptation, and pathogenesis seen in the WHsTBRF spirochete clade; plasmids likely play a significant role as well [1, 18, 29,30,31,32].
In this study, we generated complete, plasmid-resolved genomes of available isolates from the WHsTBRF spirochete clade and performed a comparative genomics analysis between these species. We focused on B. anserina BA2, B. coriaceae Co53, B. hermsii DAH and YOR, B. parkeri SLO, B. turicatae 91E135 and BTE5EL, B. puertoricensis n. sp. SUM [9], and B. venezuelensis RMA01 because they are either commonly used laboratory isolates, or their genomes have not been reported. We hypothesized that WHsTBRF spirochete genomes consist of a syntenic chromosome with plasmids grouping into distinct families, which are diverse between species. To test this hypothesis, analyses were performed to determine the relatedness of these strains by using whole genome methods including average nucleotide identity (ANI) analysis and phylogenomic analyses. Pangenome analysis was also performed to investigate gene content differences across all species. Plasmid diversity and similarities were evaluated as well through phylogenetic and dot plot analyses. We concluded our efforts by comparing the genomes of two species, B. venezuelensis and B. turicatae, which have been isolated from two different Ornithodoros species but were reported to be closely related genetically [15]. This work is important in progressing efforts to identify the molecular mechanisms of pathogenesis, vector biology, and the evolution and ecology of RF spirochetes and the Borreliaceae.
Results
Genomic evaluation of WHsTBRF spirochetes
Within the Western Hemisphere, seven TBRF species have been identified and isolated [15, 29], and a representative genome from each species was evaluated. The analysis also included a representative from each of the B. hermsii genomic groups (genomic group I: DAH, genomic group II: YOR) [33]. Furthermore, we evaluated a human isolate of B. turicatae (BTE5EL) and a commonly used laboratory isolate (91E135) (Table 1).
To aid in genome assembly of plasmids, we performed pulsed-field gel electrophoresis of genomic DNA isolated from each strain, which showed varying complexity and exemplified the variation in plasmid sizes (Fig. 1). Genomic profiles indicated that B. anserina BA2 was a relatively simple genome displaying approximately four plasmids while B. coriaceae Co53 and B. turicatae 91E135 were more complex with ~ 10 and ~ 11 visible linear plasmids, respectively. Furthermore, the high molecular weight bands on pulsed-field gels represent the presence of circular plasmids [33]. Interestingly, circular plasmids have been reported to be absent in Borrelia parkeri [34], but this analysis suggested evidence of circular plasmid(s) in the SLO isolate (Fig. 1).
Pulsed-field gel electrophoresis (PFGE) of WHsTBRF species. PFGE was performed on genomic DNA of each isolate to demonstrate the different plasmid profiles both between and within species. Isolate names have been abbreviated as follows: baBA2, B. anserina BA2; bhDAH, B. hermsii DAH; bhYOR, B. hermsii YOR; bcCo53, B. coriaceae Co53; bpuSUM, B. puertoricensis n. sp. SUM; bpSLO, B. parkeri SLO; bvRMA01, B. venezuelensis RMA01; bt91E135, B. turicatae 91E135; btBTE5EL, B. turicatae BTE5EL. The numbers to the left of the gel image correspond to DNA size in kilobases. Chr/LMP corresponds to the co-migration of the chromosome and linear megaplasmid, respectively. CP refers to circular plasmids. The full-length, uncropped gel is presented in Additional File 1: Fig S1
Genome assembly and annotation results are summarized in Additional File 2: Table S1. The dataset contained a linear chromosome for each species and a total of 128 plasmids comprised of 94 linear and 34 circular plasmids. The average number of plasmids per genome was 13.9 (range: 5–23). The average circular to linear plasmid ratio was 0.35 (range: 0.0–0.83). The smallest genome was B. anserina BA2 (1.05 Mb), which contained the least number of plasmids (four) and no circular plasmids. The largest genome was that of B. puertoricensis n. sp. SUM (1.87 Mb), which also contained the most circular plasmids, 10. The strain with the largest number of plasmids, overall, was B. coriaceae Co53 (18 linear plasmids and five circular). Interestingly, sequencing and assembly of B. parkeri SLO confirmed the presence of circular plasmids. We identified multiple circular plasmids in all genomes analyzed except B. anserina BA2 and B. venezuelensis RMA01. Borrelia anserina BA2 contained no circular plasmids while B. venezuelensis RMA01 contained only one circular plasmid (cp28). Collectively, a variety of differences in genome composition existed across the genomes of the WHsTBRF clade as exemplified by the number of plasmids, and differences in both length and topology.
ANI and phylogenomic analyses
ANI is a robust method for determining whole genome sequence identity by analyzing orthologous regions between genomes [35, 36]. Clear species boundaries have been shown for prokaryotes at intraspecies values > 95% and interspecies values < 83% [37]. Using a North American hard tick-borne relapsing fever spirochete (B. miyamotoi CT13-2396) as an outgroup, we calculated the ANI in an all-vs-all comparison of our genome assemblies (Additional File 1: Table S2). We observed an ANI between both B. turicatae isolates (91E135 and BTE5EL) of ~ 99%, which is substantially higher when compared to the ANI of the isolates representing both genomic groups of B. hermsii (DAH and YOR) at ~ 95.7%. Surprisingly, B. venezuelensis RMA01 and B. turicatae 91E135 and BTE5EL genomes indicated high ANI (~ 98.5%) despite being isolated from different Ornithodoros species located ~ 6,800 km apart from each other.
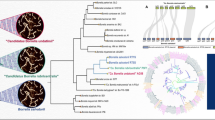
We investigated the phylogenetic relationships of the WHsTBRF spirochetes using two different species tree inference methods, concatenation and coalescence. By using the Panaroo pipeline, we identified 741 single-copy core genes between the WHsTBRF clade and B. miyamotoi CT13-2396 and performed phylogenomic analysis with this data set [38]. We tested these genes for substitution saturation as this can negatively impact the accuracy of phylogenomic analysis [39]. Saturation analysis determined that 91 genes were likely saturated and thus were removed from this dataset (Additional File 3). A maximum likelihood species tree was inferred from a concatenation of 650 single-copy core genes (720,532 total nucleotide sites) using an edge-linked proportional partition model with nucleotide sequences (Fig. 2A). The species tree of single-copy core genes recapitulated previously published tree topologies with high bootstrap support (100% in all cases) [40, 41]. To further determine if our tree was valid, we used concordance factor (CF) analysis. This analysis is important when using large datasets for phylogenomic species tree inference as there can be high bootstrap support for wrong topologies [42]. While the concatenation-inferred species tree showed high bootstrap value support for all branches, there was varying gene CF (gCF) and site CF (sCF) support. The B. hermsii and B. parkeri branches had low gCF and sCF support. We further observed that the phylogenomic analysis supported, through both bootstrap and gCF and sCF, that B. venezuelensis is a highly related sister taxon to B. turicatae. To account for potential gene tree discordance due to divergent coalescent histories we also inferred a species tree using the ASTRAL tool [43].
Phylogenomic analyses A maximum likelihood species tree was inferred from a concatenation of 650 single-copy core genes (720,532 nucleotide sites). A The tree was generated using an edge-linked proportional partition model with 1,000 ultra-fast bootstraps, and a subsequent concordance factor analysis was performed. Branch support is reported as ultra-fast bootstrap/gCF/sCF (see text for description of gCF and sCF). The scale bar is representative of substitutions per site. A coalescence-based cladogram was inferred from the 650 genes used in A. B The cladogram branch supports are reported as local posterior probability, major quartet frequency, alternative quartet frequency 1, and alternative quartet frequency 2. Red branch supports indicate alternative quartet frequencies that are significantly unequal (p < 0.001) indicating that incomplete lineage sorting alone does not explain that topology
The tree inferred by ASTRAL maintained the same topology as the concatenation tree and was well supported by local posterior probabilities at all branches (Fig. 2B). The quartet support of the ASTRAL tree also agreed with the gCF and sCF support seen in the concatenation tree. Interestingly, alternative quartet frequencies for the B. coriaceae branch were statistically different indicating that incomplete lineage sorting alone could not be attributed to driving this topology [44].
Pangenome analysis
Pangenome analysis of the WHsTBRF clade estimated there to be a core genome of 817 gene clusters and an accessory genome of 1,759 gene clusters for a total pangenome size of 2,576 gene clusters (Fig. 3A). The core genome stabilized after the addition of approximately five genomes but the pangenome size continued to increase with each genome (Fig. 3B). The continued increase in pangenome size is most likely due the expansive paralogous gene families and plasmid diversity seen in borrelial species [22, 27]. The differences between species and the unique gene clusters they contained can be appreciated in the pangenome matrix plot (Fig. 3C). Borrelia coriaceae Co53 contributed the most unique gene clusters, 484, while B. venezuelensis RMA01 contributed the least, 18. The pangenome analysis indicated a diverse and open pangenome for the WHsTBRF clade and identified a sizeable amount of unique gene content in the B. coriaceae Co53 genome.
Pangenome analysis. The core genome in this analysis is defined as gene clusters present in all nine genomes. The shell genome is defined as gene clusters present in two to eight genomes. The cloud genome is defined as gene clusters present in only 1 genome (A). A graph of the pan- and core-genomes was generated with PanGP, which indicated that the WHsTBRF clade pangenome is open and stabilizes after four genomes (B). Also shown is the pangenome matrix (C). The numbers on the right indicate the number of unique gene clusters for each isolate’s genome. In the case of multiple isolates per species (B. hermsii and B. turicatae) the species-specific unique gene clusters are reported first followed by the isolate-specific unique gene clusters (C). Taxa prefixes: btBTE5EL, B. turicatae BTE5EL; bt91E135, B. turicatae 91E135; bvRMA01, B. venezuelensis RMA01; bpSLO, B. parkeri SLO; bpuSUM, B. puertoricensis n. sp. SUM; bcCo53, B. coriaceae Co53; bhDAH, B. hermsii DAH; bhYOR, B. hermsii YOR; baBA2, B. anserina BA2
Chromosome and plasmid sequence analyses
Chromosomal sequence analysis
Previously the B. hermsii and B. turicatae chromosomes were reported to be syntenic and collinear [28], and we evaluated this across all species of the WHsTBRF borreliae. We investigated the chromosomes of our dataset first by performing ANI on chromosomal sequences only (using B. miyamotoi CT13-2396 as an outgroup). We observed a wide degree of chromosome sequence identity in the WHsTBRF spirochetes from 85.92% to 99.44% between species (Additional File 2: Table S2).
We also assessed synteny in WHsTBRF clade’s chromosomes by aligning these sequences using the Mauve aligner [45]. This analysis indicated a single, large collinear block that showed no rearrangements among the different chromosomes (Additional File 1: Fig S2). With the lack of internal structural rearrangements and the wide variation in ANI, we investigated gene content differences between chromosomes using Panaroo analysis. Panaroo was used to determine gene presence or absence differences, and to determine the core genome gene clusters that resided on the chromosome. The majority of the core genome was found in the chromosome (94%, 767 gene clusters), and largely accounts for many of the housekeeping and essential gene functions (e.g. DNA replication and repair, translation, protein synthesis, etc.). Most of the chromosomal gene content was similar, with gene presence or absence differences shown in Additional File 2: Table S3. Of the 874 gene clusters identified during the chromosomal sequence analysis, 767 gene clusters (79% of chromosomal gene clusters) were present in all isolates. The remaining 107 gene clusters were differentially present between isolates. While analyzing the gene content of the chromosome sequences, we noticed structural differences on the telomeric ends.
Both the 5’ and 3’ chromosome telomeres lacked synteny and contained gene content that differed between species. Investigation of these sequences showed the chromosome telomeres to be identical to plasmid telomere sequence (Additional File 1: Fig S3), with the exception of B. hermsii whose 5’ chromosome telomere was not found in its plasmids. Of note, we identified the presence of a variable major protein (vmp) allele on the 5’ telomere of the B. coriaceae Co53 chromosome (bcCo53_000002). The vmp alleles are important for RF spirochete pathogenesis and exist in two different but functionally synonymous types, variable large and variable small proteins [2, 46,47,48,49]. The bcCo53_000002 allele was intact and annotated as a variable large protein. Vmp alleles have not been previously reported on RF spirochete chromosomes. While sequences of RF spirochete chromosomes have been available for some time, many are missing the telomeric ends of the chromosome. Our work indicates that while most of the chromosome is syntenic, gene exchange and accumulation between chromosome and plasmid has occurred in the telomeres.
Plasmid sequence analysis
With the generated genomes, we were positioned to analyze and compare the plasmid sequences of the WHsTBRF spirochetes. We determined the number and location of putative plasmid partitioning genes for all plasmids (Fig. 4). Plasmid partitioning genes are important for the stable, heritable maintenance of plasmids in bacteria. At least five Borrelia paralogous gene families have been identified as putative plasmid partitioning genes: PF32, PF49, PF50, PF57, and PF62 [50,51,52]. The PF57 and PF62 gene families have limited similarity to other genes outside of Borrelia but are thought to be functionally redundant and have been grouped together as PF57/62 [20, 50]. Analysis of plasmid partitioning genes showed 39 plasmids contained multiple sets or copies of these genes (Fig. 4). The presence of multiple copies of partitioning genes on the same replicon suggests plasmid fusion events as each plasmid should contain a unique set or copy of the plasmid partitioning genes [50]. However, because these genes are likely involved in plasmid compatibility, containing the same set of partitioning genes on multiple plasmids would be detrimental. Such an event would result in plasmid incompatibility resulting in plasmid loss in daughter cells [21, 51]. Consequently, we would expect plasmid partitioning genes to be divergent in nucleotide identity within the same genome. To further investigate the relationships between plasmids and the partitioning genes, we performed phylogenetic analysis.
Plasmid repertoires and approximate locations of plasmid partitioning genes. The proportionate sizes of the plasmids found in each isolate are graphically depicted. The replicons are named based on topology (lp = linear plasmid, cp = circular plasmid) followed by the size to the nearest kilobase. The breaks in the megaplasmids, denoted by the discontinuity found on the left side of the megaplasmid, are of 80 kb except for B. anserina BA2 which was not broken. The approximate location of the PF32, PF49, PF50, and PF57/62 plasmid partitioning genes and their orientations are also shown with triangles facing right for positive-sense and left for negative-sense. This figure highlights the possibility of past fusion events on plasmids that have multiple plasmid partitioning gene(s) of the same type. Created with BioRender.com
Borrelial plasmids have been previously investigated and classified by phylogenetic analysis of PF32 loci [21, 22, 25], and we performed a similar analysis with our dataset (Additional File 1: Fig S4). However, we identified fourteen plasmids lacking a PF32 locus, so we performed a phylogenetic analysis of PF57/62 loci (Additional File 1: Fig S5), which was found in every plasmid in at least one copy. In these phylogenetic analyses, we included LD spirochete and B. miyamotoi CT13-2396 PF32 and PF57/62 loci (deposited genomes used are indicated in Additional File 2: Table S4) [22]. The PF32 loci phylogenetic analysis determined WHsTBRF spirochetes had a total of 30 plasmid clusters based on topology. Eleven of these plasmid clusters were related to plasmid-types found in LD spirochetes (Additional File 1: Fig S4). Seven plasmid clusters contained plasmids found in B. miyamotoi CT13-2396, while nine clusters did not contain a plasmid from either the LD spirochetes or B. miyamotoi CT13-2396 (Additional File 1: Fig S4). Within all plasmid clusters there were a variety of plasmid topologies and sizes. Due to this finding, adopting the nomenclature used in LD spirochete plasmid typing was not practical. Consequently, we designated each cluster as a plasmid family (“F” for family, F1-30) for reference in this and future analyses until more complete work can develop a harmonized nomenclature for New- and Old-World RF spirochete plasmids.
To validate the phylogenetic plasmid clusters, we performed sequence identity clustering of the PF32 loci. This analysis generally agreed with the phylogenetic analyses and demonstrated intra-cluster nucleotide sequence identities ≥ 85% (Additional File 4). There were five plasmid families where nucleotide sequence clustering of PF32 loci did not agree with the clustering in the phylogenetic analysis (F4, F5, F20, F21 and F28 families). Orphan PF32 loci that did not cluster with any plasmid family were also identified. Four orphan PF32 loci were found by phylogenetic clustering based on topology (baBA2_000862_lp12-1_ps, bcCo53_001336_lp24-1, AXH25_RS04465_cp2, and btBTE5EL_001353_lp32) (Additional File 1: Fig S4); however, only three of these loci were orphaned by the sequence identity clustering (baBA2_000862_lp12-1_ps, AXH25_RS04465_cp2, and btBTE5EL_001353_lp32) (Additional File 4).
Phylogenetic analysis of PF32 loci showed evidence of extensive inter–plasmid family fusions in both circular and linear plasmids. For example, multiple PF32 loci were found on the same plasmid but clustered in different plasmid families (e.g. bhYOR_lp93, bpSLO_lp42, bpuSUM_cp59-1, bt91E135_cp49) (Additional File 1: Fig S6). Plasmid fusion events have been described previously in borrelial genomes [20,21,22]; however, since PF32 genes are reportedly important for borrelial plasmid compatibility, no genome should contain two intact PF32 loci of the same family on different plasmids.
We identified a total of four occurrences in four assemblies where multiple copies of PF32 loci from the same family designation were found on different plasmids. Monomer and heterodimers of circular plasmids were the most common occurrences of the same PF32 loci on different plasmids in the same genome (bhYOR_000915_cp30-2 and bhYOR_001000_cp60, bpSLO_000869_cp28, bpSLO_000910_cp29, bpSLO_000951_cp58 and bpSLO_000994_cp58). We detected reads consistent with both length and sequence of monomeric and dimer forms, thus we included these replicons in the final assemblies. However, the long-term stability of such occurrences is unknown and we cannot rule out that these forms exist independently across the polyclonal population of cells.
There were two occurrences of similar PF32 loci on different linear plasmids in the same genome. The B. hermsii YOR lp73 and lp89 plasmids both had an F4 PF32 locus (bhYOR_001322 and bhYOR_001456, respectively). Further investigation of these loci found that they were less than 50% the length of other PF32 loci in the F4 family (these were not predicted to be pseudogenes). These loci were immediately upstream of PF49 pseudogenes with no PF50 or PF57/62 loci near them. It is likely that bhYOR_001322 and bhYOR_001456 are remnants from a fusion event and are not functional, thus we posit this would not cause plasmid instability. The B. venezuelensis RMA01 lp35 and lp89 plasmids both had a similar F28 PF32 locus (bvRMA01_001193 and bvRMA01_001225, respectively). These plasmids were nearly identical except for sequence near and including the 3’ telomere. This finding is part of a subsequent analysis of our dataset (manuscript in preparation). Interestingly, 13 linear plasmids only had the PF57/62 locus and no other plasmid partitioning genes (Fig. 4).
To account for the plasmids lacking a PF32 locus we performed phylogenetic analysis of the PF57/62 locus (Additional File 1: Fig S5). This analysis showed weak agreement with the PF32 phylogeny; however, phylogenetic clustering agreed for F6, F20, F27, and F28 plasmids. Interestingly, the 13 PF57/62-only plasmids clustered into a single monophyletic clade with three distinct subclades, where B. coriaceae Co53 formed a clade separate from the rest of the WHsTBRF spirochetes and B. miyamotoi CT13-2396. Linear PF57/62-only plasmids have been described before in B. miyamotoi and in LD spirochetes [20, 25]. The B. miyamotoi CT13-2396 PF57/62-only plasmids did not cluster with the WHsTBRF spirochete PF57/62-only plasmids. Lyme disease spirochete plasmids containing only PF57/62 genes (i.e. lp5) did not cluster with the WHsTBRF clade’s PF57/62-only plasmids suggesting they may be specific to TBRF spirochetes.
Dot plot analysis of the PF57/62-only plasmids demonstrated intra-clade relatedness and synteny for the non-B. coriaceae Co53 clades (Additional File 1: Fig S7). The non-B. coriaceae Co53 clades had presence of phage-related genes including a structural gene, tape measure protein, a part of the phage tail. The B. venezuelensis RMA01 lp17-1 plasmid had limited synteny to the other plasmids in the non-B.coriaceae Co53 clades and did not carry any discernable phage-related genes. The plasmids in the B. coriaceae Co53 clade were not syntenic and all but two (lp28-1 and lp29-1) carried vmp alleles. Vmp alleles were not seen in the other species’ PF57/62 clade. From these two clades several fusion events have occurred with other plasmids. Fusions appeared to have occurred in the B. coriaceae Co53 PF57/62-only plasmids but were not present in B. puertoricensis n. sp. SUM, B. parkeri SLO, or B. venezuelensis RMA01 plasmids (Additional File 1: Fig S8).
To investigate the relatedness of WHsTBRF spirochete plasmids to each other, we performed dot plot analysis of plasmid sequences in a pairwise fashion for all species. We were primarily interested in the plasmids that have remained largely syntenic across the WHsTBRF clade. Representative dot plots showing plasmid sequence synteny over a wide genetic distance is shown in Fig. 5. Dot plot analysis highlighted three plasmid families as conserved and largely syntenic across a wide evolutionary distance: the megaplasmid (F6), the small F27 plasmids, and the F28 (cp26-like plasmid). We reasoned that since these plasmids were syntenic they would harbor the majority if not all the non-chromosomal core genes. Indeed, we found that of the 50 non-chromosomal core genes, 46 of these were on the three syntenic plasmid families (Additional File 2: Table S5). Taken together, our analysis of the plasmids indicated that the WHsTBRF have a diverse plasmid repertoire that differs in PF32 and PF57/62-types compared to LD spirochetes and B. miyamotoi CT13-2396.
Representative plasmid dot plots of the WHsTBRF spirochete plasmid sequences. Dot plots were generated in a pairwise fashion of the WHsTBRF spirochete plasmids between isolates. Representative dot plots are shown between the most divergent species (B. anserina BA2 and B. turicatae BTE5EL, A), between B. hermsii DAH and B. turicatae BTE5EL (B) and B. parkeri SLO and B. turicatae BTE5EL (C), and isolates of the same species (B. turicatae 91E135 and BTE5EL, D). Synteny is represented by a purple line from the bottom left corner to the top right corner of each comparison. Inversions are indicated in blue from the top left to the bottom right of each comparison
Comparative genomics of B. venezuelensis and B. turicatae
The first isolate of B. venezuelensis was established in 2018 and a multi-locus sequence analysis placed the species in a monophyletic clade with B. turicatae. This relationship indicated a high level of sequence identity between species that were isolated from two different Ornithodoros species [15]. We initially predicted that the multi-locus sequence analysis scheme, which used rrs, flaB, and glpQ, was insufficient to resolve B. venezuelensis from B. turicatae [15]. However, both ANI (Additional File 2: Table S2) and phylogenomic analyses (Fig. 2) maintained this same relationship. The ANI results suggested that B. venezuelensis RMA01 and B. turicatae may be the same species since they are within the species cutoff of > 95% ANI (~ 98.5%) [37]. However, ANI only analyzes orthologous sequences between the two genomes being compared. If unrelated plasmids are present between the genomes being compared these sequences would not be considered during ANI analysis.
We investigated the plasmids of B. turicatae and B. venezuelensis RMA01 more closely to better understand the global genomic differences between the two species. We examined the plasmid families represented in both B. turicatae isolates and the B. venezuelensis RMA01 genomes (Additional File 2: Table S6). Twenty one plasmid families were represented with only four shared between all three genomes (F6, F20, F27, and F28). Only six plasmid families were shared between the two B. turicatae isolates and each isolate separately shared a plasmid family with B. venezuelensis RMA01. Between all three genomes, B. turicatae BTE5EL contained the greatest amount of unique plasmid families (five), whereas B. turicatae 91E135 and B. venezuelensis RMA01 each contained two unique plasmid families. In terms of the PF57/62-only plasmids, B. turicatae 91E135 contained two and both B. turicatae BTE5EL and B. venezuelensis RMA01 each contained one. These data highlight the variability in plasmid composition even between highly related isolates of the same species (B. turicatae 91E135 and BTE5EL).
We also investigated the similarity of B. venezuelensis and B. turicatae plasmid sequences using dot plot analysis. Both the ANI analysis and dot plots suggested that of the two isolates of B. turicatae, B. venezuelensis RMA01 was only modestly more related to B. turicatae 91E135 (Additional File 2: Table S2, Additional File 1: Fig S9). The similarity was due to the greater synteny of the lp17-2 plasmid of B. venezuelensis RMA01 with the lp19 plasmid in B. turicatae 91E135 as opposed to the lp46 plasmid in B. turicatae BTE5EL. However, outside of the core plasmid families (F6, F27, F28) there was little synteny between B. turicatae isolates and the B. venezuelensis RMA01 genome.
Given our observations, we analyzed the presence and absence output from Panaroo to understand gene content differences between B. turicatae and B. venezuelensis RMA01. Gene cluster analysis of the chromosomes revealed that there were only four differences between both B. turicatae isolates and B. venezuelensis RMA01 (dusA; group_368_hypothetical protein; group_563_hypothetical protein; and group_95_MATE family efflux transport, Additional File 2: Table S3). Analyzing the plasmid gene clusters, we determined that B. venezuelensis RMA01 shared 1,013 gene clusters with both B. turicatae isolates but contained 54 unique gene clusters (Fig. 6). We further investigated the gene clusters that were unique to B. venezuelensis RMA01 (54 gene clusters) and both B. turicatae isolates (138 gene clusters) (Additional File 2: Table S7). Most differentially present gene clusters were annotated as either hypothetical proteins, proteins with domains of unknown function, vmp alleles, or plasmid partitioning genes. Differences in vmp alleles were expected, given the wide variation in sequence between alleles even within the same genome [53, 54], which should result in numerous gene clusters. The difference in plasmid partitioning genes was not surprising given the variation in plasmid content between the two species. Phage-related gene clusters (mlp-family lipoprotein, Borrelia repeat proteins, tail fiber genes, etc.) were also differentially present between the two species. Moreover, gene cluster differences also reflected structural differences found between the two species. For example, gene clusters functionally annotated as “Borrelial persistence in ticks protein A” genes are located in a region of the megaplasmid that is truncated in B. venezuelensis RMA01 and the number of these genes differ between B. turicatae (5 copies) and B. venezuelensis RMA01 (2 copies). Collectively, these data demonstrate that there is extensive plasmid sequence differences between B. turicatae and B. venezuelensis RMA01 due to variation in plasmid composition and gene content.
Gene content comparison between B. turicatae isolates and B. venezuelensis RMA01. To determine the number of gene clusters shared between B. venezuelensis RMA01 and the B. turicatae isolates an area-proportional Euler diagram was generated. The gene content of the different isolates is represented by different colored circles: blue, bt91E135 = B. turicatae 91E135; black, bvRMA01 = B. venezuelensis RMA01; yellow, btBTE5EL = B. turicatae BTE5EL. The numbers in each region indicate the number of gene clusters present in that relationship
Discussion
This study generated complete chromosome and plasmid genomes from representative isolates of seven soft TBRF spirochete species found in the Western Hemisphere. With these genomes, we performed a detailed comparative genomics analysis on this RF spirochete clade. We observed plasmid diversity as they clustered into 30 families based on PF32 loci. Interestingly, only three plasmid families were syntenic between all species analyzed. Our findings supported prior observations that Borrelia species genomes are in flux with recombination occurring in plasmids and the telomeric ends of the chromosome [20,21,22, 48, 55,56,57]. Furthermore, our phylogenomic analysis identified underlying gene tree discordance.
We performed CF analysis on the concatenation-based phylogenomic tree because previous analyses have indicated unstable branches across the WHsTBRF taxa [15, 58,59,60,61,62,63]. Specifically, studies using the Borrelia multi-locus sequence typing (MLST) scheme put forth by Margos et al. are incongruent with phylogenomic analyses of the Borrelia species [40, 41, 58,59,60,61, 63, 64]. The current Borrelia MLST scheme uses eight conserved, chromosomal housekeeping genes (clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA) [58]. This approach was initially developed for LD spirochetes and later implemented with RF borreliae [59,60,61, 63]. However, when this scheme has been used with RF spirochetes a branch is formed including B. hermsii and B. anserina to the exclusion of the rest of the WHsTBRF clade, while B. miyamotoi often occurs as a sister clade to the WHsTBRF [59, 61, 63]. There was even one case where MLST phylogenetic analysis placed B. coriaceae outside of the WHsTBRF and B. miyamotoi clade [60]. In our saturation analysis of the single-copy core gene dataset, clpX and rplB were identified as having a high risk of saturation indicating that they have lost their phylogenetic signal. Importantly, previously published phylogenomic results contrasted with this MLST analysis [40, 41, 64]. They resolved B. anserina and B. hermsii into separate branches that included B. coriaceae in the WHsTBRF clade and placed the WHsTBRF and Eastern Hemisphere sTBRF spirochetes as sister clades to the exclusion of B. miyamotoi [40, 41, 64]. While we did not address the relationship of B. miyamotoi to the WHsTBRF clade here, we investigated the underlying gene tree variance in our WHsTBRF spirochete dataset using CF analysis. This showed low CF support for the B. hermsii, B. coriaceae, and B. parkeri branches. Low gCF and sCF scores are typically indicative of discordance within the individual gene trees [65]. This indicates that individual gene trees may be incongruent with the species trees and that phylogenetic analysis of one or a few loci can yield conflicting tree topologies against the species tree, as noted with the MLST phylogenetic data. Thus, one should use caution when trying to apply the clpA, clpX, nifS, pepX, pyrG, recG, rplB, and uvrA MLST scheme to the WHsTBRF taxa for phylogenetic analysis. To validate our concatenation-based tree, we also performed a coalescence-based tree inference to orthologously estimate a species tree.
The coalescence-based ASTRAL method inferred a species tree that recapitulated the concatenation-based tree topology and indicated discordance for the B. hermsii, B. coriaceae, and B. parkeri branches. ASTRAL is a gene tree summation method that estimates a species tree despite gene tree discordance due to incomplete lineage sorting and the presence of horizontal gene transfer [43, 66]. The quartet data for all branches of the ASTRAL tree had alternative topology quartet frequencies that were approximately equal (q2 = q3) and had a normalized quartet frequency of ~ 89.1%. These data suggested a low-level of incomplete lineage sorting had occurred, which may explain some of the observed gene tree discordance [43, 67]. Additionally, the B. coriaceae Co53 branch of the ASTRAL tree had a statistically significant difference in alternative frequencies indicating that incomplete lineage sorting alone could not explain this branch [67]. Sources of gene tree discordance include biological (e.g. horizontal gene transfer) and technical (gene tree estimation error and hidden paralogs) [68, 69]. Given that previous phylogenomic analyses that used different methodologies were congruent with our analysis, we suspect the gene tree discordance seen in our study was biological. Clearly, more work is needed to generate a complete and confident understanding of the evolutionary history of this WHsTBRF spirochete clade.
A pangenome analysis investigated gene diversity across the WHsTBRF clade and was reflective of the numerous paralogous families in the Borreliaceae. The WHsTBRF spirochete pangenome consisted of a total of 2,576 unique gene clusters. Previous pangenome analysis by Elbir et al. [28] on African RF borreliae (Borrelia duttonii, Borrelia recurrentis, Borrelia crocidurae, and Borrelia hispanica) estimated a pangenome size similar to ours at 2,514 gene clusters. Pangenome analysis by Mongodin et al. [70] estimated pangenome sizes of ~ 1,500 and ~ 1,859 for B. burgdorferi sensu stricto and B. burgdorferi sensu lato, respectively. However, the Borreliaceae contain approximately 175 paralogous gene families with intraspecific nucleotide identities often ≥ 60% [20, 27, 71]. Due to the low similarity of these paralogous multicopy gene families within species and between species, gene clustering algorithms will generate multiple clusters for these families. This phenomenon will inflate the overall number of gene clusters reported by pangenome analysis. Indeed, an investigation into the Panaroo output showed that the variable small protein (vsp) and variable large protein (vlp) alleles contributed 78 and 282 gene clusters, respectively. This accounted for ~ 14% of the entire pangenome. This caveat should be considered when analyzing pangenome data from Borrelia species.
Given that prior work with B. turicatae and B. hermsii indicated that the chromosomes were largely collinear and had similar gene content, a chromosomal analysis was conducted across the WHsTBRF spirochete clade [28]. Mauve alignment analysis indicated that the chromosomes of the WHsTBRF spirochetes were collinear with no large, internal structural changes. Since we produced full-length chromosomes, the Mauve analysis allowed us to detect variable gene content located at the telomeres. Variable gene content in the 3’ end of LD spirochete chromosomes has been previously reported [21, 57]. In contrast, both the 5’ and 3’ chromosome ends in WHsTBRF spirochetes were not syntenic across all species. For most of the isolates we examined, the gene content on the chromosome ends was also shared with plasmid telomeric sequence.
We investigated plasmid diversity to further appreciate the genomic complexity of RF spirochetes compared to the more characterized LD spirochete genomes. The average number of plasmids found in the WHsTBRF clade (13.9) was higher than that reported for B. burgdorferi sensu lato (12.8) but lower than that of B. burgdorferi sensu stricto (16.9) [22, 55]. The ratio of circular to linear plasmids for WHsTBRF spirochetes was 0.35 (range: 0–0.83), which is low compared to the range found in LD spirochetes [22, 55]. On average, the circular to linear plasmid ratio of B. burgdorferi sensu lato was 0.68 and 0.92 for B. burgdorferi sensu stricto [22]. The presence and sizes of circular plasmids were variable between species and even within species.
Our phylogenetic analysis of the 128 plasmids generated in this study identified 30 plasmid families, which are similar in classification to the LD spirochete plasmid-types [22]. Currently there are 30 recognized plasmid types in LD spirochetes based on the PF32 gene [22]. Our analysis found that only 11 RF plasmid families had a related LD spirochete plasmid type. This indicates that there are PF32 types unique to both RF and LD spirochetes based on this classification scheme. We found that, in agreement with previous work done in LD spirochetes and B. miyamotoi [20, 22, 25, 50, 55], no genome (with noted exceptions) contained intact PF32 loci of the same family on different plasmids. We identified 13 plasmids that lacked a PF32 locus and only had the PF57/62 locus. Plasmids lacking the PF32 gene have been observed in LD spirochetes (e.g. lp5 and cp9 plasmid-types) [22]. These LD plasmid-types did not phylogenetically cluster with the RF spirochete PF57/62-only loci. This further suggests unique plasmid types in both the WHsTBRF and LD spirochetes. Based on the gene content of the WHsTBRF PF57/62-only plasmids, we hypothesize that they are either remnants of or intact Borrelia prophages.
Deeper analysis of both circular and linear plasmids indicated the presence or remnants of prophage genomes. We identified prophage associated genes including blyA and blyB holin genes, mlp family lipoproteins, and phage recombinase genes. Variation in phage associated plasmids between the strains and species evaluated suggested differences in phage exposure. This variation is noted in B. parkeri SLO. This isolate contained multiple circular plasmids encoding phage-related genes. Interestingly, prior work with other B. parkeri strains (which did not include the SLO strain) found only linear plasmids [34]. The geographic location, reservoir host use, and variations between tick populations could drive differences in phage-related plasmid composition through different phage exposures [72,73,74]. Prophages have been documented in LD spirochetes [72, 74, 75], and these viruses have been isolated and used to mediate transduction between B. burgdorferi isolates [74, 76, 77]. Future work should investigate whether there are inducible phages in RF spirochete species and whether these viruses can mediate transduction or horizontal gene transfer. This would provide a mechanism for genome plasticity of RF spirochetes.
We concluded our analyses by comparing the B. venezuelensis RMA01 genome to that of B. turicatae 91E135 and BTE5EL. The genomes of B. venezuelensis RMA01 and B. turicatae 91E135 and BTE5EL were within the species cutoff for prokaryotes (> 95%) [37]. Interestingly, previous reports for borrelial genomes have identified species with similarly high ANI scores. Elbir et al. [28] and Adeolu and Gupta [78] reported ANI scores between Borrelia (Borreliella) garinii and Borrelia (Borreliella) bavariensis (~ 98%), B. recurrentis and B. duttonii (~ 99%), B. duttonii and B. crocidurae (~ 99%), and B. crocidurae and B. recurrentis (~ 99%). While B. crocidurae, B. duttonii, and B. recurrentis had high pairwise ANIs, these species utilize different tick or insect vectors for their life cycle [3, 79,80,81,82,83]. ANI does not consider differences in plasmid composition between species.
A biological feature of RF spirochetes that has been used to speciate the pathogens is vector specificity, which should be considered with B. venezuelensis and B. turicatae. With vector specificity, a given spirochete species colonizes and is transmitted by a given species of argasid tick [84,85,86,87]. We report here that there is a high level of sequence identity between B. turicatae and B. venezuelensis RMA01 despite being isolated from two different tick Ornithodoros species whose known distributions are separated by at least 2,300 km [88, 89]. However, prior to considering B. turicatae and B. venezuelensis as the same species, vector competence studies are needed to determine whether Ornithodoros rudis (the vector of B. venezuelensis) can maintain and subsequently transmit B. turicatae. Similarly, studies would be needed with O. turicata (the vector of B. turicatae) and B. venezuelensis. Collectively, these findings would better define the biological and ecological differences between B. venezuelensis and B. turicatae.
Limitations of this work include the small number of species in this clade and shortcomings with the software used in the analyses. RF has been reported in Mexico [90,91,92], Central America [93,94,95], and throughout South America [96,97,98,99], yet we only had two representative species from Latin America [9, 15]. Furthermore, the genome assembly software used was not designed necessarily for covalently closed linear replicons [100]. As a result, borrelial genome assembly is labor intensive and can result in errors [24, 101, 102]. We mitigated assembly errors by implementing quality assessment steps utilized by Kuleshov and co-workers [25], and incorporated a new tool, Merqury [103]. Annotation was also complicated by the lack of characterization for many borrelial proteins resulting in many genes being classified as “hypothetical proteins” or “domain of unknown function-containing proteins”. Another annotation/gene classification shortcoming was found in the classification of pseudogenes (e.g. B. hermsii lp73 and lp89, bhYOR_001322 and bhYOR_001456) and subsequently when analyzing the InterProScan data for pseudogenes. There were instances where InterProScan analysis failed to classify plasmid partitioning pseudogenes that the annotation software had classified. Lastly, the number and diversity of the borrelial paralogs complicates proper estimation of the core and accessory genomes. Regardless, the completed chromosome and plasmid genomes enabled an informative comparative genomics analysis on the WHsTBRF clade.
TBRF spirochetes are a globally distributed yet neglected disease, and the genomes generated by this study will provide needed resources to study pathogenesis, vector biology, and for the development of diagnostics. With only two known isolates of TBRF spirochete from Latin America, the genomes of B. venezuelensis RMA01 and B. puertoricensis n. sp. SUM provides the opportunity to commence vector competence studies and to determine the clinical manifestation of B. venezuelensis. In Brazil, a Lyme-like illness (Baggio-Yoshinari Syndrome) circulates in the country but there is no epidemiological or ecological support for B. burgdorferi [104, 105]. Similarly, in the southwestern United States B. turicatae can present with neurological symptoms similar to Lyme disease and is often misdiagnosed [14, 106]. Studies are now possible to ascertain the pathogenesis of B. venezuelensis and to determine whether it is phenotypically like B. turicatae. Furthermore, there are likely more species to be found throughout the entirety of the RF Borrelia clade, some of which may be important species in the evolutionary history of this group. As Borrelia spp. isolation becomes more feasible [107], efforts are needed to gain a more complete understanding of their genome evolution, plasmid diversification, and how this may impact pathogenesis and vector-host use.
Conclusions
Given the unique genomic structure of spirochetes in the Borreliaceae family and the paucity of completed chromosome and plasmid genomes for RF spirochetes, this work addressed important knowledge gaps and provides needed resources. This work tripled the number of complete, plasmid-resolved genomes for soft tick-borne RF Borrelia species found in the Western Hemisphere and we provided a comparative genomics analysis. We investigated outstanding questions in the field regarding intra- and inter-species plasmid relationships and the high degree of sequence identity between B. turicatae and B. venezuelensis, which were isolated from two different Ornithodoros species. From these analyses we have identified substantial diversity in the plasmid sequences between the WHsTBRF spirochetes, as well as a core group of plasmids found in all isolates analyzed. Furthermore, while B. turicatae and B. venezuelensis RMA01 are highly similar at the chromosome-level, at the plasmid-level there were substantial differences in composition, structure, and gene content. Collectively, our findings will be foundational for future endeavors to identify the plasmid genetic elements that play a role in vector and host adaptation for both relapsing fever spirochetes and the Borreliaceae as a whole.
Methods
Borrelial strains and culturing
Borrelia spp. and isolates used in this study are referenced in Table 1. The seven species we selected are the only soft tick-borne relapsing fever Borrelia species isolated in the Western Hemisphere. The particular isolates were chosen because they are either the only isolates available of that species (B. puertoricensis n. sp. SUM and B. venezuelensis RMA01), conventional lab strains (B. hermsii DAH, B. hermsii YOR, B. coriaceae Co53, B. parkeri SLO and B. turicatae 91E135), or are from a species with limited isolates available (B. anserina BA2). The B. turicatae BTE5EL was also included because 1) it is the only human isolate and 2) from our previous work analyzing B. turicatae isolates, BTE5EL and 91E135 are from different rrs-rrlA intergenic spacer genotypes. All of the isolates were low passage (< 10), polyclonal isolates grown in either mBSK with 12% heat-inactivated rabbit serum, at either 35° or 37 °C in a 5% CO2 environment or BSK-R (B. puertoricensis n. sp. SUM and B. venezuelensis RMA01) at 34° C with caps sealed in a microaerobic environment [107]. Initial cultures were grown in 4 mL of media in sealed 8 mL polystyrene tubes (B. puertoricensis n. sp. SUM and B. venezuelensis RMA01 were cultured in 4.5 mL of media in sealed 5 mL polystyrene tubes) to ~ 107 cells/mL and were then used to inoculate 45 mL of media in a 50 mL polypropylene tube and cultured until ~ 107 cells/mL.
Genomic DNA isolation and sample quality control
Genomic DNA was extracted from the 45 mL of culture (see above) using phenol–chloroform, as previously described [108]. The resulting DNA pellet was allowed to air dry and was resuspended in 1 × TE buffer and stored at 4 °C. After ~ 48 h at 4 °C the genomic DNA was quantified using a NanoDrop 2000 (Thermo Fisher Scientific, USA). Genomic DNA samples were assessed for quality using pulsed-field gel electrophoresis to determine the integrity of the chromosome and plasmids, as previously described [33].
Sequencing
Oxford nanopore technologies library preparation
All strains were sequenced on an Oxford Nanopore Technologies (ONT) MinION Mk1B sequencer using the R9.4.1 flow cell and the SQK-RBK004 rapid barcoding library preparation kit. Four different library preparation strategies were attempted to yield larger read sizes while decreasing the amount of free adapter sequence. Initial library preps (library prep# 1) were done without prior cleanup of genomic DNA however we found that any genomic DNA that did not have an A260/280 or A260/230 ≥ 1.8 yielded little sequencing data. This result was likely due to interference of salts and/or organics in the transposase barcoding reaction. Therefore, in subsequent library preps (library prep# 2 and 3) all genomic DNA was further purified using one volume of magnetic beads (Omega Biotek, USA), washed twice with 70% ethanol, and eluted in pre-warmed (56 °C) elution buffer (Buffer EB; Qiagen, Germany). The specific library preparation strategies utilized for each strain is indicated in Additional File 2: Table S8.
Illumina sequencing
Bead-purified genomic DNA (discussed above) was provided for each strain to the Microbial Genome Sequencing Center (MiGS Center, USA) for Illumina sequencing. Sequencing performed using the Illumina Nextera paired-end 2 × 150 bp library prep kit on a NextSeq 550 sequencer. Demultiplexed FASTQ files were provided by the MiGS Center.
Sequence analysis
Example commands for each software are given in Additional File 5.
Sequence data processing
ONT data was basecalled using ONT’s Guppy v3.6.0 or 4.0.14 yielding FASTQ files of raw, basecalled data. The raw, basecalled data was demultiplexed using guppy_barcoder with the “–detect_mid_strand_barcodes” option. The FASTQ files after demultiplexing were initially processed using NanoFilt (v2.7.0) to remove all reads less than 1 kb and quality scores less than seven [109]. Summary statistics for each isolate’s ONT data were generated using NanoStat (v1.2.0) (Additional File 2: Table S9) [109]. Illumina data was provided by the MiGS Center as FASTQ files and filtered using Fastp (v0.20.1) to remove reads below Q20, trim adapters, and perform read correction (Additional File 2: Table S9) [110]. The Illumina data was merged for short-read polishing but was also processed as separate reads which were used for the Merqury tool (v1.1) for quality assessment and genome refinement (discussed later) [103]. Sequencing read data were deposited in NCBI’s Sequence Read Archive (SRA) as FASTQ files for both ONT and Illumina datasets. These data were deposited under the BioSample accession number associated with each isolate (see Additional File 2: Table S1).
Assembly and polishing
Whole genome assemblies were generated following the bioinformatics pipeline developed by Kuleshov et al. [25]. In short, filtered ONT data was assembled with the Canu (v2.0) assembler [100]. Following assembly, circular contigs were trimmed by using the apc.pl script to remove overlapping ends [111]. Contigs were then assessed for redundancy using BLASTn (v2.11.0) and by self-vs-self dot plot assessment using FlexiDot (v1.06) [112, 113], and redundant contigs were removed. Inverted telomeric ends of linear replicons were trimmed using Unipro UGENE (v35) for visualization and sequence manipulation, and the online EMBOSS einverted tool (http://emboss.toulouse.inra.fr/cgi-bin/emboss/einverted) was used to identify the hairpin sequence [114]. After inverted telomeric ends were removed, assemblies were polished using both ONT and Illumina data. ONT polishing was done using three iterative rounds of Racon (v1.4.16) polishing and one round of Medaka (v1.0.3) polishing using the g3.6.0 model [115, 116]. The ONT polished assembly was further polished with the merged Illumina data using iterative rounds of Pilon (v1.23) using the “all” option [117]. Pilon was run iteratively until no changes were made, typically two to four rounds though B. coriaceae Co53 required eight iterations.
Genome assembly quality control and refinement
Final genome assemblies fit the following quality criteria: 1) linear replicons must be from telomere to telomere, circular replicons must have no telomeres and be contiguous from end to end; 2) all replicons must be complete (i.e. 1 replicon = 1 contig); and 3) all assemblies must have a ≥ Q40 Phred-quality score and a ≥ 99.5% completeness score as calculated by Merqury [103].
Assemblies were initially assessed by Quast (v5.0.2) for mapping percentage of the Illumina and ONT datasets [118]. Coverage was determined for each of the replicons via Mosdepth (v0.2.9), and mapping via Minimap2 (v2.17-r974) with visualization in Geneious (v2020.1.2; Biomatters, New Zealand) [119, 120]. Each replicon’s sequencing coverage and depth was calculated to ensure there were no plasmid fusions or assembly errors. However, read mapping as a quality metric of completeness can be misleading. Currently there are no high-quality, complete reference genomes for many of the species we investigated which could be used to determine the completeness and quality of our assemblies. Thus, we employed the reference-free quality assessment tool Merqury. Merqury utilizes unmerged paired-end Illumina data to assess assembly completeness as well as quality at the individual contig and whole-assembly levels. Completeness is determined by how much of the unique k-mer fraction is contained in the assembly of interest; subsequently, quality is determined by measuring the amount of unique k-mer identity clashes in the assembly.
Merqury was also used for assembly refinement in situations where completeness was shown to be low (< 99.5%). Illumina reads containing unique k-mers not found in the assembly were extracted. After extracting these reads, they were assembled into scaffolds using the SPAdes assembler (v3.13.1). These scaffolds were used as a reference to map the ONT read dataset for that isolate using Minimap2 [121]. The resulting BAM file was visualized with Geneious and reads were assessed to find plasmids that may not have been assembled in the initial assembly. Often, missing plasmids were very similar to a plasmid already in the assembly, which was typical for many of the circular plasmids. We observed that often assemblies that were missing plasmids had to undergo greater than three Pilon iterations until no changes were made. After adding the missing plasmid(s), the number of Pilon iterations required usually decreased whereas completeness and quality, as assessed by Merqury, increased. Assemblies were generally considered refined, complete, and of high-quality if there was no plasmid that could be readily identified from the SPAdes scaffolds of unassembled unique k-mer reads, the completeness score was ≥ 99.5%, and had an assembly quality score ≥ Q40.
Genome annotation, InterProScan analysis, and replicon orientation
The plasmids of the final assembly were named based on topology (lp = linear plasmid, cp = circular plasmid) and size (nearest kilobase). In instances where the size was the same between two plasmids the plasmid was further numbered such as lp28-1, lp28-2, etc. The assemblies post-Merqury were annotated via a local copy of NCBI’s prokaryotic genome annotation pipeline (PGAP, v2020-09–24.build4894) to identify and annotate all coding sequences and genome features and InterProScan supplemented this with protein classification [122, 123]. The resulting GFF file was used to extract the transcripts from the assembly FASTA file using GFFread (v0.12.2) [124]. The extracted transcripts were used as input to a local copy of InterProScan (v5.47–82.0) to further classify transcripts using the CDD, Pfam, and Superfamily databases [125,126,127]. To facilitate standardization of replicon orientation, we identified the PF57/62 plasmid partitioning gene that was intact and had the best hit from InterProScan (Pfam, PF02414) on each replicon and oriented all plasmid replicons to put this gene in the positive-sense. The megaplasmid was kept with the PF57/62 gene in the negative-sense because previous studies and analyses have been conducted with the PF57/62 gene in the negative-sense [128, 129]. The chromosome only had PF32 plasmid partitioning genes where three were put in the positive-sense which put the remaining gene in the negative-sense, this serendipitously matched previously published chromosomal sequence orientations. Circular plasmids were reoriented to start with the best hit PF57/62 gene oriented in the positive-sense. Re-oriented genomes were then annotated with PGAP and analyzed via InterProScan as before. All subsequent work was completed using the final, re-oriented genomes.
Average nucleotide identity
Average nucleotide identity was determined by FastANI (v1.32) using an all-versus-all approach on the genome and chromosome FASTA files for all the strains being investigated [37]. The –matrix option was used to generate an identity matrix.
Pangenome analysis
Pangenome analysis was carried out using the Panaroo pipeline (v1.2.3) [38]. We modified Panaroo parameters to account for particular aspects of our dataset. To account for analyzing multiple species that at the lowest pairwise ANI was ~ 85% we adjusted the –threshold parameter to 0.8. To account for the high number of paralogous gene families we used the –merge_paralogs options. To account for our assemblies’ complete and non-fragmented linear replicons, we modified –min_trailing_support and –trailing_recursive to 0 to prevent erroneous trimming of the replicon ends. All refound pseudogenes (indicated in the output by refound….._stop) were manually removed from all analyses. Visualization of the Panaroo output was done using the roary_plot.py from Roary (v3.13.0) using the Newick tree generated from our concatenation-based phylogenomic tree (see Phylogenomics and Concordance Factor Analysis) and the gene_presence_absence_roary.csv file from Panaroo [130]. To visualize the pangenome and core genome line graph, PanGP (v1.0.1) was used with the gene_presence_absence.Rtab file from Panaroo [131]. The gene presence/absence file that lists the gene loci for each isolate in each gene cluster is found in Additional File 6.
Phylogenomics and concordance factor analysis
Concatenation-based phylogenomic analysis was performed using the Panaroo pipeline including our WHsTBRF spirochete dataset and the B. miyamotoi CT13-2396 genome. From the MAFFT alignments generated by Panaroo of the core genes, we analyzed the 741 single-copy core genes for substitution saturation using PhyloMAd [132]. Genes identified by PhyloMAd as having a high risk of substitution saturation in the phylogenetically informative sites (Risk_Entropyvar) were removed from phylogenomic analysis [39]. The remaining 650 single-copy core genes were used to infer a species tree and concordance factor analysis. See Additional File 3 for a list of the genes used for analysis (Risk_Entropyvar = low.risk) and those not used (Risk_Entropyvar = high.risk). A maximum likelihood tree was inferred with IQ-TREE2 (v2.1.2) using an edge-linked proportional partition model, automatic gene alignment concatenation, and 1,000 ultrafast bootstrap replicates [133,134,135,136]. The resulting tree and analysis were used to infer gene trees in IQ-TREE2 followed by gene and site concordance factor calculation. The final tree with the bootstrap and concordance factor metadata was analyzed using FigTree and annotated in Inkscape. Concordance factor analysis and interpretation are discussed in-depth in [65].
Coalescence-based phylogenomic analysis was performed on the 650 single-copy core gene alignments as discussed above. For this analysis however, each individual gene tree was bootstrapped with 1,000 ultra-fast bootstraps and collapsed at branches with < 10% support using IQ-TREE2. The resulting trees were used to infer a coalescence-based tree using ASTRAL [43, 66, 67, 137, 138]. The alternative quartet frequencies were evaluated using a test of proportions (prop.test) in R (v4.0.4) to assess whether the frequencies were statistically unequal. The resulting tree was visualized and annotated as a cladogram in FigTree and Inkscape, respectively.
MUMmerplot dot plot analysis
Dot plot analysis of the plasmid sequences and plasmid groups was performed using MUMmer (v4.0.0beta2) [139]. The sequences were first aligned using Nucmer with the –maxmatch option to allow for visualization of repeats and redundant sequence found within and between two sequences. The dot plots were generated using MUMmerplot with default options and the output from Nucmer to generate a PNG file.
Plasmid partitioning gene phylogenetic analysis and nucleotide sequence clustering
Plasmid partitioning genes were assessed using the results of each genome’s InterProScan analysis and PGAP’s annot.gff file. This analysis was performed using our WHsTBRF spirochete dataset, and deposited data for the LD spirochete dataset and B. miyamotoi CT13-2396 (Additional File 2: Table S4). Plasmid partitioning genes were found using the CDD and Pfam databases (PF32 = cd02038 and cd02042, PF49 = PF01672, PF50 = PF0289, PF57/62 = PF02414 based on work by Kuleshov et al. [25]) and located using the annot.gff file. Plasmid partitioning genes (PF32, PF49, PF50, PF57/62) identified by InterProScan using the above methods are available in Additional File 7. Using the PF32 or PF57/62 loci nucleotide sequences, we performed phylogenetic analysis using maximum likelihood tree inference. The nucleotide sequences of either the PF32 or PF57/62 loci were aligned using MAFFT with the “–auto" option. The alignment was used to generate a maximum likelihood tree inferred by IQ-TREE2 with 1,000 ultrafast bootstraps and the “–polytomy” option. Branches were collapsed with supports less than 50% using the “-minsupnew” option. The tree was analyzed using FigTree and annotated in Inkscape. Both phylogenies were analyzed using FigTree and annotated in Inkscape. The PF32 loci nucleotide sequences used for phylogenetic analysis were clustered using CD-HIT-EST (v4.8.1) with an 85% sequence identity threshold (-c 0.85) [140].
Euler diagram analysis
Using the binary output from Panaroo for the presence/absence analysis, we extracted the bvRMA01, bt91E135, and btBTE5EL columns. These data were used to plot a Venn diagram in R (v4.0.4) using gplots package (v3.1.1) and venn function [141]. The data from that diagram was then used to generate an area-proportional Euler diagram using the online eulerr application with the disjoint combinations option selected and a seed of 3 used [142]. The Euler diagram was annotated in Inkscape.
Mauve
ProgressiveMauve (v20150226 build 10) was used to visualize sequence similarities for the chromosome [45]. Individual GBK files for each genome’s annotated chromosome was used for alignment for the WHsTBRF spirochete dataset. ProgressiveMauve was run without assuming collinearity, with the “seed families” option selected, but otherwise with default options. Visualization was done using the Mauve program.
Availability of data and materials
Genomic DNA for the isolates used in this study is available upon request with the appropriate Material Transfer Agreement.
All datasets generated during the current are a part of the PRJNA637792 NCBI BioProject (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA637792). Individual NCBI BioSample accessions are as follows: B. anserina BA2 (SAMN18441552), B. coriaceae Co53 (SAMN18441555), B. hermsii DAH (SAMN18441553), B. hersmii YOR (SAMN18441554), B. parkeri SLO (SAMN18441556), B. puertoricensis n. sp. SUM (SAMN19000909), B. turicatae 91E135 (SAMN18441557), B. turicatae BTE5EL (SAMN18441558), and B. venezuelensis RMA01 (SAMN15156993).
NCBI GenBank accession numbers for each replicon are listed below:
B. anserina BA2.
Chromosome, CP073130.
lp10, CP073131.
lp12-1, CP073132.
lp12-2, CP073133.
lp27, CP073134.
lp88, CP073135.
B. coriaceae Co53.
Chromosome, CP075076.
cp29, CP075077.
cp30-1, CP075078.
cp30-2, CP075079.
cp30-3, CP075080.
cp48, CP075081.
lp11, CP075082.
lp188, CP075083.
lp20, CP075084.
lp23, CP075085.
lp24-1, CP075086.
lp24-2, CP075087.
lp28-1, CP075088.
lp28-2, CP075089.
lp29-1, CP075090.
lp29-2, CP075091.
lp30, CP075092.
lp31, CP075093.
lp32, CP075094.
lp33, CP075095.
lp34, CP075096.
lp38, CP075097.
lp39, CP075098.
lp60, CP075099.
B. hermsii DAH.
Chromosome, CP073136.
cp30-1, CP073137.
cp30-2, CP073138.
cp55, CP073139.
cp90, CP073140.
lp12, CP073141.
lp185, CP073142.
lp26, CP073143.
lp27, CP073144.
lp28, CP073145.
lp31, CP073146.
lp59, CP073147.
B. hermsii YOR.
Chromosome, CP073148.
cp30-1, CP073149.
cp30-2, CP073150.
cp60, CP073151.
lp11, CP073152.
lp194, CP073153.
lp23, CP073154.
lp46, CP073155.
lp73, CP073156.
lp89, CP073157.
lp93, CP073158.
B. parkeri SLO.
Chromosome, CP073159.
cp28, CP073160.
cp29, CP073161.
cp58, CP073162.
lp10, CP073163.
lp157, CP073164.
lp18, CP073165.
lp24, CP073166.
lp28, CP073167.
lp30-1, CP073168.
lp30-2, CP073169.
lp32, CP073170.
lp34-1, CP073171.
lp34-2, CP073172.
lp42, CP073173.
lp58, CP073174.
lp60, CP073175.
B. puertoricensis SUM.
Chromosome, CP075379.
cp27, CP075380.
cp28-1, CP075381.
cp28-2, CP075382.
cp29-1, CP075383.
cp29-2, CP075384.
cp30, CP075385.
cp59-1, CP075386.
cp59-2, CP075387.
cp60, CP075388.
cp61, CP075389.
lp10, CP075390.
lp110, CP075391.
lp25, CP075392.
lp29-1, CP075393.
lp29-2, CP075394.
lp35, CP075395.
lp39, CP075396.
lp41, CP075397.
lp46, CP075398.
lp47, CP075399.
lp57, CP075400.
lp65, CP075401.
B. turicatae 91E135.
Chromosome, CP073176.
cp30-1, CP073177.
cp30-2, CP073178.
cp31, CP073179.
cp49, CP073180.
lp10, CP073181.
lp158, CP073182.
lp19, CP073183.
lp24, CP073184.
lp29, CP073185.
lp36, CP073186.
lp38, CP073187.
lp40, CP073188.
lp43, CP073189.
lp48, CP073190.
lp53, CP073191.
B. turicatae BTE5EL.
Chromosome, CP073192.
cp30-1, CP073193.
cp30-2, CP073194.
cp31, CP073195.
cp60, CP073196.
lp10, CP073197.
lp160, CP073198.
lp24, CP073199.
lp28, CP073200.
lp30, CP073201.
lp32, CP073202.
lp34, CP073203.
lp37, CP073204.
lp42, CP073205.
lp45, CP073206.
lp46, CP073207.
lp49, CP073208.
B. venezuelensis RMA01.
Chromosome, CP073220.
cp28, CP073221.
lp10, CP073222.
lp127, CP073223.
lp17-1, CP073224.
lp17-2, CP073225.
lp25, CP073226.
lp30, CP073227.
lp31, CP073228.
lp35, CP073229.
lp37, CP073230.
The individual read datasets generated by this study are available in the NCBI Sequence Read Archive (SRA) (www.ncbi.nlm.nih.gov/sra/). The Illumina raw read dataset for each isolate is available at the following links:
B. anserina BA2 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006057), B. coriaceae Co53 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006046), B. hermsii DAH (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006056), B. hermsii YOR (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006047), B. parkeri SLO (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006044), B. puertoricensis n. sp. SUM (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006045), B. turicatae 91E135 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006042), B. turicatae BTE5EL (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006041)., B. venezuelensis RMA01 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006043).
The Oxford Nanopore Technologies FASTQ raw read dataset for each isolate is available at the following links:
B. anserina BA2 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006040), B. coriaceae Co53 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006053), B. hermsii DAH (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006055), B. hermsii YOR (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006054), B. parkeri SLO (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006051), B. puertoricensis n. sp. SUM (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006052), B. turicatae 91E135 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006049), B. turicatae BTE5EL (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006048)., B. venezuelensis RMA01 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR15006050).
References
Talagrand-Reboul E, Boyer PH, Bergström S, Vial L, Boulanger N. Relapsing fevers: neglected tick-borne diseases. Front Cell Infect Microbiol. 2018;8:98.
Cadavid D, Barbour AG. Neuroborreliosis during relapsing fever: review of the clinical manifestations, pathology, and treatment of infections in humans and experimental animals. Clin Infect Dis. 1998;26(1):151–64.
Trape JF, Diatta G, Arnathau C, Bitam I, Sarih M, Belghyti D, et al. The epidemiology and geographic distribution of relapsing fever borreliosis in West and North Africa, with a review of the Ornithodoros erraticus complex (Acari: Ixodida). PLoS One. 2013;8(11):e78473.
Trape JF, Godeluck B, Diatta G, Rogier C, Legros F, Albergel J, et al. The spread of tick-borne borreliosis in West Africa and its relationship to sub-Saharan drought. Am J Trop Med Hyg. 1996;54(3):289–93.
Trape JF, Godeluck B, Diatta G, Rogier C, Legros F, Albergel J, et al. Tick-borne borreliosis in West Africa: recent epidemiological studies. Rocz Akad Med Bialymst. 1996;41(1):136–41.
Dworkin MS, Schwan TG, Anderson DE Jr. Tick-borne relapsing fever in North America. Med Clin North Am. 2002;86(2):417–33, viii–ix.
Barclay AJ, Coulter JB. Tick-borne relapsing fever in central Tanzania. Trans R Soc Trop Med Hyg. 1990;84(6):852–6.
Dupont HT, La Scola B, Williams R, Raoult D. A focus of tick-borne relapsing fever in southern Zaire. Clin Infect Dis. 1997;25:139–44.
Bermúdez SE, Armstrong BA, Domínguez L, Krishnavajhala A, Kneubehl AR, Gunter SM, et al. Isolation and genetic characterization of a relapsing fever spirochete isolated from Ornithodoros puertoricensis collected in central Panama. PLoS Negl Trop Dis. 2021;15(8):e0009642.
Schwan TG, Battisti JM, Porcella SF, Raffel SJ, Schrumpf ME, Fischer ER, et al. Glycerol-3-phosphate acquisition in spirochetes: distribution and biological activity of glycerophosphodiester phosphodiesterase (GlpQ) among Borrelia spirochetes. J Bacteriol. 2003;185:1346–56.
Kurashige S, Bissett M, Oshiro L. Characterization of a tick isolate of Borrelia burgdorferi that possesses a major low-molecular-weight surface protein. J Clin Microbiol. 1990;28:1362–6.
Lane RS, Burgdorfer W, Hayes SF, Barbour AG. Isolation of a spirochete from the soft tick, Ornithodoros coriaceus: a possible agent of epizootic bovine abortion. Science. 1985;230:85–7.
Cadavid D, Thomas DD, Crawley R, Barbour AG. Variability of a bacterial surface protein and disease expression in a possible mouse model of systemic Lyme borreliosis. J Exp Med. 1994;179:631–42.
Bissett JD, Ledet S, Krishnavajhala A, Armstrong BA, Klioueva A, Sexton C, et al. Detection of tickborne relapsing fever spirochete, Austin, Texas, USA. Emerg Infect Dis. 2018;24(11):2003–9.
Muñoz-Leal S, Faccini-Martínez ÁA, Costa FB, Marcili A, Mesquita E, Marques EP Jr, et al. Isolation and molecular characterization of a relapsing fever Borrelia recovered from Ornithodoros rudis in Brazil. Ticks Tick Borne Dis. 2018;9(4):864–71.
Schwan TG, Schrumpf ME, Hinnebusch BJ, Anderson DE, Konkel ME. GlpQ: an antigen for serological discrimination between relapsing fever and Lyme borreliosis. J Clin Microbiol. 1996;34:2483–92.
Dworkin MS, Schwan TG, Anderson DE. Tick-borne relapsing fever in North America. Med Clin North Am. 2002;86:417–33.
McNeil E, Hinshaw WR, Kissling RE. A study of Borrelia anserina infection (spirochetosis) in turkeys. J Bacteriol. 1949;57(2):191–206.
Chaconas G, Kobryn K. Structure, function, and evolution of linear replicons in Borrelia. Annu Rev Microbiol. 2010;64:185–202.
Casjens S, Palmer N, van Vugt R, Huang WM, Stevenson B, Rosa P, et al. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol Microbiol. 2000;35:490–516.
Casjens SR, Mongodin EF, Qiu WG, Luft BJ, Schutzer SE, Gilcrease EB, et al. Genome stability of Lyme disease spirochetes: comparative genomics of Borrelia burgdorferi plasmids. PLoS One. 2012;7(3):e33280.
Casjens SR, Di L, Akther S, Mongodin EF, Luft BJ, Schutzer SE, et al. Primordial origin and diversification of plasmids in Lyme disease agent bacteria. BMC Genomics. 2018;19(1):218.
Elbir H, Sitlani P, Bergström S, Barbour AG. Chromosome and megaplasmid sequences of Borrelia anserina (Sakharoff 1891), the agent of avian spirochetosis and type species of the genus. Genome Announc. 2017;5(11):e00018-17.
Margos G, Hepner S, Mang C, Marosevic D, Reynolds SE, Krebs S, et al. Lost in plasmids: next generation sequencing and the complex genome of the tick-borne pathogen Borrelia burgdorferi. BMC Genomics. 2017;18(1):422.
Kuleshov KV, Margos G, Fingerle V, Koetsveld J, Goptar IA, Markelov ML, et al. Whole genome sequencing of Borrelia miyamotoi isolate Izh-4: reference for a complex bacterial genome. BMC Genomics. 2020;21(1):16.
Kingry LC, Replogle A, Batra D, Rowe LA, Sexton C, Dolan M, et al. Toward a complete North American Borrelia miyamotoi genome. Genome Announc. 2017;5(5):e01557-16.
Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, et al. Genomic sequence of a Lyme disease spirochaete Borrelia burgdorferi. Nature. 1997;390(6660):580–6.
Elbir H, Abi-Rached L, Pontarotti P, Yoosuf N, Drancourt M. African relapsing fever borreliae genomospecies revealed by comparative genomics. Front Public Health. 2014;2:43.
Lopez JE, Krishnavahjala A, Garcia MN, Bermudez SE. Tick-borne relapsing fever spirochetes in the Americas. Veterinary sciences. 2016;3(16):1–18.
LeFebvre RB, Perng GC. Genetic and antigenic characterization of Borrelia coriaceae, putative agent of epizootic bovine abortion. J Clin Microbiol. 1989;27:389–93.
Johnson RC, Burgdorfer W, Lane RS, Barbour AG, Hayes SF. Hyde FW. Borrelia coriaceae sp. nov.: putative agent of epizootic bovine abortion. Int J Syst Bacteriol. 1987;37:72–4.
DaMassa AJ, Adler HE. Avian spirochetosis: natural transmission by Argas (Persicargas) sanchezi (Ixodoidea: Argasidae) and existence of different serologic and immunologic types of Borrelia anserina in the United States. Am J Vet Res. 1979;40(1):154–7.
Porcella SF, Raffel SJ, Anderson DE Jr, Gilk SD, Bono JL, Schrumpf ME, et al. Variable tick protein in two genomic groups of the relapsing fever spirochete Borrelia hermsii in western North America. Infect Immun. 2005;73:6647–58.
Schwan TG, Raffel SJ, Schrumpf ME, Policastro PF, Rawlings JA, Lane RS, et al. Phylogenetic analysis of the spirochetes Borrelia parkeri and Borrelia turicatae and the potential for tick-borne relapsing fever in Florida. J Clin Microbiol. 2005;43:3851–9.
Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 2005;102(7):2567–72.
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(Pt 1):81–91.
Jain C, Rodriguez RL, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9(1):5114.
Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21(1):180.
Duchêne DA, Mather N, Van Der Wal C, Ho SYW. Excluding loci with substitution saturation improves inferences from phylogenomic data. Syst Biol. 2022;71(3):676–89. https://doi.org/10.1093/sysbio/syab075.
Binetruy F, Garnier S, Boulanger N, Talagrand-Reboul É, Loire E, Faivre B, et al. A novel Borrelia species, intermediate between Lyme disease and relapsing fever groups, in neotropical passerine-associated ticks. Sci Rep. 2020;10(1):10596.
Gofton AW, Margos G, Fingerle V, Hepner S, Loh SM, Ryan U, et al. Genome-wide analysis of Borrelia turcica and “Candidatus Borrelia tachyglossi” shows relapsing fever-like genomes with unique genomic links to Lyme disease Borrelia. Infect Genet Evol. 2018;66:72–81.
Kumar S, Filipski AJ, Battistuzzi FU, Kosakovsky Pond SL, Tamura K. Statistics and truth in phylogenomics. Mol Biol Evol. 2012;29(2):457–72.
Zhang C, Rabiee M, Sayyari E, Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19(Suppl 6):153.
Allman ES, Degnan JH, Rhodes JA. Identifying the rooted species tree from the distribution of unrooted gene trees under the coalescent. J Math Biol. 2011;62(6):833–62.
Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5(6):e11147.
Barbour AG, Tessier SL, Stoenner HG. Variable major proteins of Borrelia hermsii. J Exp Med. 1982;156:1312–24.
Pennington PM, Cadavid D, Barbour AG. Characterization of VspB of Borrelia turicatae, a major outer membrane protein expressed in blood and tissues of mice. Infect Immun. 1999;67(9):4637–45.
Pennington PM, Cadavid D, Bunikis J, Norris SJ, Barbour AG. Extensive interplasmidic duplications change the virulence phenotype of the relapsing fever agent Borrelia turicatae. Mol Microbiol. 1999;34(5):1120–32.
Mehra R, Londoño D, Sondey M, Lawson C, Cadavid D. Structure-function investigation of vsp serotypes of the spirochete Borrelia hermsii. PLoS One. 2009;4(10):e7597.
Chaconas G, Norris SJ. Peaceful coexistence amongst Borrelia plasmids: getting by with a little help from their friends? Plasmid. 2013;70(2):161–7.
Eggers CH, Caimano MJ, Clawson ML, Miller WG, Samuels DS, Radolf JD. Identification of loci critical for replication and compatibility of a Borrelia burgdorferi cp32 plasmid and use of a cp32-based shuttle vector for the expression of fluorescent reporters in the Lyme disease spirochaete. Mol Microbiol. 2002;43(2):281–95.
Stewart PE, Chaconas G, Rosa P. Conservation of plasmid maintenance functions between linear and circular plasmids in Borrelia burgdorferi. J Bacteriol. 2003;185(10):3202–9.
Hinnebusch BJ, Barbour AG, Restrepo BI, Schwan TG. Population structure of the relapsing fever spirochete Borrelia hermsii as indicated by polymorphism of two multigene families that encode immunogenic outer surface lipoproteins. Infect Immun. 1998;66:432–40.
Restrepo BI, Kitten T, Carter CJ, Infante D, Barbour AG. Subtelomeric expression regions of Borrelia hermsii linear plasmids are highly polymorphic. Mol Microbiol. 1992;6:3299–311.
Casjens SR, Gilcrease EB, Vujadinovic M, Mongodin EF, Luft BJ, Schutzer SE, et al. Plasmid diversity and phylogenetic consistency in the Lyme disease agent Borrelia burgdorferi. BMC Genomics. 2017;18(1):165.
Dai Q, Restrepo BI, Porcella SF, Raffel SJ, Schwan TG, Barbour AG. Antigenic variation by Borrelia hermsii occurs through recombination between extragenic repetitive elements on linear plasmids. Mol Microbiol. 2006;60:1329–43.
Casjens S, Murphy M, DeLange M, Sampson L, van Vugt R, Huang WM. Telomeres of the linear chromosomes of Lyme disease spirochaetes: nucleotide sequence and possible exchange with linear plasmid telomeres. Mol Microbiol. 1997;26(3):581–96.
Margos G, Gatewood AG, Aanensen DM, Hanincová K, Terekhova D, Vollmer SA, et al. MLST of housekeeping genes captures geographic population structure and suggests a European origin of Borrelia burgdorferi. Proc Natl Acad Sci U S A. 2008;105(25):8730–5.
Han HJ, Liu JW, Wen HL, Li ZM, Lei SC, Qin XR, et al. Pathogenic new world relapsing fever borrelia in a Myotis bat, Eastern China, 2015. Emerg Infect Dis. 2020;26(12):3083–5.
Kingry LC, Anacker M, Pritt B, Bjork J, Respicio-Kingry L, Liu G, et al. Surveillance for and discovery of Borrelia species in US patients suspected of tickborne illness. Clin Infect Dis. 2018;66(12):1864–71.
Iwabu-Itoh Y, Bazartseren B, Naranbaatar O, Yondonjamts E, Furuno K, Lee K, et al. Tick surveillance for Borrelia miyamotoi and phylogenetic analysis of isolates in Mongolia and Japan. Ticks Tick Borne Dis. 2017;8(6):850–7.
Li Z-M, Xiao X, Zhou C-M, Liu J-X, Gu X-L, Fang L-Z, et al. Human-pathogenic relapsing fever Borrelia found in bats from central China phylogenetically clustered together with relapsing fever borreliae reported in the New World. PLoS Negl Trop Dis. 2021;15(3):e0009113.
Takano A, Toyomane K, Konnai S, Ohashi K, Nakao M, Ito T, et al. Tick surveillance for relapsing fever spirochete Borrelia miyamotoi in Hokkaido, Japan. PloS one. 2014;9(8):e104532.
Gupta RS. Distinction between Borrelia and Borreliella is more robustly supported by molecular and phenotypic characteristics than all other neighbouring prokaryotic genera: response to Margos’ et al. “the genus Borrelia reloaded” (plos one 13(12): e0208432). PloS One. 2019;14(8):e0221397.
Minh BQ, Hahn MW, Lanfear R. New methods to calculate concordance factors for phylogenomic datasets. Mol Biol Evol. 2020;37(9):2727–33.
Davidson R, Vachaspati P, Mirarab S, Warnow T. Phylogenomic species tree estimation in the presence of incomplete lineage sorting and horizontal gene transfer. BMC Genomics. 2015;16 Suppl 10(Suppl 10):S1.
Sayyari E, Mirarab S. Fast coalescent-based computation of local branch support from quartet frequencies. Mol Biol Evol. 2016;33(7):1654–68.
Maddison WP. Gene trees in species trees. Syst Biol. 1997;46(3):523–36.
Degnan JH, Rosenberg NA. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol Evol. 2009;24(6):332–40.
Mongodin EF, Casjens SR, Bruno JF, Xu Y, Drabek EF, Riley DR, et al. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics. 2013;14:693.
Yang XF, Hübner A, Popova TG, Hagman KE, Norgard MV. Regulation of expression of the paralogous Mlp family in Borrelia burgdorferi. Infect Immun. 2003;71(9):5012–20.
Brisson D, Zhou W, Jutras BL, Casjens S, Stevenson B. Distribution of cp32 prophages among Lyme disease-causing spirochetes and natural diversity of their lipoprotein-encoding erp loci. Appl Environ Microbiol. 2013;79(13):4115–28.
Eggers CH, Samuels DS. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J Bacteriol. 1999;181(23):7308–13.
Zhang H, Marconi RT. Demonstration of cotranscription and 1-methyl-3-nitroso-nitroguanidine induction of a 30-gene operon of Borrelia burgdorferi: evidence that the 32-kilobase circular plasmids are prophages. J Bacteriol. 2005;187(23):7985–95.
Eggers CH, Samuels DS. Molecular evidence for a new bacteriophage of Borrelia burgdorferi. J Bacteriol. 1999;181:7308–13.
Eggers CH, Gray CM, Preisig AM, Glenn DM, Pereira J, Ayers RW, et al. Phage-mediated horizontal gene transfer of both prophage and heterologous DNA by ϕBB-1, a bacteriophage of Borrelia burgdorferi. Pathog Dis. 2016;74(9):ftw107.
Eggers CH, Kimmel BJ, Bono JL, Elias AF, Rosa P, Samuels DS. Transduction by phiBB-1, a bacteriophage of Borrelia burgdorferi. J Bacteriol. 2001;183(16):4771–8.
Adeolu M, Gupta RS. A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie Leeuwenhoek. 2014;105(6):1049–72.
Vial L, Durand P, Arnathau C, Halos L, Diatta G, Trape J-F, et al. Molecular divergences of the Ornithodoros sonrai soft tick species, a vector of human relapsing fever in West Africa. Microbes Infect. 2006;8(11):2605–11.
Baltazard M, Bahmanyar M, Mofidi C. Ornithodoros erraticus et fièvres récurrentes. Bull Soc Pathol Exot. 1950;43:595–601.
Burgdorfer W. Analyse des Infektionsverlaufes bei Ornithodorus moubata (Murray) und der naturlichen Uebertragung von Spirochaeta duttoni. Acta Trop. 1951;8:194–262.
Morel PC. . Les tiques d’Afrique et du Bassin Méditerranéen. Paris: Bulletin de l'Académie Vétérinaire de France; 1968.
Mackie FP. The part played by Pediculus corporis in the transmission of relapsing fever. BMJ. 1907;2:1706–9.
Davis GE. Species unity or plurality of the relapsing fever spirochetes. In: Moulton FR, editor. A symposium of relapsing fever in the Americas. Washington: American Association for the Advancement of Science; 1942. p. 41–7.
Brumpt E. Étude du Spirochaeta turicatae, n. sp., agent de la fièvre récurrente sporadique des Etats-Unis trasmise par Ornithodoros turicata. CR Soc Biol, Paris. 1933;113:1369.
Skrynnik A. The specificity of ticks of Ornithodoros species as vectors of the causal agents of tick-borne spirochaetosis. Parazitologiya. 1968;2(1):3–9.
Schwan TG. Vector specificity of the relapsing fever spirochete Borrelia hermsii (Spirochaetales: Borreliaceae) for the tick Ornithodoros hermsi (Acari: Argasidae)involves persistent infection of the salivary glands. J Med Entomol. 2021;58(4):1926–30.
Guzmán-Cornejo C, Herrera-Mares A, Robbins RG, Rebollo-Hernández A. The soft ticks (Parasitiformes: Ixodida: Argasidae) of Mexico: species, hosts, and geographical distribution. Zootaxa. 2019;4623(3):zootaxa.4623.3.3.
Fairchild GB, Kohls GM, Tipton VJ. The ticks of panama (Acarina: Ixodoidea): Field Museum of Natural History. 1966.
Brumpt E, Brumpt LC. Identite du spirochete des fievres recurrentes a tiques des plateaux mexicains et du Spirochaeta turicatae agent de la fievre recurrente sporadique des Etats-Unis. Ann Parasitol Hum Compar. 1939;17:287–98.
Vazquez-Guerrero E, Adan-Bante NP, Mercado-Uribe MC, Hernandez-Rodriguez C, Villa-Tanaca L, Lopez JE, et al. Case report: A retrospective serological analysis indicating human exposure to tick-borne relapsing fever spirochetes in Sonora, Mexico. PLoS Negl Trop Dis. 2019;13(4):e0007215.
Colunga-Salas P, Sánchez-Montes S, Volkow P, Ruíz-Remigio A, Becker I. Lyme disease and relapsing fever in Mexico: an overview of human and wildlife infections. PloS One. 2020;15(9):e0238496.
Darling ST. The relapsing fever of Panama. Arch of Intern Med. 1909;4:150–85.
Bates LB, Dunn LH, St. John JH. Relapsing fever in Panama: The human tick, Ornithodoros talaje, demonstrated to be the transmitting agent of relapsing fever in Panama by human experimentation. Am J Trop Med Hyg. 1921;1(4):183–210.
Dunn LH, Clark HC. Notes on relapsing fever in Panama with special reference to animal hosts. Am J Trop Med Hyg. 1933;13(2):201–9.
Davis GE. Observations on the biology of the argasid tick, Ornithodoros brasiliensis Aragao, 1923; with the recovery of a spirochete, Borrelia brasiliensis, n. sp. J Parasitol. 1952;38(5):473–6.
Dunn LH. Studies on the South American tick, Ornithodoros venezuelensis Brumpt, in Colombia. Its prevalence, distribution, and importance as an intermediate host of relapsing fever. J Parasitol. 1927;13(4):249–55.
Ciceroni L, Bartoloni A, Guglielmetti P, Paradisi F, Barahona HG, Roselli M, et al. Prevalence of antibodies to Borrelia burgdorferi, Borrelia parkeri and Borrelia turicatae in human settlements of the Cordillera Province, Bolivia. J Trop Med Hyg. 1994;97(1):13–7.
Parola P, Ryelandt J, Mangold AJ, Mediannikov O, Guglielmone AA, Raoult D. Relapsing fever Borrelia in Ornithodoros ticks from Bolivia. Ann Trop Med Parasitol. 2011;105(5):407–11.
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–36.
Wick RR, Holt KE. Benchmarking of long-read assemblers for prokaryote whole genome sequencing. F1000Res. 2019;8:2138.
Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. 2016;32(22):3380–7.
Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020;21(1):245.
Mantovani E, Costa IP, Gauditano G, Bonoldi VL, Higuchi ML, Yoshinari NH. Description of Lyme disease-like syndrome in Brazil. Is it a new tick borne disease or Lyme disease variation? Braz J Med Biol Res. 2007;40(4):443–56.
Yoshinari NH, Oyafuso LK, Monteiro FG, de Barros PJ, da Cruz FC, Ferreira LG, Lyme disease, et al. Report of a case observed in Brazil. Rev Hosp Clin Fac Med Sao Paulo. 1993;48(4):170–4.
Campbell SB, Klioueva A, Taylor J, Nelson C, Tomasi S, Replogle A, et al. Evaluating the risk of tick-borne relapsing fever among occupational cavers-Austin, TX, 2017. Zoonoses Public Health. 2019;66(6):579–86.
Replogle AJ, Sexton C, Young J, Kingry LC, Schriefer ME, Dolan M, et al. Isolation of Borrelia miyamotoi and other Borreliae using a modified BSK medium. Sci Rep. 2021;11(1):1926.
Lopez JE, Schrumpf ME, Raffel SJ, Policastro PF, Porcella SF, Schwan TG. Relapsing fever spirochetes retain infectivity after prolonged in vitro cultivation. Vector Borne Zoonotic Dis. 2008;8(6):813–20.
De Coster W, D’Hert S, Schultz DT, Cruts M, Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34(15):2666–9.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Fass J, LeClari R, Seeman T. A perfect circle (apc) [Available from: https://github.com/jfass/apc.
Seibt KM, Schmidt T, Heitkam T. FlexiDot: highly customizable, ambiguity-aware dotplots for visual sequence analyses. Bioinformatics. 2018;34(20):3575–7.
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421.
Okonechnikov K, Golosova O, Fursov M. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–7.
Vaser R, Sović I, Nagarajan N, Šikić M. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res. 2017;27(5):737–46.
medaka: Sequence correction provided by ONT Research [Available from: https://github.com/nanoporetech/medaka.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5.
Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34(18):3094–100.
Pedersen BS, Quinlan AR. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34(5):867–8.
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77.
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24.
Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–40.
Pertea G, Pertea M. GFF utilities: GffRead and GffCompare. Res. 2020;9:ISCB Comm J-304.
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, et al. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–8.
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47(D1):D427–32.
Pandurangan AP, Stahlhacke J, Oates ME, Smithers B, Gough J. The SUPERFAMILY 2.0 database: a significant proteome update and a new webserver. Nucleic Acids Res. 2019;47(D1):D490-d4.
Miller SC, Porcella SF, Raffel SJ, Schwan TG, Barbour AG. Large linear plasmids of Borrelia species that cause relapsing fever. J Bacteriol. 2013;195(16):3629–39.
Wilder HK, Raffel SJ, Barbour AG, Porcella SF, Sturdevant DE, Vaisvil B, et al. Transcriptional profiling the 150 kb linear megaplasmid of Borrelia turicatae suggests a role in vector colonization and initiating mammalian infection. PLoS One. 2016;11(2):e0147707.
Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3.
Zhao Y, Jia X, Yang J, Ling Y, Zhang Z, Yu J, et al. PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics. 2014;30(9):1297–9.
Duchêne DA, Duchêne S, Ho SY. PhyloMAd: efficient assessment of phylogenomic model adequacy. Bioinformatics. 2018;34(13):2300–1.
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35(2):518–22.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–4.
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9.
Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 2016;65(6):997–1008.
Rabiee M, Sayyari E, Mirarab S. Multi-allele species reconstruction using ASTRAL. Mol Phylogenet Evol. 2019;130:286–96.
Yin J, Zhang C, Mirarab S. ASTRAL-MP: scaling ASTRAL to very large datasets using randomization and parallelization. Bioinformatics. 2019;35(20):3961–9.
Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018;14(1):e1005944.
Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–2.
Galili T. JA, Pilosov M. gplots [Available from https://github.com/talgalili/gplots]. 2020.
Larsson J. eulerr: area-proportional Euler and Venn diagrams with ellipses [Available from : https://cran.r-project.org/package=eulerr]. R package. 2020;version 6.1.0.
Acknowledgements
The authors would like to thank Konstanin Kuleshov for his help with the borrelial genome assembly pipeline and Tom G. Schwan for the B. anserina BA2, B. hermsii DAH and YOR, B. coriaceae Co53, B. parkeri SLO, and B. turicatae 91E135 isolates.
Funding
These studies were supported by AI137412-01 (JEL), AI148219-01 (JEL), AI123652-01 (JEL), FAPESP #2018 /02521–1 (SML) and FAPESP #2019/17960–3 (SML).
Author information
Authors and Affiliations
Contributions
A.R.K conceived the idea, performed the sequencing, data analysis, prepared all figures, and wrote the main manuscript text. A.K. helped perform experiments. S.M.L, A.J.R, L.C.K., S.E.B., M.B.L. provided reagents, and critiqued the manuscript. J.E.L conceived the idea, obtained funding, and critiqued and edited the manuscript. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 3.
PhyloMAd saturation test output.
Additional file 4.
CD-HIT-EST nucleotide sequence identity cluster output of PF32 gene loci.
Additional file 5.
Example commands.
Additional file 6.
Panaroo presence/absence output.
Additional file 7.
Plasmid Partitioning Gene Loci Interproscan Information.
Rights and permissions
This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kneubehl, A.R., Krishnavajhala, A., Leal, S.M. et al. Comparative genomics of the Western Hemisphere soft tick-borne relapsing fever borreliae highlights extensive plasmid diversity. BMC Genomics 23, 410 (2022). https://doi.org/10.1186/s12864-022-08523-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08523-7