Abstract
Background
Classification of parasitic bopyrids has traditionally been based on morphological characteristics, but phylogenetic relationships have remained elusive due to limited information provided by morphological data and tendency for loss of morphological features as a result of parasitic lifestyle. Subfamily Argeiinae was separated from Bopyrinae based on morphological evidence, although the assignment of all genera has not been phylogenetically evaluated. Bopyroides hippolytes has been traditionally classified in Bopyrinae, but divergent morphological characters make this assignment questionable. To investigate the relationship of bopyrines, we sequenced the complete mitochondrial genome of B. hippolytes and four mitochondrial genes of two other Bopyrinae.
Results
The phylogenetic trees based on separate and combined cox1and 18S sequence data recovered Bopyridae as robustly monophyletic, but Bopyrinae as polyphyletic. Bopyroides hippolytes was a close sister to Argeia pugettensis, type species to Argeiinae. Mitochondrial phylogenomics also suggested that B. hippolytes was close to Argeiinae. We also found a novel gene order in B. hippolytes compared to other isopods.
Conclusions
Bopyroides hippolytes should be excluded from the Bopyrinae and has a close affinity with Argeia pugettensis based on molecular and morphological data. The conserved syntenic blocks of mitochondrial gene order have distinctive characteristics at a subordinal level and may be helpful for understanding the higher taxonomic level relationships of Isopoda.
Similar content being viewed by others
Introduction
Bopyridae Rafinesque, 1815 is a parasitic family of isopod crustaceans, with 10 subfamilies, 208 genera and 634 species (Boyko et al., 2008 onwards) [1]. Bopyrids stunt the growth and reduce the reproductive abilities of their hosts [2]. Molecular phylogenetic research on Bopyridae has been scarce, partly because of the challenge of assembling sequenceable material. Molecular phylogenetic analyses began only recently. Boyko et al. [3] assessed phylogenetic relationships among Bopyridea Rafinesque, 1815 and Cryptoniscoidea Kossmann, 1880 using 18S sequence data, and recovered Bopyridae as monophyletic. Yu et al. [4] and An et al. [5] sequenced and analyzed the mitochondrial genome of the bopyrids Gyge ovalis (Shiino, 1939) and Argeia pugettensis Dana, 1853, respectively, but the paucity of comparative sequence data prevented resolution of bopyrid relationships at the subfamily level.
With some exceptions, members of each bopyrid subfamily are restricted to hosts from one decapod infraorder [6]. Bopyrinae, Argeiinae and Pseudioninae infest the branchial chambers of caridean shrimp. Shiino [7] grouped a series of bopyrid genera, including Argeia Dana, 1853, Stegoalpheon Chopra, 1923, Bopyrosa Nierstrasz & Brender à Brandis, 1923, Parargeia Hansen, 1897 into his Bopyrus-group, later recognized as the Bopyrinae. Markham [8] placed these four genera together with Argeiopsis, into his newly erected subfamily Argeiinae. He noted that Argeiinae can be distinguished from Bopyrinae in that the female of the latter has a head that is not oval or fusiform and is usually fused with the pereon, an oval or deltoid body outline, some or all pleomeres fused at least on one side, lateral plates and uropods that are greatly reduced or absent, and pleopods that are generally biramous.
Bopyroides Stimpson, 1864 is currently placed in the Bopyrinae but does not fit well there. Females have an oval head, distinctly separated from pereon, all pleomeres are distinct without any fusion, have prominent dorsolateral bosses and tergal projections, and pleopods are reduced or uniramous tuberculiform [9,10,11]. Thus, there remains uncertainty about the boundaries between the Bopyrinae and Argeiinae, and the validity of Argeiinae has also been questioned [12].
Bopyroides includes three valid species, all carid parasites in Palearctic waters. Bopyroides hippolytes (Kröyer, 1838) parasitizes several shrimp species in the families Thoridae and Pandalidae in the cold, northern waters of the Pacific, Atlantic, and Arctic Oceans [5, 13], and has an unusually great range among Bopyridae. Kröyer [10] described this species in Bopyrus, infesting Lebbeus olaris (Sabine, 1824) in Greenland. Bate & Westwood [14] moved the species to Gyge, then Sars [15] transferred it to Bopyroides, established by Stimpson [16] for Bopyroides acutimarginatus Stimpson, 1864. Bonnier [17] described Bopyroides sarsi infesting L. olaris from the Arctic Ocean. Richardson [18] regarded B. acutimarginatus and B. sarsi Bonnier, 1900 as synonyms of B. hippolytes. These synonymies and the placement of Bopyroides in Bopyrinae have been accepted by subsequent authors [6, 13, 19,20,21,22,23], even though several characters conflict with that subfamilial assignment as noted above. Scott [9] described Pleuroctypta cluthae infesting Pandalina brevirostris (Rathke, 1843) (Pandalidae) from Clyde Sea (Scotland). Bourdon [21] transferred it to Bopyroides and compared it with B. hippolytes in detail. Rybakov& Avdeev [11] described B. shiinoi from the Russian Pacific, based on a female with five pairs of uniramous pleopods. Boyko [23] suggested that Shiino’s [19] and Kim & Kwon’s [22] specimens of B. hippolytes from Japan and Korea are also referable to B. shiinoi, as all have female with five pairs of uniramous pleopods. Classification history and synonym for the three Bopyroides species are shown in Table 1.
Key to species of Bopyroides:
-
1
Pleomere 6 of female with lateral, biramous extension…………………B. cluthae
-
Pleomere 6 of female not laterally extended, round or truncate……………2
-
-
2
Female with four pairs of uniramous pleopods, head of male with straight posterior margin…B. hippolytes
-
Female with five pairs of uniramous pleopods, head of male with curved posterior margin…B. shiinoi
-
We sequenced the mitogenome and 18S rRNA of three species currently assigned to the Bopyrinae: Bopyroides hippolytes, Bopyrella malensis Bourdon, 1980, and Parabopyrella cf. mortenseni (Bourdon, 1980), and analyzed their phylogenetic placements among bopyrids based on this and other available sequence data. We also compared gene arrangements across all available mitogenomes of the Isopoda and describe a novel mitochondrial gene order in bopyrids.
We addressed the following questions: (1) Is the Bopyrinae monophyletic? (2) What is the phylogenetic position of Bopyroides hippolytes? (3) How has mitochondrial gene rearrangement in the Isopoda?
Results
Sequence alignment and data partitions
Sequence information for the four datasets after alignment and trimming are shown in Table 2. The subset partitions and best-fit models from Partition Finder and Model Finder are presented in Table S1.
Phylogenetic position of B. hippolytes and molecular phylogeny of Bopyridae
The six phylogenetic trees obtained based on separate and combined cox1 and 18S sequence data with ML and BI were congruent (Fig. 1). All analyses recovered most species currently assigned to the Bopyrinae (Probopyrus pandalicola, P. pacificiensis, P. buitendijki, Parabopyrella mortenseni, Bopyrella malensis) as robustly monophyletic, except for Bopyroides hippolytes, which was always a close sister to Argeia pugettensis. Our Bopyroides hippolytes sequence matches others in GenBank, further confirming the identification. The relationship of Argeiinae + Hemiarthrinae had strong support.
Mitochondrial phylogenomics of Isopoda
Phylogenetic analysis based on AA sequences of the 13 PCGs recovered Phreatoicidea, then Asellota, as sister to all other sampled suborders in the Isopoda, albeit with modest support. All suborders except Cymothoida were monophyletic, although the polyphyly of Cymothoida also lacked robust support (Fig. 2). Bopyroides hippolytes was recovered as sister to Argeia (Argeiinae), distant from the other two Bopyrinae species.
Novel mitochondrial gene arrangements
Bopyroides hippolytes has a unique mitochondrial gene order that differs from that of all other known in Isopoda (Fig. 3). The circular mitochondrial genome of B. hippolytes is 15,329 bp, with C + G content of 39.9%. It possesses the standard 13 PCGs and two rRNA genes (12S and 16S), but only 19 tRNA genes, as three tRNA genes (trnK, trnW and trnI) are missing compared to the standard animal mitogenome (Fig. 3). Nineteen genes are encoded on the +strand (light strand), whereas 4 protein-coding genes, 10 tRNAs and one rRNAs are located on the -strand (heavy strand). In base composition, B. hippolytes exhibits negative AT skew (− 0.129) and positive GC skew (0.039).
Comparison of gene arrangement of the complete mitogenomes for Isopoda mapped on a simplified isopod tree inferred from CAT-GTR. The most conserved syntenic blocks at the suborder level are marked with a blue background box. Gene duplications and deletions are marked with red triangles and white rectangles, respectively. The novel gene order in B. hippolytes is marked with the star. CR: control region
Isopods have undergone numerous gene duplication and deletion events in the mitogenome among taxa sampled to date, except for the basal Phreatoicidea, which retains a standard mitochondrial genome of 37 genes. While only tRNA genes were lost or duplicated in most isopod clades, PCGs were also duplicated in Argeia pugettensis. Expect Limnoria quadripunctata, the putative CR (control region) of all available isopod mitogenomes is located between the rrnS (+strand) and cob (−strand) genes.
Discussion
Phylogenetic relationships and taxonomic implications
Markham [8] separated the Argeiinae from the Bopyrinae based on several morphological features, including the shape of the head and pleopods, but considered these subfamilies to be closely related. Bopyroides hippolytes was originally described in Bopyrus, the type genus of Bopyrinae (Kröyer, 1838). While this widespread and common species has received substantial attention, and was transferred to its own genus, its subfamilial classification has not been discussed and remained accepted as Bopyrinae [6, 11, 13, 17,18,19,20, 22, 23].
The Bopyrinae was polyphyletic in all our phylogenetic analyses, with Bopyroides hippolytes separating from other bopyrine genera, but close to Argeia pugettensis. We suggest that Bopyroides hippolytes should be excluded from Bopyrinae and has a close affinity with Argeia pugettensis. This conclusion is also supported by some morphological data, such as the dorsolateral boss and tergal projection of Bopyroides hippolytes, described by Shiino [19], Allen [20], and Rybakov and Avdeev [11] that are characteristics of Argeiinae, not Bopyrinae. The two other species assigned to Bopyroides, B. cluthae and B. shiinoi, also have distinct pleomeres, prominent dorsolateral bosses and tergal projections [9, 11, 20] and likely are indeed congeneric. So, the boundary between Bopyrinae and Argeiinae is obscure, and the correct rank of Bopyroides need more data.
Our phylogenetic results are congruent with Boyko et al’s [3] analysis of bopyrids using 18S sequence data but extends it with greater sampling of Bopyrinae and Argeiinae. These two subfamilies are well separated on the phylogeny suggesting that their similarities perceived by Markham [24] are the result of convergence. There remain limited sequence data for this large family of parasitic isopods and further work will likely lead to additional changes in their classification.
Gene rearrangement
Mitochondrial gene rearrangement provides useful information for understanding relationships at higher taxonomic levels [25, 26]. Duplication and deletion of tRNA are common in the rearrangement of mitochondrial genes in metazoans [27,28,29], likely caused by the replication slippage mechanisms [30, 31].
Most of the available isopod mitochondrial genomes in GenBank are partial or incomplete, with only 11 mitogenomes being complete (as of 30 Dec. 2020). We evaluated gene rearrangements by assembling the 12 available complete isopod mitogenomes, including Bopyroides hippolytes sequenced in this study. Among all complete isopod mitogenomes analyzed, tRNA deletions occur in all species except Limnoriidea and Phreatoicidea. These suborders are early cladogenesises among isopods and appear to retain the ancestral complement of tRNAs. Two general changes in the tRNA genes of all isopods include a reduction or loss of trnI and reduction of trnC. trnI was missing in both Epicaridea and Cymothoida (Fig. 3), and while it was retained in the other suborders, its cloverleaf structure was incomplete (loss of D-loop or TΨC). The D-loop region was lacking in the predicted secondary structures of trnC for all mitogenome analyzed. This phenomenon was also described by Kilpert and Podsiadlowski [25], who considered that this feature might be a putative autapomorphy of Isopoda. It is noticeable that an unusual deletion of trnK appears in B. hippolytes, reported for the first time in isopods. The area between trnR and trnH was considered a ‘hot spot’ of mitochondrial gene rearrangement in Isopoda [32, 33]. As the most parsimonious explanation of gene order change in this region, Kilpert and Podsiadlowski [25] assumed multiple translocation events. Because of the mixture of inversions and genome shuffling, tandem-duplication/random loss models were not a better way to explain the gene rearrangement [25, 34]. Crustacean taxa usually exhibit negative overall GC skews and positive AT skews on the heavy strand, but many studies have found that the bias for this strand is inversed in isopod mitogenomes [32,33,34,35]. B. hippolytes mitogenome in this study also exhibits negative AT skews and positive GC skews. It is considered to be the result of the architectural hypervariability and frequent inversions of the origin of mitochondrial replication (ORI) located in the control region (CR), where the changed replication order of two mitochondrial DNA strands consequently resulted in an inversed strand asymmetry [33, 35].
Comparison of mitochondrial gene order across Isopoda shows relative stability within suborders, but substantial differences among suborders (Fig. 4). In Epicaridea, we speculated three small conserved syntenic blocks of the mitogenomes. Both deletion and duplication of genes occurred in this suborder, particularly Argeia pugettensis. Whereas in Cymothoida and Oniscidea, we supposed two large conserved blocks. Comparison of conserved regions of different suborders can indicate that mitogenomes gene order is an especially useful tool for higher taxonomy of isopod species. Sequencing of additional isopod mitogenomes promises to be a fertile ground for research.
Conclusions
Our phylogenetic analyses based on cox1, 18S sequence and mitochondrial genome suggest that Bopyrinae is polyphyletic and Bopyroides hippolytes should be excluded from the Bopyrinae. We found a novel gene order in B. hippolytes compared to other isopods. The comparison of mitochondrial gene order shows that conserved syntenic blocks have distinctive characteristic at a subordinal level, and may be helpful for understanding the higher taxonomic level relationships of Isopoda.
Materials and methods
Taxon sampling
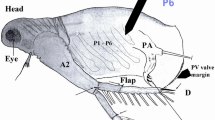
Bopyroides hippolytes (Fig. 4) (UF Arthropoda 45,428), infesting Pandalopsis dispar Rathbun, 1902 (UF Arthropoda 45,427), from the USA, Washington State, San Juan Islands, San Juan Channel, 80-120 m (48.578° N, 123.048° W, 17 Oct, 2015), collected by Gustav Paulay.
Parabopyrella cf. mortenseni (UF Arthropoda 44,587), infesting Lysmata sp. (UF Arthropoda 46,079), from Panama, Bocas del Toro Province, Cayo Hermanas, 3-3.5 m (9.268°, − 82.352°, 30 May 2016), collected by Matthieu Leray, Francois Michonneau and Robert Lasley. This specimen matches Parabopyrella mortenseni, described from Djibouti morphologically, but is unlikely to be conspecific given the great geographic separation.
Specimen of Bopyrella malensis (UF Arthropoda 46,906), infesting Synalpheus sp. (UF Arthropoda 46,905), from New Caledonia, Province Sud, Noumea lagoon, IIot St Marie, 1-10 m. (− 22.309°, 166.484°, 16 Nov, 2017), collected by Gustav Paulay, Daisuke Uyeno and Leonid Moroz.
Voucher specimens are deposited in the Florida Museum, University of Florida (UF).
Available 18S and cox1 sequences of bopyrids were obtained from GenBank (Tables S2, S3).
DNA extraction, amplification, sequencing, and annotation
Total genomic DNA was extracted from eggs or the pereon of female specimens using the genomic DNA rapid extraction kit (Aidlab Biotechnologies Co., Ltd) according to the manufacturer’s instructions. 18S rRNA gene region was amplified and sequenced using the same primers from Boyko et al. [3]. PCR conditions were: denaturation at 98 °C for 2 min, 40 cycles of 98 °C for 10 s, 50 °C for 15 s, and 68 °C for 1 min, and a final extension of 72 °C for 10 min. The complete mitochondrial genome was amplified by PCR using 12 pairs of primers (Table S5). The mitochondrial genome was sequenced and annotated following our previous study [4].
The complete mitogenome was obtained from Bopyroides hippolytes (GenBank accession number: MK905237). Because of the low quality of DNA extraction, only partial mitochondrial genomes were obtained from Parabopyrella mortenseni and Bopyrella malensis.
Gene arrangement comparisons
PhyloSuite [35] was used to batch-download the 11 complete isopod mitochondrial genomes available from GenBank, and to assess genomic features and gene order. Phylograms and gene orders were visualized in iTOL [36]. For the purposes of visualization, we arbitrarily designated the beginning of the cox1 gene as position 1 in each genome (pointing in the direction of cox2).
Phylogenetic analyses
Most of the available sequence data for Bopyridae are mitochondrial genes and 18S rRNA. We constructed four datasets to assess phylogenetic relationships of bopyrids (Tables S2, S3, S4): (1) cox1 dataset; (2) 18 s rRNA dataset; (3) combined cox1 + 18 s rRNA dataset; (4) mitogenome dataset. Nucleotide sequences were used in the first three datasets and amino acid sequences were used in the mitogenome analyses. Missing sequences were treated as missing data. MAFFT [37, 38] was used to align sequences: nucleotide and amino acid sequences were aligned in batches (using codon and normal-alignment modes, respectively) with “–auto” strategy, whereas 18S rRNA gene was aligned using Q-INS-i algorithm, which takes secondary structure information into account. We used Gblocks v0.91b (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) [39] to eliminate the ambiguous sequences after alignment, as they impact phylogenetic analyses [40]. Parameters were set as follows: type of sequence was set to codons for PCGs alignments; the minimum length of a block was set to 3 bp for PCGs and 2 bp for rRNA genes, and gap positions (−b5) were allowed with half. DNAsp v5 [41] and MEGA 5.03 [42] were used to calculate sequence composition and variability.
We tested the performance of homogeneous substitution models (using Maximum Likelihood (ML), and Bayesian Inference (BI)) for the single-gene dataset and COI-18S dataset. Data were partitioned by gene and codon position for the BI and ML analyses. PartitionFinder ver. 2.1.1 [43] was used to select partition schemes for BI, and ModelFinder [44] was used to select these for ML analysis in IQ-TREE, using the corrected Akaike Information Criterion (AICc). MrBayes ver. 3.2.6 [45] was used for BI, with four simultaneous runs with four chains each run for 10 million generations, sampling every 1000 trees. The first 25% of these trees were discarded as burn-in when computing the consensus tree (50% majority rule). Sufficient mixing of the chains was considered to be reached when the average standard deviation of split frequencies was below 0.01. ML analyses were conducted in IQ-TREE [46] with 1000 ultrafast BS [47].
Bayesian Inference with the CAT-GTR model based on mitogenome amino acid (AA) data outperform partitioned homogeneous models in isopods [35]. Consequently, we tested the performance of heterogeneous CAT-GTR model for mitogenome dataset, using AA sequence of 13 protein-coding genes (13PCGs). The CAT-GTR inference was implemented in PhyloBayes-MPI 1.7a on the beta version of the Cipres server [48], with default parameters (burnin = 500, invariable sites automatically removed from the alignment, two MCMC chains), and the analysis was stopped when the conditions indicated that a good run was reached (PhyloBayes manual: maxdiff < 0.1 and minimum effective size > 300).
Availability of data and materials
All datasets are stored in NCBI. The accessions of the datasets are MW535162, MW535163, MK905237, MW540887, MW540885, MW540884. The analyzed data during this study are included in this published article and its supplementary information files.
References
Boyko CB, Bruce NL, Hadfield KA, Merrin KL, Ota Y, Poore GCB, et al. (Eds) (2008 onwards). World Marine, Freshwater and Terrestrial Isopod Crustaceans database. Bopyridae Rafinesque, 1815. Accessed through: World Register of Marine Species at: http://marinespecies.org/aphia.php/aphia.php?p=taxdetails&id=1195 on 2021-06-04.
Somers I, Kirkwood G. Population ecology of the grooved tiger prawn, Penaeus semisulcatus, in the North-Western Gulf of Carpentaria, Australia: growth, movement, age structure and infestation by the bopyrid parasite Epipenaeon ingens. Mar Freshw Res. 1991;42:349–67.
Boyko CB, Moss J, Williams JD, Shields JD. A molecular phylogeny of Bopyroidea and Cryptoniscoidea (Crustacea: Isopoda). Syst Biodivers. 2013;11:495–506.
Yu J, An J, Li Y, Boyko CB. The first complete mitochondrial genome of a parasitic isopod supports Epicaridea Latreille, 1825 as a suborder and reveals the less conservative genome of isopods. Syst Parasitol. 2018;95:465–78.
An J, Zheng W, Liang J, Xi Q, Chen R, Jia J, et al. Disrupted architecture and fast evolution of the mitochondrial genome of Argeia pugettensis (Isopoda): implications for speciation and fitness. BMC Genomics. 2020;21:1–14.
Markham JC. A review of the bopyrid isopods infesting caridean shrimps in the northwestern Atlantic Ocean, with special reference to those collected during the hourglass cruises in the Gulf of Mexico. Mem Hourglass Cruises. 1985;7(3):1–156.
Shiino SM. Phylogeny of the genera within the family Bopyridae. Bull Muséum Natl d’Histoire Nat Paris. 1965;37(3):462–5.
Markham JC. Description of a new western Atlantic species of Argeia Dana with a proposed new subfamily for this and related genera (Crustacea, Isopoda, Bopyridae). Zool Meded. 1977;52:107–23.
Scott T. Notes on Scottish Crustacea. Ann Mag Nat Hist. 1902;7(10):1–5.
Kröyer H. GrönlandsAmfipoder. Det Konigelige Danske Videnskabernes Selskabs Naturvidenskabelige og Mathematiske Afhandlinger, series 4, vol. 7; 1838. p. 229–326.
Rybakov A, Avdeev V. Two Bopyroides species (Isopoda, Bopyridae) from the far-eastern shrimps. Parazitologiâ. 1991;25:167–72.
Adkison DL, Heard RW, Clark GT. Description of the male and notes on the female of Argeiopsis inhacae (Crustacea: Isopoda: Bopyridae). Proc Biol Soc Wash. 1982;95:334–7.
An J, Boyko CB, Li X. A review of bopyrids (Crustacea: Isopoda: Bopyridae) parasitic on caridean shrimps (Crustacea: Decapoda: Caridea) from China. Bull Am Mus Nat Hist. 2015;399:1–85.
Bate CS, Westwood JO. A History of the British Sessile-eyed Crustacea. London: John van Voorst; 1868. p. lvi + 536.
Sars OG. An account of the crustacea of Norway with short descriptions and figures of all species. Bergen: Isopoda, Bergen Museum. ALB. Cammermeyers Forlag, Christiania; 1899.
Stimpson W. Descriptions of new species of marine invertebrates from Puget Sound, collected by the naturalists of the north-west boundary commission, AH Campbell, Esq., commissioner. In: Proceedings of the Academy of Natural Sciences of Philadelphia; 1864. p. 153–61.
Bonnier J. Contribution à l’étude des épicarides. Les Bopyridae. Travaux de l'lnstitut Zoologique de Lille et du Laboratoire de Zoologie Maritime de Wimereux, vol. 8; 1900. p. 1–478.
Richardson H. Isopods from the Alaska salmon investigation. Bulletin of the U.S. bureau of. Fisheries. 1905;24:211–21.
Shiino SM. Some additions to the bopyrid fauna of Japan. Annotationes Zoologicae Japonenses. 1937;6:293–300.
Allen J. Bopyroidessarsi (Bonnier) and Pleurocryptacluthae Scott (Isopoda, Epicaridea). Crustaceana. 1965;9:90–2.
Bourdon R. Les Bopyridae des mersEuropéennes. Mém Muséum Natl Hist Nat Paris Nouv Sér A. 1968;50(2):77–424.
Kim HS, Kwon DH. Bopyrid isopods parasitic on decapod crustaceans in Korea. Anim Syst Evol Divers. 1988;2:199–221.
Boyko CB. The Bopyridae (Crustacea: Isopoda) of Taiwan. Zool Stud. 2004;43:677–703.
Markham JC. Further notes on the Isopoda Bopyridae of Hong Kong. In: Morton B, editor. Proceedings of the second international marine biological workshop: the marine flora and fauna of Hong Kong and southern China, Hong Kong, 1986, vol. 2. Hong Kong: Hong Kong University Press; 1990. p. 555–68.
Kilpert F, Podsiadlowski L. The Australian fresh water isopod (Phreatoicidea: Isopoda) allows insights into the early mitogenomic evolution of isopods. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:36–44.
Hua CJ, Li WX, Zhang D, Zou H, Li M, Jakovlić I, et al. Basal position of two new complete mitochondrial genomes of parasitic Cymothoida (Crustacea: Isopoda) challenges the monophyly of the suborder and phylogeny of the entire order. Parasites Vectors. 2018;11:628.
Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–216.
Kumazawa Y, Ota H, Nishida M, Ozawa T. Gene rearrangements in snake mitochondrial genomes: highly concerted evolution of control region-like sequences duplicated and inserted into a tRNA gene cluster. Mol Biol Evol. 1996;13:1242–54.
Campbell NJ, Barker SC. The novel mitochondrial gene arrangement of the cattle tick, Boophilus microplus: fivefold tandem repetition of a coding region. Mol Biol Evol. 1999;16(6):732–40.
Zhang DX, Hewitt GM. Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol. 1997;25:99–120.
Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol Biol Evol. 1997;14:91–104.
Kilpert F, Podsiadlowski L. The complete mitochondrial genome of the common sea slater, Ligia oceanica (Crustacea, Isopoda) bears a novel gene order and unusual control region features. BMC Genomics. 2006;7:241.
Kilpert F, Held C, Podsiadlowski L. Multiple rearrangements in mitochondrial genomes of Isopoda and phylogenetic implications. Mol Phylogenet Evol. 2012;64:106–17.
Zou H, Jakovlić I, Zhang D, Chen R, Mahboob S, AI-Ghanim KA, et al. The complete mitochondrial genome of Cymothoa indica has a highly rearranged gene order and clusters at the very base of the Isopoda clade. PLoS One. 2018;13:e0203089.
Zhang D, Gao F, Jakovlić I, Zou H, Wang GT. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 2020;20(1):348–55.
Letunic I, Bork P. Interactive tree of life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8.
Katoh K, Toh H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics. 2008;9:212.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52.
He B, Su T, Wu Y, Xu J, Huang D, Yue BS. Phylogenetic analysis of the mitochondrial genomes in bees (Hymenoptera: Apoidea: Anthophila). PLoS One. 2018;13:e0202187.
Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2.
Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20.
Lanfear R, Calcott B, Ho S, Guindon S. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 2012;29(6):1695–701.
Kalyaanamoorthy S, Minh BQ, Wong TK, Haeseler AV, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9.
Ronquist F, Teslenko M, Mark P, Ayres DL, Darling A, Hhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42.
Nguyen LT, Schmidt HA, Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Hoang DT, Chernomor O, Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–22.
Miller MA, Pfeiffer W, Schwartz T. The CIPRES science gateway: a community resource for phylogenetic analyses. In: Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery; 2011. p. 1–8.
Acknowledgements
We are indebted to all collectors of specimens in the Florida Museum of Natural History.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 32070512]; Program of Ministry of Science and Technology of the People’s Republic of China [grant number 2015FY210300]; and Natural Science Foundation of Shanxi Province [grant number 201901D111274].
Author information
Authors and Affiliations
Contributions
AJ conceived the study; AJ and WR designed the study; AJ and PG procured the samples; AJ, GR, XQ conducted the lab work; AJ, WR, and PG analyzed and interpreted the data; AJ, GR, WR and PG drafted the article; all authors revised the article critically for important intellectual content, gave final approval for publication, and agree to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
As the animal handling included only unprotected invertebrates (crustaceans), no special permits were required to retrieve and process the sample.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Partitioning strategies and best models from PartitionFinder and Morderfinder for the datasets.
Additional file 2: Table S2.
Species and GenBank accession numbers (cox1 gene) in the phylogenetic analyses.
Additional file 3: Table S3.
Species and GenBank accession numbers (18S rRNA) in the phylogenetic analyses.
Additional file 4: Table S4.
Species and GenBank accession numbers (mitochondrial genome) in the phylogenetic analyses.
Additional file 5: Table S5.
Primers used for PCR amplification of the mitochondrial genome of Bopyroides hippolytes.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wu, R., Guo, R., Xi, Q. et al. Phylogenetic position of Bopyroides hippolytes, with comments on the rearrangement of the mitochondrial genome in isopods (Isopoda: Epicaridea: Bopyridae). BMC Genomics 23, 253 (2022). https://doi.org/10.1186/s12864-022-08513-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-022-08513-9