Abstract
Background
While recent advances in genomics has enabled vast improvements in the quantification of genome-wide diversity and the identification of adaptive and deleterious alleles in model species, wildlife and non-model species have largely not reaped the same benefits. This has been attributed to the resources and infrastructure required to develop essential genomic datasets such as reference genomes. In the absence of a high-quality reference genome, cross-species alignments can provide reliable, cost-effective methods for single nucleotide variant (SNV) discovery. Here, we demonstrated the utility of cross-species genome alignment methods in gaining insights into population structure and functional genomic features in cheetah (Acinonyx jubatas), snow leopard (Panthera uncia) and Sumatran tiger (Panthera tigris sumatrae), relative to the domestic cat (Felis catus).
Results
Alignment of big cats to the domestic cat reference assembly yielded nearly complete sequence coverage of the reference genome. From this, 38,839,061 variants in cheetah, 15,504,143 in snow leopard and 13,414,953 in Sumatran tiger were discovered and annotated. This method was able to delineate population structure but limited in its ability to adequately detect rare variants. Enrichment analysis of fixed and species-specific SNVs revealed insights into adaptive traits, evolutionary history and the pathogenesis of heritable diseases.
Conclusions
The high degree of synteny among felid genomes enabled the successful application of the domestic cat reference in high-quality SNV detection. The datasets presented here provide a useful resource for future studies into population dynamics, evolutionary history and genetic and disease management of big cats. This cross-species method of variant discovery provides genomic context for identifying annotated gene regions essential to understanding adaptive and deleterious variants that can improve conservation outcomes.
Similar content being viewed by others
Background
As natural habitats and ecosystems are increasingly impacted by anthropogenic events, a growing number of species require some form of ex situ management to prevent extinction. Big cat species are amongst the most vulnerable, having experienced dramatic population declines as result of illegal poaching, trade and habitat destruction. All 38 wild felid species have a negative global population trend [1]. Despite these challenges, wild felids are revered as cultural symbols and are important flagship species for engendering public interest in conservation programmes. As keystone species, strategies for the protection of large felids also benefit their ecosystems [2]. Allelic variation is essential to preserving species’ genetic integrity and maintaining functioning ecosystems, as genetically diverse populations tend to have higher fitness and adaptive capacity [3]. Given their conservation status, many big cats are the subjects of Species Survival Plans (SSP) [4, 5]; internationally coordinated programmes that manage ex situ breeding and aim for healthy, self-sustaining and genetically diverse populations. Retaining genetic diversity in a closed, captive population is challenging and expensive [6, 7]. Reductions in genetic diversity due to inbreeding, genetic drift and selection can lead to an accumulation of deleterious mutations in a captive-bred population, threatening their long-term viability [8, 9].
With genomic resources becoming increasingly accessible for a wide diversity of species, single nucleotide variant (SNV)-based genetic analyses offer conservationists higher resolution for measuring diversity and addressing conservation questions than previously [10]. In model species, SNVs are the genetic marker of choice for the advancement of functional, quantitative and evolutionary research [11,12,13]. Their high frequency across coding and non-coding regions, low typing error rates and ease of comparability across datasets, make them a favourable alternative or complement to low density markers such as microsatellites, allozymes and mtDNA [14]. Genome-wide SNV discovery methods have enabled the transition from estimates of inbreeding using pedigree data and microsatellite markers, to the direct quantification of inbreeding via genome-wide scans of individual homozygosity [15,16,17]. The use of genome-wide SNV data in wild cats has illuminated patterns of genetic variation relating to population history, physiological adaptation and speciation and the extent of genomic synteny among Felidae [18,19,20,21,22]. However, there remains a notable gap between these studies and the integration of genomics into the conservation management of these species. Recent reviews have highlighted several barriers to the widespread uptake of genomic data in conservation, including: high costs associated with sequencing and sampling, lack of computational infrastructure, need for specialist bioinformatic expertise, and the absence of genomic resources (i.e. high-quality reference data) for non-model species [23, 24]. Here, ‘non-model species’ refers to those with limited genomic resources, specifically reference genomes.
A shortage of high-quality reference genomes is often cited as a barrier to SNV discovery and genotyping in non-model species [24,25,26]. Reference-guided SNV discovery is more computationally efficient than methods that do not use a reference, offering higher accuracy at lower sequencing depths, as well as the ability to physically map and determine linkage disequilibrium between SNVs. The genomic context provided by a reference genome allows for identifying annotated gene regions essential to understanding any potential adaptive or deleterious variant consequences. Compared with many other carnivorous genera, felid genomics has been extensively studied: a number of domestic cat (Felis catus) reference assemblies have been developed since 2007 [27], followed by draft assemblies of tiger (Panthera tigris) [20], cheetah (Acinonyx jubatus) [18], leopard (Panthera pardus) [28], jaguar (Panthera onca) [21] and lion (Panthera leo) [29], among others. Compared with its wild counterparts, the most recent domestic cat reference assembly (felCat9) is a chromosome-level build that offers fewer gaps at a higher sequencing depth. Generating these reference genomes required considerable financial, computational and bioinformatic resources, rarely available to conservation managers. High-quality genome assemblies are differentiated from draft-quality assemblies by their lower error rates, fewer gaps (e.g. chromosome-level assembly), and high-quality annotations. Recently, a correction of the tiger draft assembly was published [30], highlighting the potential for inadequately validated draft assemblies to bias the outcomes of genomic observations and biological conclusions.
Cross-species reference-guided SNV calling and genotyping methods [31,32,33,34], reduced-representation libraries [19, 35, 36] and pooled sequencing (poolseq) [37, 38] have successfully been used to study genomics in non-model species, circumventing the costs, computational resources and extensive sampling required to develop a high quality reference genome. A high degree of genomic synteny has been observed across felid species using cytological and genomic approaches [18, 20, 29, 39,40,41]. Here, we took advantage of this conserved genomic architecture among felids and performed cross-species whole genome sequence (WGS) alignment and SNV discovery using the domestic cat reference genome assembly and WGS data of three big cat species; Sumatran tiger (Panthera tigris sumatrae), snow leopard (Panthera uncia) and cheetah. We compare the utility of non-barcoded pooled versus individual WGS data for cross-species alignment and SNV discovery. We present an annotated catalogue of high-quality variants from seventeen whole genome sequences comprising twenty-six individuals across the three species. We show that reference genomes from distantly related species can be successfully used for SNV discovery to inform conservation management. Gene enrichment and gene diversity analyses provided a proof of principle of the effectiveness of cross-species application of the domestic cat reference genome in variant calling in big cat species.
Results
Cross-species WGS alignment and variant calling using the felCat9 reference assembly
Genomic DNA of four Sumatran tigers, four cheetahs and four snow leopards was pooled by species in equimolar ratios and sequenced. Whole genome sequences for seven Sumatran tigers, six cheetahs and one snow leopard were downloaded from sequence read archive (SRA) [42]. Individual and pooled samples were aligned to the felCat9 [43] reference assembly. The number of reads in each species pool was 830 M for cheetah, 960.7 M for snow leopard and 896.7 M for Sumatran tiger. Overall sequencing performance is summarised in Table S1 and results for alignments and variant calling for all individuals and pools are presented in Table 1. An average of 170 M cheetah reads, 627 M snow leopard reads, and 251 M Sumatran tiger reads were mapped to the Felis_catus_9.0 (felCat9; GCA_000181335.4) reference assembly. On average, 94 % of cheetah, 93 % of snow leopard and 95 % of Sumatran tiger reads were properly paired and mapped to felCat9 chromosomes (Figure S1). The proportion of successfully paired and mapped reads was lower for Sumatran tiger and cheetah pools compared with their individually sequenced counterparts. The read depth for all individual samples ranged between 5.56x and 35.91x. Genome coverage was highest over a greater portion of bases for all species pools compared with their SRA counterparts (Figure S2). Read coverage for species pools ranged from 25.78x to 50.14x and alignment of species pools to the felCat9 reference assembly resulted in greater than 90% coverage of the reference at a minimum depth of 20x compared with a depth of ~ 5x, ~7x and 10x in cheetah, Sumatran tiger and snow leopard individual samples, respectively.
To compare the utility of non-barcoded pooled versus individual WGS, variant calling was performed in diploid mode for all samples. Across Sumatran tiger and cheetah samples, fewer biallelic variants were called for individuals than for their pooled counterparts, while the snow leopard pool and individual samples showed approximately equal numbers of variants (Table 1). Individual nucleotide diversity (π) is reported under the assumption that both parental sets of chromosomes have been sequenced to equivalent coverage depths. This statistic was similar among pools and individuals for Sumatran tigers and snow leopards, however the cheetah pool displayed markedly higher π compared to all sequenced individuals and compared to pools for the other two species. Despite having the lowest average read coverage, cheetahs had the highest proportion of reads mapped and high-quality SNVs called against the felCat9 reference assembly (Table 1). Cheetahs had a higher density of SNVs per kilobase across all felCat9 chromosomes than Sumatran tigers and snow leopards. The cheetah and Sumatran tiger pools had a significantly higher density of SNVs across all chromosomes compared with individuals of the respective species. In total, 13,414,953, 15,504,143 and 38,839,061 biallelic SNVs passed quality filtering in Sumatran tiger, snow leopard and cheetah, respectively. Of these, 10,472,528 in Sumatran tigers, 9,124,699 in snow leopards and 26,430,702 in cheetahs were transitions (Ts) and 5,030,622 in Sumatran tigers, 4,285,891 in snow leopards and 12,258,571 in cheetahs were transversions (Tv). Ts/Tv ratios for pooled samples were higher than individual samples (Table 1). Sumatran tiger individuals had a mean Ts/Tv ratio of 1.7 (σ = 0.06) while the Sumatran tiger pool reported a Ts/Tv ratio of 2.09. Cheetah individuals similarly reported a lower mean Ts/Tv ratio of 1.8 (σ = 0.02) compared with their pooled counterparts, which had a Ts/Tv ratio of 2.1, potentially indicating a higher rate of false variants in the individual sample variant sets. Ts/Tv ratios for species variant sets ranged from 2.08 to 2.15.
Population structure and demographic statistics
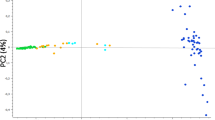
Population structure and demographic statistics were calculated using minor allele frequency (MAF) filtered datasets for all species. Multi-dimensional scaling (MDS) was used to partition total genomic variation among individuals in each species group (Fig. 1). There was clear differentiation between individuals and pools in all three species, and among individuals of different geographical provenance. The first component (C1) corresponded to the axis of differentiation among Tanzanian and Namibian cheetahs, with the pool distinctly clustering with Namibian cheetahs. Among Sumatran tigers, the second component (C2) accounted for differentiation between Indonesian (IDN) and American (USA), with two distinct clusters of Indonesian individuals separated by C1.
Distinct clustering of samples within species groups based on multi-dimensional scaling (MDS). a Cheetah individuals are labelled as either Namibian (CHEETAH_NAM) or Tanzanian (CHEETAH_TZA) based on geographic provenance, b Snow leopard pool and individual samples and c Sumatran tiger individuals were also labelled as either American (SUM_USA) or Indonesian (SUM_IND) based on geographic provenance
Estimates of genetic variability and pairwise similarity were measured in Sumatran tiger and cheetah individuals (only one individual sample was available for snow leopard) (Table 2). Co-ancestry coefficients (ϴ) were calculated as the probability of finding identical alleles when randomly sampling one allele from each heterozygous individual and suggested unrelated kinship among all Sumatran tiger individuals. Expected (He) and observed heterozygosity (Ho) were calculated to measure genetic diversity of each population. Ho was lower than He across all samples, indicating a deviation from Hardy-Weinberg equilibrium (HWE) and possible inbreeding (non-random mating). Mean individual inbreeding coefficients (F) among cheetahs was 0.346 ± 0.062 (range:0.259–0.419) and 0.585 ± 0.033 (range:0.233–0.273) among Sumatran tigers. These results may reflect close relationships among samples within each species, however this cannot be confirmed as data regarding their pedigree was not made available. The proportion of pairwise identity by state (IBS) allelic similarity among cheetahs ranged from 0.472 to 0.394 (µ = 0.437 ± 0.028). Mean IBS among Namibian cheetahs was 0.412 and 0.398 among Tanzanian cheetahs. Among Sumatran tigers IBS ranged from 0.392 to 0.521 (µ = 0.5 ± 0.026).
Functional annotation of genomic variants
Variant datasets were annotated using Variant Effect Predictor (VEP) in order to identify coding variants of potential functional significance. VEP assigned 29,059,874, 25,194,581 and 73,915,269 functional classes to SNVs in snow leopards, Sumatran tigers and cheetahs, respectively, based on the felCat9 reference assembly annotation. The number of functional classes defined by VEP is higher than the total number of SNVs, because some sites have multiple annotations. The quantity of quality-filtered, fixed and MAF-filtered variants annotated by VEP varied among species, however the number of transcripts and genes overlapped were consistent across all species (Table 3) and functional annotation of transcript-associated variants was similar across all three species, with over 60 % of coding variants labelled as synonymous for all species (Fig. 2). A summary of functional annotation of MAF-filtered and fixed SNVs is provided for each species dataset (Table S2).
Cheetah MAF-filtered variants were enriched for over 90 terms that were reduced to best representative terms: macromolecular localisation (GO:0033036; Padj=5.67 × 10− 9), regulation of biological quality (GO:0065008; Padj=7.36 × 10− 8), cytoskeleton organisation (GO:0007010; Padj=5.09 × 10− 9), developmental process (GO:0032502; Padj=7.91 × 10− 17) and cytoskeleton organisation (GO:0005856, Padj=9.20 × 10− 12) (Table S3a). In Sumatran tigers, variants passing the MAF-filter were enriched for protein binding (GO:0005515; Padj=3.54 × 10− 4), cilium (GO:0005929; Padj=1.53 × 10− 4), and collagen-containing extracellular matrix (GO:0062023; Padj=3.53 × 10− 4) (Table S3b). The snow leopard MAF-filtered dataset was not found to be functionally enriched. Calling variants from one snow leopard individual and a pool called in diploid mode has likely confounded the number of fixed and MAF-filtered variants in snow leopards. In snow leopard, a lack of significant functional enrichment among genes containing MAF-filtered variants may be attributed to small sample size.
Genomic signatures of adaptation among big cats
Signatures of selection within each species were tested using nucleotide and gene diversity of synonymous and non-synonymous SNVs. Most genes had a pairwise nucleotide diversity (πN/πS) < 1 while 54 genes in cheetahs, six genes in Sumatran tigers and one gene in snow leopards revealed signs of positive selection (Table S4). All genes displaying signatures of positive selection in tiger and snow leopard were involved in olfaction. In cheetahs, genes showing signs of selection were involved in olfaction (LOC101084218, LOC101085032, LOC101086178, LOC101095034 and LOC101101377) and immune responses (TLR3, SYTL2, OAS3 and RAB44). Genes under positive selection also included dynein axonemal heavy chain genes DNAH2 and DNAH6, involved in flagellum-dependent cell motility (GO:0001539) and SGCG and XIRP1, expressed exclusively in skeletal muscle.
SNVs fixed in each species group were collected and annotated to identify genes potentially involved in species-specific phenotypic signatures of adaptation. This also served to highlight phenotypic differences between the domestic cat and big cat species. In cheetahs, genes harbouring homozygous SNVs related to KEGG pathways were: HEXB, HARS1, PPP2CA, TGDS, ALG11 and PDE4D. Fixed missense alleles in cheetahs also occurred in ACTN3, previously associated with athletic performance in humans and horses [44, 45]. When reduced to the most representative subset of GOterms by semantic similarity, a total of 144 representative GO terms were analysed for fixed SNVs in Sumatran tigers and 95 in snow leopards (Table S5a,b). In Sumatran tigers, genes harbouring fixed variants were enriched for metabolic pathways (KEGG:01100; Padj=1.94 × 10− 17). Fixed non-synonymous alleles shared by snow leopards and Sumatran tigers were enriched for growth (GO:0040007), locomotion (GO:0040011), and developmental process (GO:0032502). These included missense variants in genes previously associated with body size: LCORL that were unique to Panthera species (Table S6). Pathway enrichment analysis was performed to gain mechanistic insight into genes containing fixed variants. Fixed non-synonymous variants in 418 genes common to all three big cat species were annotated for cadherin signalling (Padj=1.51 × 10− 5) and Wnt signalling (Padj=0.001) pathways. Genes in the protocadherin family displayed fixed allelic differences between all big cats and the domestic cat genome (Table S7).
Genetic insights into heritable conditions affecting big cats
To identify genes potentially underlying conditions of clinical importance in each species, a non-redundant list of genes categorised by GO terms relevant to known heritable conditions, immune and reproductive function was collected for each species. Genes containing deleterious variants (SIFT score 0-0.05) were collected from MAF-filtered datasets and observed in 201 genes annotated for relevant GO terms in cheetah, in six genes in snow leopards and in 44 genes in Sumatran tigers (Table S8). In cheetahs, these genes were annotated for spermatogenesis (GO:0007283), cilium assembly (GO:0060271), sperm flagellum (GO:0036126), B cell mediated immunity (GO:0019724) and embryo development (GO:0009790) (Figure S3a). In cheetahs, genes housing variants previously associated with known disease relating to ciliary dysfunction included PCDH15, HOMER2, SPEF2, NAGLU, PHGDH, ATR and ABL1. In Sumatran tigers, genes containing deleterious variants were restricted to terms relating to cilium structure and assembly (GO:0060271, GO:0032420) (Figure S3b) and ATR was also the only gene included in Sumatran tiger phenotype-associated genes. Snow leopard genes containing deleterious variants were: ADAM29, ADCY10, CCNYL1, CCT3, KIF20B and LAMB1.
Discussion
The cross-species application of the domestic cat reference presented here takes advantage of the high degree of synteny between Felidae genomes. Previously, cheetah [18] and tiger [20, 22] genomes have shown a high level of conserved synteny and repeat composition when compared with the domestic cat. This similarity is supported by the high alignment quality of all samples included here. Successful alignment of big cat samples and pools resulted in > 99 % coverage of the felCat9 reference assembly at varying sequence depths. Despite differences in phylogenetic distances between Acinonyx and Panthera lineages and domestic cats, the proportion of reads that aligned for each species did not appear to decrease with phylogenetic distance. The high affinity alignment presented here indicates strong genomic conservation within Felidae. This likely reflects the relatively contemporary speciation of modern felids, occurring < 11MYA [46]. Genomic synteny between species provides opportunities to interpret genomic structure and gene function in an evolutionary context. The high degree of synteny observed here is of crucial importance in studying diversity among felid species and mechanisms underlying local adaptation that differ among them [21, 47, 48]. Additionally, the highly conserved synteny of the cat genome with that of humans and other mammalian species has given insight into ancestral genome organisation [49,50,51], supporting the cat as a valuable biomedical model for heritable human diseases [52,53,54,55].
Estimates of heterozygosity are vulnerable to reference bias
Demographic parameters based on estimates of heterozygosity were inconsistent with previous studies. This may result from small sample sizes, unknown relatedness among samples and the cross-species genome alignment method employed here. While felid genomes may be highly homologous, and previous reports indicate no large-scale chromosomal rearrangements among felids [56], reference sequence bias can substantially impact estimates of heterozygosity and downstream population genomic analyses [57, 58]. Cross-species alignments have been found to bias heterozygosity estimates while correctly measuring population structure [29, 59]. Cheetahs were previously found to have lower rate of heterozygous SNVs when aligned to the draft cheetah assembly (0.0019–0.0021 [18], compared with 0.012 when aligned to the domestic cat reference herein). A similar pattern was also observed for the snow leopard (0.0002 [20] compared with 0.004 herein). In genomic studies, reference-guided variant calling will always be biased toward the properties of the reference genome, rather than those shared across a population. Reference genomes are idiosyncratic, type-specimens and preferential alignment of genomic sequences to the reference alleles, results in underestimating the level of variation in aligned samples from different populations. This problem is demonstrated by the higher SNV frequency in cheetahs (more closely related to the domestic cat), compared with snow leopards and Sumatran tigers. When aligned to its own species’ reference assembly, the cheetah genome displays lower overall SNV frequency and a significantly higher proportion of homozygous stretches than the domestic cat [18]. Any demographic statistics that rely on low frequency variants, may be affected by this bias which likely accounts for high inbreeding coefficients and low heterozygosity observations. Population structure analyses driven by common variants are largely unaffected by reference bias, as MDS showed samples of the same geographic provenance clustered together. Multi-genome alignment techniques that overcome these biases are available in humans, but these resources will likely remain scarce in the context of big cat conservation as they require large sample sets [60,61,62]. We recommend against using demographic inferences based on cross-species genome alignments to inform conservation management.
Poolseq is a cost-effective means of SNV discovery
We compared sequencing of individual genomes with sequencing of pooled DNA from multiple (N = 4) individuals (poolseq). After quality filtering, pooled samples called more SNV than lower coverage individual WGS (Sumatran tiger and cheetah) and an equivalent number of SNV to high coverage individual WGS (snow leopard). Poolseq can be used to provide a high-quality approach for genotyping the collective genomic profile of a population, comparable to population-level allele frequency estimates of individual WGS [63]. Poolseq is a cost-effective method for assessing population structure and genome-wide patterns of variation [64, 65], but does present statistical challenges in deriving estimates of demographic and other inferences that rely on individual heterozygosity. Estimating allele frequencies from poolseq is vulnerable to experimental noise and bias at a number of protocol stages, from pooling equimolar ratios of DNA [66] through to library construction [67], sequencing and analysis. Poolseq is a less efficient technique for discovering SNVs than individual WGS when coverage is low and sample sizes are small [63]. Previous studies have shown that effects of experimental error are greater when a pool is small (N < 10) and sequencing depth is low [68]. Suitably large sample sizes (N > 50) are often unfeasible for conservationists working with endangered species, particularly large carnivores that exist in low densities in both the wild and ex situ management settings. To compensate for small pool size (N = 4), pools were sequenced at a high coverage, quality-based filtering and MAF-based filtering of SNVs was performed. MAF thresholds for each species group were used to exclude variants present in a heterozygous state in one individual only. This approach can be used to reliably characterise the allele frequency spectrum, however the downside is that attempts to control for high error rates will have excluded low-frequency or rare alleles. As a result of excluding these alleles, we did not include poolseq samples in estimations of demographic parameters. Choosing poolseq under these circumstances is to accept the trade-off of losing information about rare alleles, in favour of a cost-effective estimation of genome-wide allele frequency of a population.
Genomic signatures into big cat hypercarnivory
Considering our data in the genomic and phenotypic context of the domestic cat has highlighted a suite of defining characteristics of domestication. These characteristics typically include neurological and behavioural changes associated with tameness, a shift toward a polyoestrous reproductive cycle, altered dietary habits and morphological changes [69]. Fixed variants presented here highlight differences in metabolic function, body size and neurological processes in wild and domestic cats. Changes in neural crest-related genes are believed to underlie the evolution of tameness across a range of domesticated species [48, 70,71,72,73]. The expression of cell adhesion proteins including cadherins and protocadherins during neural crest cell development is regulated by Wnt signalling [74, 75]. Fixed differences in protocadherin genes is consistent with comparative studies in wildcats (Felis silvestris) [48] and domesticated foxes [76]. As hyper-carnivorous ambush predators, cats share physiological traits essential for hunting and endogenous glucose demands [72, 77, 78]. Fixed variants in big cats reveal adaptive physiological functions essential to their evolutionary success as carnivorous species [79, 80]. In cheetahs this included genes involved in spatiotemporal awareness (HARS1) [81] and skeletal muscle function (ACTN3, SACS, MEGF10, SGCG and XIRP1) [44, 82,83,84]. Among Sumatran tiger and snow leopards these included unique missense variants in LCORL, a gene previously associated with body size in domestic mammals [13, 85, 86]. These results reflect species-specific genetic adaptations associated with hyper-carnivory, highlighting candidate genes underlying species-specific adaptative mechanisms integral to the evolutionary success of big cats. Carnivorous diets are associated with increased metabolism, faster growth rates and higher fecundity [28, 87] and is dependent on an abundance of prey species. Habitat loss and prey depletion threaten the ecological niche occupied by big cats as keystone species [88]. Ongoing genomic studies of these traits can highlight mechanisms by which big cats interact with their ecosystems, complementing ecological studies and serve as an essential component of holistic management plans [89,90,91,92].
Genomic insights into the pathogenesis of diseases affecting big cats
Adding to the ecological complexities of big cat conservation is the impact of infectious and heritable diseases [93, 94]. Captive cheetahs, snow leopards and Sumatran tigers have historically presented a range of infectious and degenerative diseases, while their wild counterparts have remained unaffected [93, 95, 96]. Captive breeding programmes typically operate to maintain within-population genetic diversity, however for many threatened species, population bottlenecks in the wild have resulted in genetically depauperate populations that display impaired fitness and increased susceptibility to infectious diseases [15, 97]. Domestic animals are increasingly used to model complex and simple genetic diseases in humans [52, 54, 98]. The methods employed by these studies, including genome-wide association studies, can be adapted by conservationists studying the genetic basis of heritable diseases. As an example, candidate genes relevant to health and reproductive success studied here can be used to inform studies of the genetic basis of documented conditions of ciliary dysfunction including CVD in Sumatran tigers [95], poor spermatozoan quality in cheetahs [99] and chronic respiratory infections [93, 94], by testing whether any of these variants are associated with clinical findings.
Sumatran tiger cubs in Australian zoos have been reportedly affected by CVD with a heritable component since 1990. Pedigree and segregation analyses suggested an autosomal dominant mode of inheritance with incomplete penetrance of CVD within the Australian population [95]. Stereocilia in the vestibular system play a functional role in spatial navigation and self-motion perception [100] and their dysfunction can cause neurological symptoms, such as those observed in the Sumatran tiger. Genes containing deleterious variants of potential clinical significance identified here included: SPEF2 previously implicated in primary ciliary dyskinesia [101] and vestibular stereocilia function [102], HOMER2 a stereociliary scaffolding protein, essential for normal hearing and vestibular function in humans and mice [103, 104] and PCDH15 implicated in Usher syndrome in humans and balance disorders and deafness in mice [105]. Similar enrichment of ciliary genes was observed in the cheetah with deleterious variants observed in seven genes (IFT140, CPLANE1, DYNC2H1, CCDC39, CC2DA, RPGRIP1L) involved in cilium development and structure. Studies in mice and humans have shown defects in ciliary structure and dysmotility are typically present from birth, with affected individuals suffering recurrent respiratory infections and poor fertility [106, 107]. Respiratory infections have long been observed as a significant cause of mortality in captive cheetahs [93, 108]. Data presented here can be used to pinpoint mechanistic bases of ciliary dysfunction in mucociliary clearance of the respiratory tract, paranasal sinuses and middle ear during respiratory infections. Eighteen genes known to cause primary cilia dyskinesia in humans [109] were found to contain missense variants in the cheetah dataset, of these CCDC39, DNAH8 LRRC6 contained deleterious variants. These genes are highly conserved between felids and humans and as such may serve as valuable candidates for understanding the pathogenesis of reproductive, respiratory and vestibular diseases present in big cat populations and aid in improved diagnosis and treatment by veterinarians.
Conclusions
We have demonstrated the utility of cross-species genome alignments in gaining insights into population structure and functional genomic features in big cat species. The datasets presented here provide a useful resource for future studies into population dynamics, evolutionary history and genetic and disease management of big cats. The high degree of synteny among felid genomes enabled the successful application of the domestic cat reference for visualising population structure, discovering variants associated with adaptive traits, genes under selection and pathogenesis of heritable diseases. Importantly however, this method is limited in its capacity to adequately quantify heterozygosity and low-frequency variants. Poolseq proved a low-cost method for genotypic profiling of each species. This cross-species method of variant discovery provides genomic context for identifying annotated gene regions essential to understanding the genomic landscape underpinning traits and diseases that can be used to improve conservation outcomes.
Methods
Animals and DNA sequencing
Whole genome sequences for seven Sumatran tigers, six cheetahs and one snow leopard were downloaded from Sequence Read Archive (SRA) in fastq format (Table S9). Sumatran tiger samples originated from American and Indonesian facilities, the snow leopard individual originated from Korea and Cheetah samples originated from free-ranging individuals in Nambia and Tanzania. Whole blood samples from four snow leopards, four cheetahs and four Sumatran tigers currently housed in Australian zoos were collected as a part of routine health examinations by registered veterinarians employed by each zoo and submitted to the University for infectious disease screening. Individuals comprising each species pool were chosen from a broader cohort based on pairwise relatedness (siblings and parent-offspring pairs were excluded) and individual inbreeding coefficients (FIS) calculated using International Studbook data. Genomic DNA was isolated from whole blood by phenol-chloroform extraction and pooled by species in equimolar ratios (4 individuals/species pool). Library preparation and whole genome sequencing of species pools was performed by Ramaciotti Centre for Genomics, University of New South Wales (Kensington, Australia). Illumina paired-end libraries were prepared and sequenced on the Illumina HiSeq 2000 platform with 150 bp paired-end reads.
Reference genome alignment, variant calling and filtering
All samples were aligned to the felCat9 reference assembly using Burrows Wheeler Aligner algorithm, BWA-mem [110]. Aligned reads were sorted using Samtools 1.9 [111]. Base quality score recalibration was performed using Genome Analysis ToolKit (GATK) [112]. Summary statistics of alignments were collected using Samtools stats function. GATK best practices [113] were used for SNV and short indel calling. For each sample, GATK’s HaplotypeCaller tool was used to call variants from the recalibrated bam file. GVCF files containing unfiltered SNVs and short indel calls for all sites were then submitted for joint genotyping with GATK’s GenotypeGVCF tool. A VCF was generated for each species cohort and then passed to VariantFiltration tool for hard filtering according to GATK recommendations: QUAL > 40.0, QD > 2.0, MQ > 50.0, FS < 50 and then to SelectVariants tool to remove indel calls. Summary statistics of quality filtered VCF datasets and diversity estimates for individuals and species cohorts including: nucleotide diversity (π), TsTv ratio, and individual heterozygosity (Hs) were performed using VCFTools –site-pi, --tstv and –het flags [114]. Species-specific SNVs (fixed) were collected from hard-filtered species cohort VCFs using minor allele frequency (MAF) thresholds (Table 2) with BCFtools view function [115]. Population structure within species was detected by MDS using PLINK [116] with MAF-filtered SNV datasets. To examine whether any individuals were closely related, relatedness among individuals in each species cohort were calculated using VCFTools –relatedness2 function.
Variant annotation and gene enrichment analysis
Variant datasets were annotated using Ensembl Variant Effect Predictor (VEP) tool [117] and the NCBI annotation release 104 of the felCat9 genome build. Gene annotation and enrichment analyses were performed for fixed and MAF-filtered datasets on protein change variants annotated by VEP with the following impact terms: missense_variant, start_lost, stop_gained, stop_lost, stop_retained_variant, splice_acceptor_variant, splice_donor_variant. Genes with an accelerated rate of non-synonymous to synonymous substitutions (dN/dS) and nucleotide diversity of non-synonymous to synonymous coding sites (πN/πS) within each species were calculated using SNPGenie v1.2 [118]. Enrichment analysis of GO terms and pathways was performed using gPofiler [119]. The gProfiler gOSt function gathers functional annotation terms from various annotation sources including gene ontology terms, biological pathways, protein databases and human phenotype ontology. Enrichment terms were considered significant if they passed a significance threshold of P < 0.001, corrected for multiple testing. This was done for fixed variant sets for each species to highlight genes of potential significance in felid evolution. It was also performed for MAF-filtered SNV datasets to highlight genes of potential significance within each species. GO enrichment output of fixed and MAF-filtered datasets from gProfiler were reduced to their most specific GOterms using REVIGO [120]. Network pathway enrichment analysis was performed using WebGestalt [121] overrepresentation analysis and EnrichNet [122] to identify network interconnectivity score (XD score) and overlap-based enrichment score (Fisher’s exact test) of genes containing fixed variants and known phenotypic annotations. Phenotypic annotation of genes containing fixed SNVs for each species to the OMIM [123] database was performed using Ensembl’s BioMart [124]. A list of genes previously associated with body size in domestic species was collected from the literature (Table S10) and cross-searched using fixed variant datasets in all big cat species. MEGA-X software [125] was used to perform multiple sequence alignment of LCORL protein, previously implicated in body size variation among mammals. LCORL protein sequences for domestic dog (Canis lupus familiaris), domestic cat, horse (Equus caballus), cow (Bos taurus) and lion were downloaded from Ensembl and aligned to consensus FASTA sequences of cheetah, Sumatran tiger and snow leopard. Multiple sequence alignment was created using the Muscle algorithm.
To identify genes potentially implicated in the reproductive success and overall health of captive-bred big cats, a list of clinically relevant genes was curated using GO annotation terms with AmiGO2 [126] in humans, dogs, pigs, cats, rats and mice (Table S11). This list comprised genes implicated in a list of heritable conditions affecting captive-bred cheetahs, snow leopard and Sumatran tigers, compiled from literature (Table S12) and genes implicated in reproduction, immunity and embryonic development. Variants in these genes were collected from each species cohort MAF-filtered VCF file and submitted for annotation analysis using GOnet [127] and Web-Gestalt using over-representation analysis and disease OMIM and GLAD4U [128] functional databases. High impact variants in these genes were included if they had been annotated as ‘deleterious’ by VEP.
Availability of data and materials
Sequence data for cheetah, Sumatran tiger and snow leopard individual samples is available on NCBI Sequence Read Archive. Cheetah samples: SRR2737540, SRR2737541, SRR2737542, SRR2737543, SRR2737544, SRR2737545 were deposited under BioProject no. PRJNA297824. Sumatran tiger samples: SRR7152379, SRR7152382, SRR7152383, SRR7152384, SRR7152385, SRR7152386, SRR7152388 were deposited under BioProject no. PRJNA437782. Snow leopard sample SRR836372 was deposited under BioProject no. PRJNA182708. Final VCF files for cheetahs is available at https://doi.org/10.6084/m9.figshare.12996920, for Sumatran tigers is available at https://doi.org/10.6084/m9.figshare.12996947 and for snow leopards is available at https://doi.org/10.6084/m9.figshare.12996977.
References
IUCN. The IUCN Red List of Threatened Species. Version 2020-2. 2020 [Available from: https://www.iucnredlist.org]
Ordiz A, Bischof R, Swenson JE. Saving large carnivores, but losing the apex predator? Biol Cons. 2013;168:128–33.
Reed DH, Frankham R. Correlation between Fitness and Genetic Diversity. Conserv Biol. 2003;17(1):230–7.
Grisham J. editor Cheetah Species Survival Plan in situ Conservation Programs. Proceedings of the 58th Annual Meeting, World Association of Zoos and Aquariums, WAZA, 16–20 November, 2003: Cooperation Between Zoos in Situ and Ex Situ Conservation Programmes; 2004: World Association of Zoos and Aquariums.
Tetzloff J. Role of Zoos in Snow Leopard Conservation: The Species Survival Plan in North America. Snow Leopards: Elsevier; 2016. pp. 301–10.
Moran E, Cullen R, Hughey FD. The costs of single species programmes and the budget constraint. Pacif Conserv Biol. 2008;14(2):108–18.
Willoughby JR, Ivy JA, Lacy RC, Doyle JM, DeWoody JA. Inbreeding and selection shape genomic diversity in captive populations: Implications for the conservation of endangered species. PLoS ONE. 2017;12(4):e0175996.
Frankham R, Loebel DA. Modeling problems in conservation genetics using captive Drosophila populations: rapid genetic adaptation to captivity. Zoo Biol. 1992;11(5):333–42.
Theodorou K, Couvet D. The efficiency of close inbreeding to reduce genetic adaptation to captivity. Heredity. 2015;114(1):38–47.
Supple MA, Shapiro B. Conservation of biodiversity in the genomics era. Genome Biol. 2018;19(1):131.
Gandolfi B, Alhaddad H, Abdi M, Bach LH, Creighton EK, Davis BW, et al. Applications and efficiencies of the first cat 63K DNA array. Sci Rep. 2018;8(1):7024.
Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat Genet. 2008;40(3):340–5.
Plassais J, Kim J, Davis BW, Karyadi DM, Hogan AN, Harris AC, et al. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat Commun. 2019;10(1):1489.
Jones AG, Small CM, Paczolt KA, Ratterman NL. A practical guide to methods of parentage analysis. Molecular Ecology Resources. 2010;10(1):6–30.
Hedrick PW, Garcia-Dorado A. Understanding Inbreeding Depression, Purging, and Genetic Rescue. Trends Ecol Evol. 2016;31(12):940–52.
Keller MC, Visscher PM, Goddard ME. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics. 2011;189(1):237–49.
Galla SJ, Moraga R, Brown L, Cleland S, Hoeppner MP, Maloney RF, et al. A comparison of pedigree, genetic and genomic estimates of relatedness for informing pairing decisions in two critically endangered birds: Implications for conservation breeding programmes worldwide. Evol Appl. 2020;13(5):991–1008.
Dobrynin P, Liu S, Tamazian G, Xiong Z, Yurchenko AA, Krasheninnikova K, et al. Genomic legacy of the African cheetah, Acinonyx jubatus. Genome Biol. 2015;16(1):277.
Natesh M, Atla G, Nigam P, Jhala YV, Zachariah A, Borthakur U, et al. Conservation priorities for endangered Indian tigers through a genomic lens. Sci Rep. 2017;7(1):9614.
Cho YS, Hu L, Hou H, Lee H, Xu J, Kwon S, et al. The tiger genome and comparative analysis with lion and snow leopard genomes. Nat Commun. 2013;4(1):2433.
Figueiró HV, Li G, Trindade FJ, Assis J, Pais F, Fernandes G, et al. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci Adv. 2017;3(7):e1700299.
Liu Y-C, Sun X, Driscoll C, Miquelle DG, Xu X, Martelli P, et al. Genome-wide evolutionary analysis of natural history and adaptation in the world’s tigers. Curr Biol. 2018;28(23):3840–9.e6.
Galla SJ, Buckley TR, Elshire R, Hale ML, Knapp M, McCallum J, et al. Building strong relationships between conservation genetics and primary industry leads to mutually beneficial genomic advances. Mol Ecol. 2016;25(21):5267–81.
Shafer ABA, Wolf JBW, Alves PC, Bergström L, Bruford MW, Brännström I, et al. Genomics and the challenging translation into conservation practice. Trends Ecol Evol. 2015;30(2):78–87.
Brandies P, Peel E, Hogg CJ, Belov K. The Value of Reference Genomes in the Conservation of Threatened Species. Genes. 2019;10(11):846.
Ouborg NJ, Pertoldi C, Loeschcke V, Bijlsma R, Hedrick PW. Conservation genetics in transition to conservation genomics. Trends Genet. 2010;26(4):177–87.
Pontius JU, Mullikin JC, Smith DR, Agencourt Sequencing T, Lindblad-Toh K, Gnerre S, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17(11):1675–89.
Kim S, Cho YS, Kim H-M, Chung O, Kim H, Jho S, et al. Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome Biol. 2016;17(1):211.
Armstrong EE, Taylor RW, Miller DE, Kaelin CB, Barsh GS, Hadly EA, et al. Long live the king: chromosome-level assembly of the lion (Panthera leo) using linked-read, Hi-C, and long-read data. BMC Biol. 2020;18(1):3.
Mittal P, Jaiswal SK, Vijay N, Saxena R, Sharma VK. Comparative analysis of corrected tiger genome provides clues to its neuronal evolution. Sci Rep. 2019;9(1):18459.
Galla SJ, Forsdick NJ, Brown L, Hoeppner MP, Knapp M, Maloney RF, et al. Reference Genomes from Distantly Related Species Can Be Used for Discovery of Single Nucleotide Polymorphisms to Inform Conservation Management. Genes. 2019;10(1):9.
Minias P, Dunn PO, Whittingham LA, Johnson JA, Oyler-McCance SJ. Evaluation of a Chicken 600K SNP genotyping array in non-model species of grouse. Sci Rep. 2019;9(1):6407.
Miller JM, Moore SS, Stothard P, Liao X, Coltman DW. Harnessing cross-species alignment to discover SNPs and generate a draft genome sequence of a bighorn sheep (Ovis canadensis). BMC Genom. 2015;16(1):397.
Robinson JA, Ortega-Del Vecchyo D, Fan Z, Kim BY, vonHoldt BM, Marsden CD, et al. Genomic Flatlining in the Endangered Island Fox. Current biology: CB. 2016;26(9):1183–9.
Blåhed I-M, Königsson H, Ericsson G, Spong G. Discovery of SNPs for individual identification by reduced representation sequencing of moose (Alces alces). PLoS ONE. 2018;13(5):e0197364.
Wright B, Farquharson KA, McLennan EA, Belov K, Hogg CJ, Grueber CE. From reference genomes to population genomics: comparing three reference-aligned reduced-representation sequencing pipelines in two wildlife species. BMC Genom. 2019;20(1):453.
Kurland S, Wheat CW, de la PC Mancera, Kutschera M, Hill VE, Andersson J. A, et al. Exploring a Pool-seq-only approach for gaining population genomic insights in nonmodel species. Ecol Evol. 2019;9(19):11448–63.
Micheletti SJ, Narum SR. Utility of pooled sequencing for association mapping in nonmodel organisms. Mol Ecol Resour. 2018;18(4):825–37.
Hsu TC, Rearden HH, Luquette GF. Karyological Studies of Nine Species of Felidae. Am Nat. 1963;97(895):225–34.
Wurster-Hill DH, Gray CW. Giemsa banding patterns in the chromosomes of twelve species of cats (Felidae). Cytogenet Cell Genet. 1973;12(6):388–97.
Perelman PL, Graphodatsky AS, Serdukova NA, Nie W, Alkalaeva EZ, Fu B, et al. Karyotypic conservatism in the suborder Feliformia (Order Carnivora). Cytogenet Genome Res. 2005;108(4):348–54.
Leinonen R, Sugawara H, Shumway M, Collaboration INSD. The sequence read archive. Nucleic Acids res. 2010;39(suppl_1):D19–21.
Buckley RM, Davis BW, Brashear WA, Farias FHG, Kuroki K, Graves T, et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. PLOS Genetics. 2020:16(10):1–28.
Pickering C, Kiely J. ACTN3: More than Just a Gene for Speed. Front Physiol. 2017;8(1080):1–9.
Ropka-Molik K, Stefaniuk-Szmukier M, Musiał AD, Piórkowska K, Szmatoła T. Sequence analysis and expression profiling of the equine ACTN3 gene during exercise in Arabian horses. Gene. 2019;685:149–55.
Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, et al. The Late Miocene Radiation of Modern Felidae: A Genetic Assessment. Science. 2006;311(5757):73.
Xu X, Dong G-X, Hu X-S, Miao L, Zhang X-L, Zhang D-L, et al. The Genetic Basis of White Tigers. Curr Biol. 2013;23(11):1031–5.
Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SMJ, et al. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci. 2014;111(48):17230–5.
Rettenberger G, Klett C, Zechner U, Bruch J, Just W, Vogel W, et al. ZOO-FISH analysis: cat and human karyotypes closely resemble the putative ancestral mammalian karyotype. Chromosome Res. 1995;3(8):479–86.
Murphy WJ, Sun S, Chen ZQ, Pecon-Slattery J, O’Brien SJ. Extensive conservation of sex chromosome organization between cat and human revealed by parallel radiation hybrid mapping. Genome Res. 1999;9(12):1223–30.
O’Brien SJ, Cevario S, Martenson JS, Thompson M, Nash WG, Chang E, et al. Comparative gene mapping in the domestic cat (Felis catus). J Hered. 1997;88(5):408–14.
Samaha G, Beatty J, Wade CM, Haase B. The Burmese cat as a genetic model of type 2 diabetes in humans. Anim Genet. 2019;50(4):319–25.
Narfström K, Holland Deckman K, Menotti-Raymond M. The Domestic Cat as a Large Animal Model for Characterization of Disease and Therapeutic Intervention in Hereditary Retinal Blindness. J Ophthalmol. 2011;2011:906943.
Gandolfi B, Alamri S, Darby WG, Adhikari B, Lattimer JC, Malik R, et al. A dominant TRPV4 variant underlies osteochondrodysplasia in Scottish fold cats. Osteoarthritis Cartilage. 2016;24(8):1441–50.
Gandolfi B, Gruffydd-Jones TJ, Malik R, Cortes A, Jones BR, Helps CR, et al. First WNK4-hypokalemia animal model identified by genome-wide association in Burmese cats. PLoS One. 2012;7(12):e53173-e.
Wurster-Hill DH, Centerwall WR. The interrelationships of chromosome banding patterns in canids, mustelids, hyena, and felids. Cytogenet Cell Genet. 1982;34(1–2):178–92.
Günther T, Nettelblad C. The presence and impact of reference bias on population genomic studies of prehistoric human populations. PLoS Genet. 2019;15(7):e1008302.
Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, et al. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14(5):R51.
Gopalakrishnan S, Samaniego Castruita JA, Sinding M-HS, Kuderna LFK, Räikkönen J, Petersen B, et al. The wolf reference genome sequence (Canis lupus lupus) and its implications for Canis spp. population genomics. BMC Genom. 2017;18(1):495.
Shukla HG, Bawa PS, Srinivasan S. hg19KIndel: ethnicity normalized human reference genome. BMC Genomics. 2019;20(1):459.
Huang L, Popic V, Batzoglou S. Short read alignment with populations of genomes. Bioinformatics. 2013;29(13):i361-70.
Rasmussen M, Guo X, Wang Y, Lohmueller KE, Rasmussen S, Albrechtsen A, et al. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334(6052):94–8.
Futschik A, Schlötterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186(1):207–18.
Dorant Y, Benestan L, Rougemont Q, Normandeau E, Boyle B, Rochette R, et al. Comparing Pool-seq, Rapture, and GBS genotyping for inferring weak population structure: The American lobster (Homarus americanus) as a case study. Ecol Evol. 2019;9(11):6606–23.
Doyle JM, Willoughby JR, Bell DA, Bloom PH, Bragin EA, Fernandez NB, et al. Elevated Heterozygosity in Adults Relative to Juveniles Provides Evidence of Viability Selection on Eagles and Falcons. J Hered. 2019;110(6):696–706.
Craig JE, Hewitt AW, McMellon AE, Henders AK, Ma L, Wallace L, et al. Rapid inexpensive genome-wide association using pooled whole blood. Genome Res. 2009;19(11):2075–80.
Kofler R, Nolte V, Schlötterer C. The impact of library preparation protocols on the consistency of allele frequency estimates in Pool-Seq data. Mol Ecol Res. 2016;16(1):118–22.
Hivert V, Leblois R, Petit EJ, Gautier M, Vitalis R. Measuring genetic differentiation from Pool-seq data. Genetics. 2018;210(1):315–30.
Wilkins AS, Wrangham RW, Fitch WT. The “domestication syndrome” in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics. 2014;197(3):795–808.
Pendleton AL, Shen F, Taravella AM, Emery S, Veeramah KR, Boyko AR, et al. Comparison of village dog and wolf genomes highlights the role of the neural crest in dog domestication. BMC Biol. 2018;16(1):64.
Sánchez-Villagra MR, Geiger M, Schneider RA. The taming of the neural crest: a developmental perspective on the origins of morphological covariation in domesticated mammals. R Soc Open Sci. 2016;3(6):160107.
Frantz LAF, Schraiber JG, Madsen O, Megens H-J, Cagan A, Bosse M, et al. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet. 2015;47(10):1141–8.
Salinas PC. Wnt signaling in the vertebrate central nervous system: from axon guidance to synaptic function. Cold Spring Harb Perspect Biol. 2012;4(2):a008003.
Simões-Costa M, Bronner ME. Establishing neural crest identity: a gene regulatory recipe. Development. 2015;142(2):242–57.
Cousin H. Cadherins function during the collective cell migration of Xenopus Cranial Neural Crest cells: revisiting the role of E-cadherin. Mech Dev. 2017;148:79–88.
Wang X, Pipes L, Trut LN, Herbeck Y, Vladimirova AV, Gulevich RG, et al. Genomic responses to selection for tame/aggressive behaviors in the silver fox (Vulpes vulpes). Proc Natl Acad Sci. 2018;115(41):10398–403.
Eisert R. Hypercarnivory and the brain: protein requirements of cats reconsidered. J Comp Physiol B. 2011;181(1):1–17.
Sicuro FL, Oliveira LFB. Skull morphology and functionality of extant Felidae (Mammalia: Carnivora): a phylogenetic and evolutionary perspective. Zool J Linnean Soc. 2011;161(2):414–62.
Depauw S, Hesta M, Whitehouse-Tedd K, Vanhaecke L, Verbrugghe A, Janssens GP. Animal fibre: the forgotten nutrient in strict carnivores? First insights in the cheetah. J Anim Physiol Anim Nutr. 2013;97(1):146–54.
Winchester B. Lysosomal metabolism of glycoproteins. Glycobiology. 2005;15(6):1R–15R.
Safka Brozkova D, Deconinck T, Beth Griffin L, Ferbert A, Haberlova J, Mazanec R, et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain. 2015;138(8):2161–72.
Parfitt DA, Michael GJ, Vermeulen EGM, Prodromou NV, Webb TR, Gallo J-M, et al. The ataxia protein sacsin is a functional co-chaperone that protects against polyglutamine-expanded ataxin-1. Hum Mol Genet. 2009;18(9):1556–65.
Logan CV, Lucke B, Pottinger C, Abdelhamed ZA, Parry DA, Szymanska K, et al. Mutations in MEGF10, a regulator of satellite cell myogenesis, cause early onset myopathy, areflexia, respiratory distress and dysphagia (EMARDD). Nat Genet. 2011;43(12):1189–92.
Hudson PE, Corr SA, Payne-Davis RC, Clancy SN, Lane E, Wilson AM. Functional anatomy of the cheetah (Acinonyx jubatus) forelimb. J Anat. 2011;218(4):375–85.
Saif R, Henkel J, Jagannathan V, Drögemüller C, Flury C, Leeb T. The LCORL Locus Is under Selection in Large-Sized Pakistani Goat Breeds. Genes. 2020;11(2):1–10.
Metzger J, Schrimpf R, Philipp U, Distl O. Expression Levels of LCORL Are Associated with Body Size in Horses. PLoS ONE. 2013;8(2):e56497.
Muñoz-Garcia A, Williams JB. Basal metabolic rate in carnivores is associated with diet after controlling for phylogeny. Physiol Biochem Zoology. 2005;78(6):1039–56.
Wolf C, Ripple WJ. Prey depletion as a threat to the world’s large carnivores. R Soc Open Sci. 2016;3(8):160252.
Durant SM, Mitchell N, Groom R, Pettorelli N, Ipavec A, Jacobson AP, et al. The global decline of cheetah Acinonyx jubatus and what it means for conservation. Proc Natl Acad Sci. 2017;114(3):528.
Chapron G, Miquelle DG, Lambert A, Goodrich JM, Legendre S, Clobert J. The impact on tigers of poaching versus prey depletion. J Appl Ecol. 2008;45(6):1667–74.
Alexander JS, Gopalaswamy AM, Shi K, Hughes J, Riordan P. Patterns of snow leopard site use in an increasingly human-dominated landscape. PLoS One. 2016;11(5):e0155309.
Luskin MS, Albert WR, Tobler MW. Sumatran tiger survival threatened by deforestation despite increasing densities in parks. Nat Commun. 2017;8(1):1783.
Terio KA, Mitchell E, Walzer C, Schmidt-Küntzel A, Marker L, Citino S. Diseases Impacting Captive and Free-Ranging Cheetahs. Cheetahs: Biology and Conservation. 2018:349–64.
Ostrowski S, Gilbert M. Diseases of Free-Ranging Snow Leopards and Primary Prey Species. Snow Leopards: Elsevier; 2016. pp. 97–112.
Wheelhouse JL, Hulst F, Beatty JA, Hogg CJ, Child G, Wade CM, et al. Congenital vestibular disease in captive Sumatran tigers (Panthera tigris ssp. sumatrae) in Australasia. Vet J. 2015;206(2):178–82.
Herrin KV, Allan G, Black A, Aliah R, Howlett CR. Stifle osteochondritis dissecans in snow Leopards (< i > UNCIA UNCIA). J Zoo Wildlife Med. 2012;43(2):347–54.
Smallbone W, van Oosterhout C, Cable J. The effects of inbreeding on disease susceptibility: Gyrodactylus turnbulli infection of guppies, Poecilia reticulata. Exp Parasitol. 2016;167:32–7.
Karlsson EK, Sigurdsson S, Ivansson E, Thomas R, Elvers I, Wright J, et al. Genome-wide analyses implicate 33 loci in heritable dog osteosarcoma, including regulatory variants near CDKN2A/B. Genome Biol. 2013;14(12):R132.
Crosier AE, Wachter B, Schulman M, Lüders I, Koester DC, Wielebnowski N, et al. Chapter 27 - Reproductive Physiology of the Cheetah and Assisted Reproductive Techniques. In: Nyhus PJ, Marker L, Boast LK, Schmidt-Küntzel A, editors. Cheetahs: Biology and Conservation: Academic Press; 2018. pp. 385–402.
de Lahunta A, Glass E. Vestibular System: Special Proprioception. Veterinary Neuroanatomy Clin Neurol. 2009:319–47.
Sironen A, Kotaja N, Mulhern H, Wyatt TA, Sisson JH, Pavlik JA, et al. Loss of SPEF2 function in mice results in spermatogenesis defects and primary ciliary dyskinesia. Biol Reprod. 2011;85(4):690–701.
Scheffer DI, Shen J, Corey DP, Chen Z-Y. Gene Expression by Mouse Inner Ear Hair Cells during Development. The Journal of Neuroscience. 2015;35(16):6366.
Azaiez H, Decker AR, Booth KT, Simpson AC, Shearer AE, Huygen PLM, et al. HOMER2, a Stereociliary Scaffolding Protein, Is Essential for Normal Hearing in Humans and Mice. PLOS Genetics. 2015;11(3):e1005137.
Azaiez H, Decker AR, Booth KT, Simpson AC, Shearer AE, Huygen PLM, et al. HOMER2, a stereociliary scaffolding protein, is essential for normal hearing in humans and mice. PLoS Genet. 2015;11(3):e1005137-e.
Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet. 2001;27(1):99–102.
Tilley AE, Walters MS, Shaykhiev R, Crystal RG. Cilia dysfunction in lung disease. Annu Rev Physiol. 2015;77:379–406.
Sironen A, Shoemark A, Patel M, Loebinger MR, Mitchison HM. Sperm defects in primary ciliary dyskinesia and related causes of male infertility. Cell Mol Life Sci. 2020;77(11):2029–48.
Bell K. Morbidity and Mortality in Hand Reared Cheetah Cubs Animal Keeper’s Forum2005. p. 306 – 14.
Lobo J, Zariwala MA, Noone PG. Primary ciliary dyskinesia. Semin Respir Crit Care Med. 2015;36(2):169–79.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303.
DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8.
Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8.
Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–93.
Chang CC, Chow CC, Tellier LCAM, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4(1):7.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122.
Nelson CW, Moncla LH, Hughes AL. SNPGenie: estimating evolutionary parameters to detect natural selection using pooled next-generation sequencing data. Bioinformatics. 2015;31(22):3709–11.
Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019;47(W1):W191-W8.
Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLOS ONE. 2011;6(7):e21800.
Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–205.
Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012;28(18):i451-i7.
Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids research. 2005;33(suppl_1):D514-D7.
Kinsella RJ, Kähäri A, Haider S, Zamora J, Proctor G, Spudich G, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011(bar030):1–9.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9.
Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–9.
Pomaznoy M, Ha B, Peters B. GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinformatics. 2018;19(1):470.
Jourquin J, Duncan D, Shi Z, Zhang B. GLAD4U: deriving and prioritizing gene lists from PubMed literature. BMC Genomics. 2012;13(S8):20.
Acknowledgements
The authors acknowledge the University of Sydney’s high-performance computing cluster Artemis for providing the high-performance computing resources that have contributed to the research reported within this manuscript. We would also like to acknowledge Dr Jacqueline Norris, Zoos Victoria, Melbourne Zoo, Mogo Zoo, Weribee Open Range Zoo and Taronga Conservation Society Australia for providing whole blood samples of big cats included in poolseq datasets.
Funding
The research undertaken in this project was funded by the Jenna O’Grady Donley Fund, Sydney School of Veterinary Science.
Author information
Authors and Affiliations
Contributions
All authors conceptualised the study design. B.W and C.M.W were responsible for the provision of resources. Experimental work, bioinformatic analyses and visualisation were conducted by G.S under the supervision of B.H and C.M.W. C.E.G provided valuable advice on statistical analysis. G.S drafted the original manuscript which all authors edited. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Research was conducted at The University of Sydney, under Animal Ethics Committee approval no: N00/9–2009/3/5109, 24 September 2009. Whole blood samples were collected under Zoos Victoria ethics approval ZV09006 and Taronga Conservation Society approval #R12B127.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Sequencing and alignment performance for all individual samples and species pools. Cheetah_pool, tiger_pool and snowleopard_pool refer to species pools consisting of 4 multiplexed individuals each. Figure S1. Mapped and total reads across WGS samples and pools. Total number of mapped reads (black) includes singletons and pairs. Number of paired reads mapped indicated by the grey line. Tiger_zoo, snowleopard_zoo and cheetah_zoo refer to multiplexed pool samples. Figure S2. Plots of genome coverage for each sample .bam file aligned to the felCat9 reference assembly for a. cheetah, b. Sumatran tiger and c. snow leopard cohorts. On each panel, the key indicates coloured line of each sample and their sequencing depth in brackets. Table S2. Functional annotation of all fixed and within-species SNPs for each species. Table S3a. Top 20 gene ontology terms (GOterms) enriched for species-specific SNV within cheetahs. P-values were adjusted for multiple testing using Benjamini-Hochberg false discovery rate. Table S3b. Top 20 gene ontology terms (GOterms) enriched for species specific SNVs within Sumatran tigers. P-values were adjusted for multiple testing using Benjamini-Hochberg false discovery rate. Table S4. Genes under positive selection identified as those displaying elevated πNπSratios across all three species. Table S5a. Top 20 gene ontology terms (GOterms) enriched across fixed SNVs in snow leopards. P-values were adjusted for multiple testing using Benjamini-Hochberg FDR (false discovery rate). Table S5b. Top 20 gene ontology terms (GOterms) enriched across fixed SNVs in Sumatran tigers. P-values were adjusted for multiple testing using Benjamini-Hochberg FDR (false discovery rate). Table S6. Multi-species alignment of LCORL (ENSFCAG00000029474) revealed Panthera-specific conservation of six missense variants. Cheetah, snow leopard and Sumatran tiger refer to samples aligned to the domestic cat (felCat9) reference assembly. Protein positions are reported relative to the Ensembl transcript ENSFCAT00000081895.1. Table S7. Protocadherin genes containing fixed non-synonymous SNVs common to all big cat species relative to the domestic cat (felCat9) reference assembly. Table S8. Genes containing deleterious SNVs implicated in heritable conditions affecting big cats grouped by species. Figure S3a. Gene ontology annotation of clinically significant GOterms in cheetahs. Deleterious variants were observed in 201 genes included in custom list of GO terms relevant to known heritable conditions, immune and reproductive function. These genes were annotated for terms relevant to reproductive and immune function. Network interaction graph produced by GOnet (https://tools.dice-database.org/GOnet/). Figure S3b. Gene ontology annotation of clinically significant GOterms in Sumatran tigers. Deleterious variants were observed in 44 genes included in custom list of GO terms relevant to known heritable conditions, immune and reproductive function. These genes were annotated for terms relevant to cilium structure. Network interaction graph produced by GOnet (https://tools.dice-database.org/GOnet/). Table S9. Samples downloaded from Sequence Read Archive (SRA) comprised six cheetahs, one snow leopard and seven Sumatran tigers. Table S10.Genes associated with size in domestic species identified from a literature search. Table S11. Selected GO enrichment terms used to classify deleterious variants potentially implicated in reproductive success and overall health of captive bred big cats. Table S12. Known heritable conditions affecting captive bred big cat species and relevant GO terms.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Samaha, G., Wade, C.M., Mazrier, H. et al. Exploiting genomic synteny in Felidae: cross-species genome alignments and SNV discovery can aid conservation management. BMC Genomics 22, 601 (2021). https://doi.org/10.1186/s12864-021-07899-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-021-07899-2