Abstract
Background
The impossibility of closing the life cycle of the European eel (Anguilla anguilla) in captivity troubles the future of this critically endangered species. In addition, the European eel is a highly valued and demanded resource, thus the successful closing of its life cycle would have a substantial economic and ecological impact. With the aim of obtaining the highest gamete quality, the study of the effects of environmental factors, such as temperature, on reproductive performance may prove valuable. This is especially true for the exposure to cold water, which has been reported to improve sexual development in multiple other Actinopterygii species.
Results
European eel males treated with cold seawater (10 °C, T10) for 2 weeks showed an increase in the proliferation and differentiation of spermatogonial cells until the differentiated spermatogonial type A cell stage, and elevated testosterone and 11-ketotestosterone plasma levels. Transcriptomes from the tissues of the brain-pituitary-gonad (BPG) axis of T10 samples revealed a differential gene expression profile compared to the other experimental groups, with clustering in a principal component analysis and in heat maps of all differentially expressed genes. Furthermore, a functional analysis of differentially expressed genes revealed enriched gene ontology terms involved in the regulation of circadian rhythm, histone modification, meiotic nuclear division, and others.
Conclusions
Cold seawater treatment had a clear effect on the activity of the BPG-axis of European eel males. In particular, our cold seawater treatment induces the synchronization and increased proliferation and differentiation of specific spermatogonial cells. In the transcriptomic results, genes related to thermoception were observed. This thermoception may have caused the observed effects through epigenetic mechanisms, since all analysed tissues further revealed differentially expressed genes involved in histone modification. The presented results support our hypothesis that a low temperature seawater treatment induces an early sexual developmental stage in European eels. This hypothesis is logical given that the average temperature experienced by eels in the early stages of their oceanic reproductive migration is highly similar to that of this cold seawater treatment. Further studies are needed to test whether a cold seawater treatment can improve the response of European eels to artificial hormonal treatment, as the results suggest.
Similar content being viewed by others
Background
The decrease in the wild population of the European eel (Anguilla anguilla) has led to this species being listed as critically endangered by the International Union for Conservation of Nature [1]. Although it is possible to induce sexual development in eel using exogenous hormones [2,3,4,5], these treatments are long (several months), expensive, result in highly variable rates of fertilization and hatching, and have never resulted in an adult F1 generation. Therefore, it is necessary to improve these procedures in order to reproduce eels in captivity, which in turn would reduce the fishing pressure. Spawning European eels have never been observed in the wild, and the precise environmental conditions under which maturation happens are thus unknown. However, it is commonly hypothesized that maturation is initiated early on in the oceanic reproductive migration, and first comes to completion after or in the late stages of this migration [6, 7]. During the last decade, satellite pop-up tag experiments have shown that, after the eels leave the continental shelf (> 10° West longitude), they all migrate at similar temperatures (average of ~ 10 °C), ranging from up to ~ 12 °C at night and down to ~ 8 °C during the day, due to daily vertical migration [7, 8]. The hypothetical spawning area, the Sargasso Sea, is believed to be ~ 20 °C at the predicted time of spawning [9]. Therefore, most artificial European eel maturation experiments have been performed at ~ 20 °C [3, 4, 10, 11], but the apparent discrepancy between artificial maturation temperatures and the natural temperatures eels experience during early sexual development is an obvious candidate for investigation. Furthermore, cold temperatures in particular, can be beneficial for Actinopterygii sexual maturation, e.g. in white sturgeon (Acipenser transmontanus) 3 months of cold water at 10–12 °C, instead of 16–24 °C, have been shown to improve late oogenesis, prevent ovarian regression and increase blood estradiol (E2) and 11-ketotestosterone (11KT) levels [12, 13]. Also, lower fecundity and egg quality in wild Actinopterygii populations have been observed in years with warmer winters, e.g. seen in striped bass (Morone saxatilis; [14]), white sturgeon [12, 13], and others (for review see [15, 16]). Cold water treatments have additionally been shown to be a modulating factor of sexual developmental induction in several teleosts; Particularly in wolffish (Anarhichas lupus; [17]), cod (Gadus morhua; [18, 19]), pollack (Pollachius pollachius; [20]), European sea bass (Dicentrarchus labrax; [21, 22]), Eurasian perch (Perca fluviatilis; [23]), and yellow perch (Perca flavescens; [15]).
Previous temperature studies on European eel have illustrated the importance of this environmental factor [24,25,26,27,28,29]. E.g. in hormonally treated females, higher E2 plasma levels and follicle-stimulating hormone beta subunit (fshb) expression was registered at low temperatures (10 °C; [25]), meanwhile in hormonally treated males, a temperature higher than 10 °C was necessary in order to achieve complete gonad maturation [26]. In all these studies, a combination of temperature and hormonal treatment effects were studied. If a cold seawater treatment alone, in fact, stimulates natural early sexual development, this should be tested without the administration of hormonal treatments, which bypass the natural endocrine control of sexual development.
Thus, we hypothesized that a thermal treatment of low temperatures would be able to stimulate early sexual development in the European eel, and could possibly improve current artificial maturation procedures. In order to test the effect of a cold seawater treatment, we exposed European eel males to 3 different temperature regimes, including a constant low temperature (10 °C; T10), a constant high temperature (20 °C; T20) and a variable temperature (Tvar) over the course of 2 weeks. From these fish, we analyzed changes in biometric characteristics (length, weight, fin index, eye index, and hepatosomatic index), key male sex-steroids: namely testosterone (T) and 11KT, pituitary gonadotropin protein levels, the effect on transcriptomes of the BPG axis (brain, pituitary and testis), and histologically identified and quantified spermatogonial cells. Due to the short treatment period (2 weeks), few morphological changes were expected.
Results
Biometric parameters
No differences between the Control and the treated groups were seen in total length, total weight, fin color, eye index, or hepatosomatic index. However, significantly shorter fin lengths were found in the T10 and T20 groups (18.08 ± 0.36 and 18.19 0.36 mm, respectively) compared to the Control and Tvar groups (19.37 ± 0.38 and 19.31 ± 0.39 mm, respectively; Table 1). These differences also resulted in significantly lower fin indices (P-value = 0.008) in the T10 and T20 groups (4.75 ± 0.11 and 4.79 ± 0.07, respectively) compared to the Control and Tvar groups (5.25 ± 0.07 and 5.07 ± 0.10, respectively; Table 1).
Gonad histology
SPGAund* (Fig. 1) composed a significantly higher average proportion of the cells identified in the Control samples (11.7 ± 1.4% cells per field) compared to all the 3 treatment groups (< 1% cells per field; Fig. 2). The Control samples also contained a higher average proportion of SPGAund (Fig. 1) cells per field (52.1 ± 1.9%) compared to Tvar, T20, and T10 (28.7 ± 1.2, 26.8 ± 1.3, and 9.3 ± 0.4%, respectively). The proportion of SPGAund cells had an inverse relationship with the proportion of SPGAdiff cells with the T10 samples contained a significantly higher average proportion of SPGAdiff cells per field (69.6 ± 1.0%) compared to the T20, Tvar and Control groups (57.1 ± 1.5, 52.3 ± 1.5, and 23.8 ± 1.9%, respectively). Undefined cells were also registered in higher proportions in the T10 samples. Although all the experimental groups contained relatively low average proportions of early SPGB cells (Fig. 1), the T20 and Tvar groups (7.5 ± 1.1 and 8.2 ± 1.2%, respectively) reported a higher proportion than the T10 and Control groups (4.7 ± 0.5 and 4.5 ± 0.8%, respectively). Finally, the average total number of SPG cells per field was significantly higher in the T10 samples (189.9 ± 3.0 cells per field), compared to the T20, Tvar, and Control groups (134.0 ± 3.7, 130.7 ± 3.6, and 117 ± 3.8 cells per field, respectively), while the average total number of SPG cells per field was significantly lower in the Control samples compared to T20 and Tvar (Fig. 2).
Identified spermatogonia types. Forty times magnification fields of selected histological sections representing the spermatogonia stages: the most undifferentiated spermatogonia type A (SPGAund*; panel a), the second most undifferentiated spermatogonia type A (SPGAund; panel b), differentiated spermatogonia type A cells (SPGAdiff; panel c), and early spermatogonia type B cells (SPGB; panel d). These identifiable characteristics are further labelled with arrows: Blood vessels (Bv), nucleus (Nu), nucleoli (No), Sertoli cells (Sc), Sertoli cell cytoplasmic extensions (Scce), heterochromatin (Hech) and nuage (Nuage)
Histological cell counts. Boxplots of cell counting results for the T10, T20, Tvar and Control groups. The panels show the percentage proportion of the most undifferentiated spermatogonia type A cells (SPGAund*), the second most undifferentiated spermatogonia type A cells (SPGAund), the differentiated spermatogonia type A cells (SPGAdiff), and the early spermatogonia type B cells (Early SPGB) in each group. The panels labelled “Undefined cells” presents the percentage proportion of cells in each group, which were identified as spermatogonial cells but could not be distinguished between the specific spermatogonial cell types. The panels labelled “Total cells” presents the accumulated cells count of all identified cell types in each group. Letters indicate significant differences
Steroid analysis
The plasma T analysis revealed a basal level of 0.99 ± 0.12 ng/ml in the Control group, which increased significantly to 2.32 ± 0.17 ng/ml after rearing the fish for 2 weeks at 10 °C. No differences were observed in the rest of the experimental groups (Fig. 3). The basal 11KT plasma level was 1.67 ± 0.31 ng/ml, which increased significantly after 2 weeks of rearing to 4.46 ± 0.43 ng/ml and 3.37 ± 0.30 ng/ml at 10 or 20 °C, respectively (Fig. 3).
Radioimmunoassay steroid results. Boxplots of radioimmunoassay steroid results from the blood of fish from the T10, T20, Tvar and control groups. Significant differences are indicated with letters. Panel a shows the testosterone (T) results, while panel b shows the 11-ketotestosterone (11KT) results
Gonadotropin analysis
Due to their biological relevance, fshb and luteinizing hormone beta subunit (lhb) were chosen as target genes for additional analysis. In particular, the transcripts per million (TPM) for these genes in the pituitary transcriptome were retrieved (see Additional file 1: Figure S1) and the variance of the means between the groups were analysed with a one-way analysis of Variance (ANOVA). The analysis indicated that temperature caused a significant change to the lhb (P-value = 0.0296) expression, while the differences observed in fshb were insignificant (P-value = 0.0746).
Furthermore, additional fish were treated in a different experimental run, under the same conditions, and the pituitaries of these fish were sampled. Fshβ and Lhβ immunofluorescence labelling were performed on these pituitaries, except the pituitaries of Tvar treated fish, which were excluded due to the TPM results of these samples (see Additional file 1: Figure S1). Due to loss of pituitary tissue during laboratory analysis (e.g. pituitary drying during incubation or breaking during sectioning), initial “n” (10) decreased to 6, 5, and 4 for T10, Control, and T20, respectively, for the Lhβ immunofluorescence labelling, and to 2, 2, and 3 for T10, Control, and T20 respectively for the Fshβ immunofluorescence labelling. All remaining pituitaries were successfully labelled with both Fshβ and Lhβ; however, no reliable difference could be observed from the Fshβ immunofluorescence labelling (see Additional file 1: Figure S2), possibly due to the low n. On the other hand, a consistently stronger Lhβ signal was seen in the pituitaries of T10 treated fish compared to either Control or T20 (Fig. 4).
Pituitary Lhβ histochemical identification. Confocal images of the immunofluorescence labelled European eel (Anguilla anguilla) male pituitaries, which showed the strongest Lhβ signal from each of the analyzed groups: the 2 week 10 °C pretreated group (T10; panel d, e, and f), the 2 week 20 °C pretreated group (T20; panel g, h, and i), and Control (Panel a, b, and c). “DAPI” indicates pictures filtered to only reveal fluorescents labeled to 4,6-diamidino-2-phenylindole dihydrochloride (Panel a, d, and g). “Lhβ” indicates pictures filtered to only reveal fluorescents labeled to luteinizing hormone beta subunit protein (Panel b, e, and h). “DAPI / Lhβ” indicates pictures filtered to reveal both fluorescents labelled to luteinizing hormone beta subunit protein and 4,6-diamidino-2-phenylindole dihydrochloride (Panel c, f, and i)
RNA-sequencing
Our raw Illumina data contained between 48 and 75 million 101 bp paired-end sense strand reads per library. Using BWA-MEM [30] to map our transcripts to our de novo transcriptome resulted in a mapping percentage range of 89 to 96%. Out of the 77,247 transcripts in our transcriptome, 25,368 protein encoding genes were predicted. Using DEseq, differentially expressed transcripts (FDR < 0.05) were determined between the 3 treatment groups (T10, T20, and Tvar) and the Control group, in all combinations and for the 3 tissues: brain, pituitary, and testes. Approximately half of the differentially expressed transcripts could be annotated with a gene symbol and assigned gene ontology (GO) terms, with little variation between groups (Table 2). All differentially expressed genes have been accumulated into 3 heat maps (one per tissue), which can be found in the Additional file 1: Figure S3-S5).
Differential testes gene expression
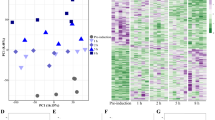
Differentially expressed transcripts were more frequently found in T10 testes samples relative to any the testes of any other treatment group. Compared to T10, the Tvar testes samples had the highest number of differentially expressed transcripts (Table 2 and see Additional file 1: Figure S6). Additionally, all the differentially expressed genes, found in the testes, were hierarchically clustered in a heat map, which is presented in the Additional file 1: Figure S3. This analysis revealed clusters of T10 testes samples. Furthermore, in a PCA analysis, clusters of T10 samples were observed from the normalized expression data of the testis samples.
Among the differentially expressed genes found in the testes, some reproduction-related genes were found to be upregulated in the T10 group relative to Control e.g. follicle-stimulating hormone receptor (fshr; FDR = 0.020), EH domain-containing protein 1 (ehd1; FDR = 0.002; see Additional file 1: Figure S3 and S7), and several growth factor related genes including platelet-derived growth factor receptor beta (pgfrb; FDR = 0.005) or vascular endothelial growth factor C (vegfc; FDR = 0.036; see Additional file 1: Figure S3 and S7). Meanwhile other interesting reproduction-related genes were found to be downregulated in the testes of the T10 group specifically relative to Tvar e.g. OB domain-containing protein (meiob; FDR = 0.025), synaptonemal complex protein 2 (Sycp2; FDR = 0.039), testis expressed protein 11 (tex11; FDR = 0.016), bromodomain testis-specific protein (brdt; FDR = 0.005), and bromodomain-containing protein 2 (brd2; FDR = 0.001 see Additional file 1: Figure S3 and S7).
Differential brain and pituitary gene expression
Similar to the testes transcriptome results, differentially expressed transcripts were more frequently found in T10 samples relative to any other group in the pituitary samples and compared to T10, the Tvar pituitary samples had the highest number of differentially expressed transcripts (Table 2 and see Additional file 1: Figure S6). Additionally, the hierarchical cluster of all differentially expressed genes, found in the pituitary (Additional file 1: Figure S4), revealed clusters of T10 pituitary samples. Also similar to the testis samples, clusters of T10 samples were observed from the normalized expression data of the pituitary in a PCA analysis.
In the brain samples, the quantities of differentially expressed transcripts between groups relative to T10 were more similar; however, T20 showed the highest number of differentially expressed genes (see Additional file 1: Figure S6). Additionally, the hierarchical cluster of all differentially expressed genes, found in the brain (Additional file 1: Figure S5), revealed clusters of both T10 and Tvar brain samples. Furthermore, no cluster could be seen among the brain samples in a PCA analysis (Fig. 5).
Principal component analysis of expression data. Principal component 2 (PC2) over principal component 1 (PC1) from 3 principal component analysis of all normalized expression data from all transcriptomes of the pituitary, testes, and brain samples. Hollow circles labeled “Cont.”, “T20”, and “Tvar” represents transcriptomes of Control, T20 and Tvar samples, respectively. T10 samples are marked with red filled circles, and labelled “T10”
Some of the genes that were found to be upregulated in the brain and pituitary of the T10 group are known for their involvement in thermoception e.g. heat shock protein HSP 90-alpha 1 (h90a1; FDR = 0.00006) and transient receptor potential cation channel subfamily V member 1 (trpv1;; FDR = 0.001; See Additional file 1: Figure S4 and S5). Several genes involved in reproduction were also found to be differentially expressed in the T10 group including, dopamine receptor drd4 (FDR = 0.017) and the estrogen receptor esr1 (FDR = 0.00006; see Additional file 1: Figure S4 and S5). The FDR of the above examples are from the Pituitary T10 samples relative to Control.
Functional annotation
The Fisher’s exact test found enriched GO terms among the differentially expressed genes (Tables 3, 4, 5, 6 and 7 and Figs. 6 and 7, also see Additional file 1: Table S2 and S3). Most notably, the enriched terms found in the brain and pituitary included several GO terms related to immune response. However, several terms related to epigenetic alterations were also found to be enriched among the differentially expressed genes from all the tissues (Tables 4, 6, and 7, Fig. 7 and see Additional file 1: Table S2 and S3). In particular, various functions and processes related to histone modification e.g. “positive regulation of histone H3-K9 methylation”, “histone H3 deacetylation”, “chromatin binding”, or “histone displacement” (Table 2) were found to be enriched. In the pituitary and testes GO terms related to circadian rhythm were also found to be significantly enriched as a result of the T10 treatment (Table 4; Fig. 6 and see Additional file 1: Table S2). Other GO terms found to be enriched among the differentially expressed genes in the testes included the term “male meiotic nuclear division” and “stem cell differentiation”. Specifically, the differentially expressed genes found between T10 and T20 in the pituitary only included 1 enriched GO term “neurohypophyseal hormone activity”, while the biological process of “response to steroid hormone” was highly significant (p = 0.00006) before FDR correction but not after (FDR = 0.16181).
Treemap of BP GO terms found between T10 and Tvar in the pituitary. Treemap of the significantly enriched biological process GO term from the significantly differentially expressed genes found between the T10 and Tvar groups from the pituitary samples. Each rectangle represents a single cluster of related terms. Loosely related single cluster rectangles are clustered together in superclusters of the same color. The size of each cluster is adjusted to reflect the false discovery rate corrected P-value (FDR) of the enrichment of the GO tem (larger rectangles indicates lower FDR)
Treemap of BP GO terms found between T10 and Control in the brain. Treemap of the significantly enriched biological process GO terms from the significantly differentially expressed genes found between the T10 and Control groups from the brain samples. Each rectangle represents a single cluster of related terms. Loosely related single cluster rectangles are clustered together in superclusters of the same color. The size of each cluster is adjusted to reflect the false discovery rate corrected P-value (FDR) of the enrichment of the GO terms (larger rectangles indicates lower FDR)
Discussion
Histological results, biometric parameters and plasma levels of androgens
The highest proportion of SPGAdiff cells was observed as a consequence of the cold seawater treatment (Fig. 2), which together with significantly increased total cell counts (Fig. 2) indicates that the cold seawater treatment promoted the highest rate of SPG differentiation into SPGAdiff cells and SPG proliferation. However, the lower proportion of SPGAund* and SPGAund, and the higher total cell counts of all the treatments compared to the Control, indicate that prolonged housing after saltwater acclimation promotes spermatogonial differentiation and proliferation in European eel testes, regardless of the housing temperature (between 20 and 10 °C; Fig. 2). These processes are likely induced by steroids [31, 32] and are thus an expected consequence of the increases in plasma steroid levels. Several studies on European eel males [26, 33, 34] have documented increased plasma steroid levels as a result of saltwater acclimation alone, and it is, therefore, likely that even the Control group in this experiment represents a state of elevated steroid levels compared to freshwater housed eels, with resulting SPG proliferation and differentiation.
However, the histological data also indicate that the T10 treatment in particular promotes differentiation and proliferation of SPGAund cells into SPGAdiff cells and that the T10 treatment is the only treatment for which prolonged housing did not promote a significant increase in the proportion of SPGB cells (Fig. 2). In an experiment where complete sexual maturation of European eel males was induced through weekly hormonal injections, plasma 11KT levels increased from 1.14 ± 0.5 ng/ml to 4.7 ± 0.37 ng/ml after 1 week of treatment and did not change significantly after the second week [10]. In comparison, our cold seawater treatment induced an increase in the average blood plasma level from 1.67 ± 0.31 ng/ml to 4.46 ± 0.43 ng/ml 11KT. This similarity may suggest that a similar process is initially induced by both types of treatment; however, while the hormonally injected eels all contained spermatocytes in their testis after 2 weeks of treatment, the fish from the T10 group did not contain cells of more advanced developmental stages than SPGB. Therefore, the androgen levels observed, together with the histological results, suggest that the effect of androgens on European eel SPG cell differentiation beyond the SPGAdiff cell stage is being inhibited, during cold seawater treatment. This phenomenon has previously been observed during hormonally induced sexual development of European eel males at 10 °C [26].
Interestingly, while the T10 treatment may induce elevated Lhβ protein levels, the observed histological stage of the T10 samples is highly similar to the stages which eel testes could not surpass during recombinant Lhβ injections [2].
Together, increased total SPG cell counts, increased SPGAdiff abundance, and decreased SPGB abundance indicate that a cold seawater treatment promotes synchronization and increased proliferation of SPG cells at the SPGAdiff stage. It seems reasonable to assume that the synchronization and proliferation of SPG cells inferred here also occur in nature, given that the environmental temperature eels experience during their early oceanic migration [8] is approximately the same as that of our T10 treatment [7, 8].
The inferred proliferation might have been stimulated by androgens since the T10 group contained significantly higher amounts of T and 11KT than the Control group. Furthermore, higher plasma androgen levels have been correlated with European eel SPGA proliferation in previous studies [26]. As mentioned above, increases in steroid levels have been reported in European eel after salinity acclimation [24, 26, 33]; however, the duration of these increases has not been reported. Since the Control and T20 groups share all the same parameters except exposure time, the observed increase in 11KT levels (Fig. 3) may well be the result of a continued increase in 11KT levels rather than a specific increase that happens in the 2nd and 4th weeks of seawater housing.
Few significant differences were registered in terms of the measured biometric parameters. This was to be expected due to the short treatment period, and lack of hormonal injections. Only lower fin index was observed, which has been traditionally attributed to a less mature sexual developmental stage of European eels [10, 35], although fin length has not been found to correlate with maturity stage in other studies [36]. Interestingly, the T20 group also showed a significantly lower fin index compared to the Control group, and therefore this change may not be a result of the temperature treatment, but rather a result of the prolonged fasting or housing in seawater during the experiment. A similar tendency was seen in the Tvar group, although without significant differences.
Gonadotropins
Pituitary gonadotropins stimulate testicular spermatogenesis and steroidogenesis. In both European eel males and females, lhb and fshb were shown to be differentially expressed in the pituitary during gametogenesis, with an maximum of fshb expression occurring early on in the sexual developmental process, and a later maximum of lhb expression [10, 37]. In eels, both gonadotropins have further been shown to induce 11KT and T production from the testes [2, 38]. While 11KT can induce complete spermatogenesis in vitro [39, 40], this is not the case in vivo [41]; however, 11KT has been shown to stimulate the expression of the Fsh receptor (fshr) [42] and thereby Fsh sensitivity and activity [43, 44]. In Japanese eel, Fsh is an important factor in spermatogonial proliferation; however, only in combination with steroidogenesis [39]. Furthermore, a positive feedback loop of sex steroids on gonadotropins appears to exist in European eels, as androgens have been shown to stimulate Lhb expression from the pituitary [45]. Our immunofluorescence labeling of Fsh did indicate that Fsh was present in all collected pituitaries, therefore it is plausible that Fsh was a mediating factor of the observed steroid increase and/or the documented proliferation in the T10 group. We draw this conclusion based on the expression of fshr in the testes, which was up-regulated by our cold seawater treatment (see Additional file 1: Figure S3 and S7), thus Fsh sensitivity and activity in the testes may have been enhanced. Our immunofluorescence labelling of Lhβ indicates that our cold seawater treatment induces a consistently stronger Lhβ signal in the pituitary. Higher Lh levels could, in turn, also be a stimulating factor generating the observed increases in androgens and thereby fshr expression and proliferation; however, since androgens can also stimulate Lhβ production, the question remains as to which factor came first. Furthermore, since Lhβ release was not analyzed in this study it is possible that the lower Lhβ levels suggested in the T20 and Control groups are in fact a result of a higher Lh release. However, increased lhb expression supports a hypothesis of increased protein production in the pituitaries of the eels from the T10 group.
The FDR correction applied in this study to all gene expression analyses is rather conservative, making the significant results obtained after FDR correction reliable [46]. This claim is further supported by the immunofluorescence labelling results of Lhβ, as the lhb gene expression differences observed were not significant after FDR correction, yet strong enough for a consistently stronger Lhβ signal. Furthermore, the concurrence of the immunofluorescence labelling results and DEseq analysis results suggests that our findings are reproducible, at least for lhb, since the fish used for immunofluorescence labelling were treated in the second experimental run, while those used for the transcriptome analysis were treated in the first experimental run. Of course, as only Lhβ was successfully analysed, this result only provide evidence for the Lhβ signal itself. Furthermore, due to the nature of immunofluorescence labelling and the use of an objective “quantitative” measuring technique these results only provides suggestive evidence of the reproducibility of our experiment and of the validity of the FDR correction applied.
Transcriptomic analysis
Differential brain and pituitary gene expression
The significant effect of the cold seawater treatment indicates that some mechanisms of thermoception were activated. Specifically, some genes found differentially expressed in the brain and pituitary are known for their involvement in thermoception e.g. h90a1 and trpv1 (See Additional file 1: Figure S4 and S5) and thus these genes may be involved in the registration of temperature differences, which could be the driver of the changes observed in this study.
Although the GO term “response to steroid hormone” was not significantly enriched after FDR correction, some interesting significantly differentially expressed genes were assigned this term. Among these were drd4 and esr1. It has been suggested that dopamine may be involved in the sexual developmental blockage of puberty in European eels [47] and the D2-like receptor, drd4, was significantly down-regulated in the pituitary after the cold seawater treatment (see Additional file 1: Figure S4). Although speculative, this could indicate a weakening of dopamine-mediated neuroendocrine inhibition of eel puberty. The pituitary is a major target for estrogen in European eel [33]. In our data, esr1 expression increased significantly in the pituitary after the T10 treatment (see Additional file 1: Figure S4). This result may suggest stimulation of sexual development, as the expression of esr1 has been shown to increase in the pituitary of European eel males early on in artificial maturation [33].
The most notable results from the transcriptomic data from all the tissues were the enrichment of GO terms related to the immune response in the brain and pituitary. No visual signs of infection were seen on the animals during the experiment or at sampling, which would be expected if the massive differential expression of immune related genes was caused by an infection. Additionally, the brain and pituitary are not the most likely organs to observe differential expression patterns caused by an infection. Interestingly, several studies have documented a neural function for most of the enriched immune response GO terms found in this study (reviewed by [48, 49]). E.g. cytokines [50], Toll-like receptors [50], major histone complexes (MHC) [51,52,53], and T-cell receptors [48, 54, 55] have documented functions in neural development. Specifically, T-cell receptor signaling has been shown to be conveyed through cell-cell contact through MHC [55] and it has been speculated that the pruning of synapses of the visual system [56] can be facilitated by MHC/T-cell receptor signaling [48]. As such, there seems to be a high occurrence of genes with documented and connected neural functions among the differentially expressed genes related to immune functions found in the brain and pituitary in the current study. The hypothesized involvement of these genes in the pruning of synapses of the visual system leads to the speculation that the cold seawater treatment affects the synapses of the visual system in the eel brain. Since the light environment of migrating eels is vastly different from that of yellow eels foraging in shallow freshwaters, changes to the synapses of the visual system have been hypothesized to be part of the adaptation of eels in preparation for migration [57]. Furthermore, the upregulation of genes involved in photo signal transduction and visual system development [58], and alterations to the retina, have previously been observed in developing European eels [57].
Differential gene expression in the testes
The GO terms found to be enriched among the genes differentially expressed between T10 and Tvar groups, included the term “male meiotic nuclear division”. As previously discussed, a mechanism repressing spermatogonial differentiation towards meiosis may have been activated by the T10 treatment, as a decrease in differentiation beyond the SPGAdiff cell stage, relative to T20 and Tvar groups was observed. This mechanism could be driven by an active downregulation/ inhibition of upregulation of genes involved in later sexual developmental processes, including meiosis, which could serve to optimize the synchronization of sexual development. The genes annotated to the GO term “male meiotic nuclear division”, could be involved in such a process, as the vast majority of these genes were down regulated in the T10 testis samples relative to Tvar. Some of these downregulated genes were meiob, Sycp2, tex11, brdt, and brd2 (see Additional file 1: Figure S3 and S7), all of which may, therefore, be interesting factors to analyse in future studies on the latter developmental stages of the European eel.
Epigenetic factors
The GO terms found to be enriched among the differentially expressed genes found in the testes, between the T10 and the Control groups, were often related to epigenetic alterations, similar to those seen in the pituitary and the brain (Tables 4, 6, and 7, Fig. 7 and see Additional file 1: Table S2 and S3). In particular, various functions and processes related to histone modification were found to be enriched. Histone modification can affect the alteration of transcription as a result of posttranslational modifications in the N-terminal tail of the histone proteins [59, 60]. Specifically methylation changes of H3-K9 have, interestingly, been shown to be dependent on cold temperatures in Arabidopsis thaliana [61] and regulate gametogenesis specifically at the meiotic prophase in mice [62].
Circadian rhythm factors
GO terms related to circadian rhythm were also found to be significantly enriched in the testes and pituitary as a result of the T10 treatment (Table 4; Fig. 6 and see Additional file 1: Table S2). The circadian clock is a central oscillator, which coordinates endogenous rhythms in the host. Although light is the strongest modulator, temperature has also been shown to influence the circadian rhythm system, especially in the absence of a light cycle [63, 64]. The strong regulation of the circadian rhythm system, caused by our T10 treatment, supports our hypothesis that the T10 treatment may have initiated alterations that the eels would naturally experience during early migration.
Other differentially expressed genes
Among the other significantly upregulated genes which were not related to enriched pathways were fshr, and ehd1 (see Additional file 1: Figure S3 and S7). These genes are particularly important for the stimulation of early teleost sexual development. ehd1 specifically, has been shown to be expressed in both Sertoli cells and spermatogonia, and to be vital in the pre-pubertal sexual development and spermatogenesis of mice [65]. Furthermore, the genes found to be differentially expressed in the testes between the T10 and Control groups, also included several growth factor related genes including pgfrb or vegfc (see Additional file 1: Figure S3 and S7). Growth factor related genes have been associated with early sexual development in teleost testes with decreasing expressions at later developmental stages [31].
No GO terms were found to be significantly enriched (FDR < 0.05) among the 94 differentially expressed genes found between T10 and T20 in the testes. Since the Control and the T20 groups shared all the same conditions other than exposure time, a similar array of enriched GO terms were expected to be found within the differentially expressed genes from these groups relative to T10. Notably, when expanding the significance threshold to 0.1 (FDR < 0.1), the GO terms “positive regulation of histone H3-K9 methylation”, “regulation of transcription involved in meiotic cell cycle”, “positive regulation of transcription involved in meiotic cell cycle” and “histone displacement” were found to be enriched. As the genes annotated to these GO terms are significantly differentially expressed following the same criteria as all the others, this indicates that the processes affected are similar in groups T20, Control and Tvar and therefore differ in a similar fashion to those of T10. Nevertheless, as shown in the histological results, the differences between T10 and T20 seem less pronounced than those found between T10 and Control.
Conclusion
In this study, clear effects of a cold seawater treatment were observed in European eel males, including an increase in proliferation and differentiation of SPGAund into SPGAdiff cells, decrease in the differentiation of SPGAdiff cells into early SPGB cells, changes in blood plasma steroid levels, possible increase in pituitary Lhβ protein levels, and BPG-axis transcriptomes. These results support our hypothesis that a cold seawater treatment causes a physiological transition that European eels naturally experience during the early stages of their oceanic migration. This hypothesis is logical given that the average temperature experienced by the eels in the early stages of their oceanic migration is highly similar to that of our cold seawater treatment. Apart from preparing the eels for migration, the hypothesized natural transition could improve the reproductive potential of eel males, which is indicated by the increased androgen levels [66] and by increasing spermatogonial proliferation and synchronization. However, further studies would need to be conducted to test whether the cold seawater treatment can improve the eels´ response to hormonal treatments.
Materials and methods
Fish maintenance
110 farmed European eel males (mean body weight 97.5 ± 1.97 g) were supplied by Valenciana de Acuicultura S.A. (Puzol, Valencia, Spain) and transported to the Aquaculture Laboratory at the Universitat Politècnica de València (Valencia, Spain), in 2 batches. The fish were kept in 200-L tanks, equipped with individual recirculation systems, temperature control systems (with heaters and coolers), and aeration. The fish were gradually acclimated to seawater (final salinity 37 ± 0.3‰), over the course of 2 weeks. The temperature, oxygen level and pH of rearing were 20 °C, 7–8 mg/L and ~ 8.2, respectively. The tanks were covered to keep the level of light as low as possible and to reduce fish stress. The fish were not fed throughout the experiment and were sacrificed using an overdose of anesthesia (benzocaine).
Experimental design
The following experiment was conducted twice with the same acclimation, control, and treatment but with different n’s and samples collected. The first experimental run was conducted with a total of 70 fish while the second run included 40 fish. In both runs, before the experiments began, ten fish were sacrificed at the end of the acclimation period to act as the Control group, and biometric measurements were collected. The biometric measurements included: total weight, total length, vertical and horizontal eye diameters, fin color, liver weight, and pectoral fin length. From these measurements, the eye index [35], fin index [36] and HSI were calculated as: (eye area / total length) X 100, (fin length / total length) X 100, and (liver weight / total weight) X 100, respectively. Precise gonadosomatic indexes could not be calculated due to the low testes weight, as a consequence of the early sexual developmental state.
In the first run of the experiment, blood samples from the caudal vein were taken from all sacrificed fish and kept in heparinized vials, centrifuged (3500 rpm, 15 min), and blood plasma was stored at 4 °C. Sampled pituitaries, forebrain (telencephalon, diencephalon, and olfactory bulb), and testes from 3 fish were stored in RNA-later at 4 °C for 24 h and then at − 20 °C until RNA extraction. Additional testis samples were fixed in 10% paraformaldehyde (PFA) diluted in PBS (pH 7.4; 10% PFA-PBS) for histological analysis. In the second run of the experiment, only the pituitaries were sampled and immediately fixed in ice-cold 4% PFA in PBST (PBS with 0.1% Tween 20, pH = 7.4).
After this control sampling, in both runs, the remaining fish were randomly distributed into 3 200-L tanks with the same conditions that the fish experienced after seawater and temperature acclimation. These 3 tanks were then set up to expose the fish to 3 different temperature regimes for 2 weeks. The 3 regimes included 2 with a constant temperature of 10 °C (T10) or 20 °C (T20), and 1 with a variable temperature regime (Tvar) which alternated between 10 and 20 °C every 12 h. No hormonal treatments were administered at any time. After the 2 weeks of thermal treatment, biometric measurements were collected from all the fish from both experimental runs. From the first experimental run 3 samples of brain, pituitary, and testes, were collected from 3 fish per group for transcriptome analysis and blood was collected for RIA steroid analysis from all the sacrificed fish. From the second experimental run, the pituitaries of 10 fish per group were sampled for immunofluorescence visualization of gonadotropins.
Histology
The testis samples collected from the first experimental run and fixed in 10% PFA-PBS, were dehydrated in increasing percentages of ethanol, after which the samples were embedded in paraffin. Sections 5–10 μm thick were cut with a Shandom Hypercut manual microtome and stained with hematoxylin and eosin. The slides were then observed with a Nikon Eclipse E-400 microscope, and pictures were taken with a Nikon DS-5 M camera attached to the microscope. Cell types (Fig. 1) were categorized following the description suggested by Schulz et al. [31]. As such, the most undifferentiated SPG type A cells (SPGAund*; Fig. 1) were characterized as single cells, surrounded by Sertoli cells, with irregular or convoluted nuclear envelopes, with low nuclear heterochromatin, 1 or 2 nuclei, and containing large perinuclear amounts of the electron-dense material called “nuage”. The second most undifferentiated SPG type A cells (SPGAund; Fig. 1) were characterized as single cells, surrounded by Sertoli cells, with regular nuclear envelopes, 1 prominent nucleolus, with low levels of nuclear heterochromatin, and containing low amounts of perinuclear nuage. Differentiated SPG type A cells (SPGAdiff; Fig. 1) were characterized as cells found in clusters of 2–8 cells surrounded by Sertoli cells, with regular and round or oval nuclear envelopes, 1 or more nucleolus, and with low levels of nuclear heterochromatin or perinuclear nuage. Early SPG type B cells (early SPGB; Fig. 1) were characterized as smaller cells, with little cytoplasmic volume, found in clusters of many cells, with an oval or round nucleus with large amounts of heterochromatin. Furthermore, some cells were identified as SPG cells but could not be distinguished into a specific SPG type (Undefined cells) e.g. due to unclear Sertoli cell projections, broken cells, unfocused field area etc. The number of each cell type was counted, using FIJI/ImageJ software, from 5 microscope fields per sample, and from ten samples per treatment group and the Control.
Steroid analysis
Heparinized blood plasma samples were assayed for plasma T and 11KT levels by radioimmunoassays (RIA) following the protocol described by Schulz [67]. Assay characteristics and cross-reactivities of T antisera have previously been examined by Frantzen et al. [68] and further validated for eel plasma by Mazzeo et al. [27]. The cross-reactivity of the 11KT antiserum used in the current study has previously been described by Johnsen et al. [69] and validated for European eel plasma by Baeza et al. [34]. In summary, 5 mL diethylether was used to extract free steroids from 100 to 300 μL plasma by mixing and shaking for 4 min. The aqueous phase was then frozen in liquid nitrogen and the organic phase was transferred to a glass tube. Diethylether was then evaporated in a water bath at 45 °C and the sample was then reconstituted by the addition of 3X volume of RIA-buffer (300–900 μL) and then assayed for each steroid.
RNA extraction and sequencing
Total RNA of brain, pituitary and testis samples of 3 fish were extracted using Ambion (mirVana) and Qiagen (AllPrep) columns following the protocol of Peña-Llopis and Brugarolas [70]. Resulting RNA was quality and quantity tested on a bioanalyser (Agilent Technologies, USA). RNA samples with RIN values > 8 and with > 3 μg of total RNA were selected for sequencing. Total RNA samples were shipped to the company Macrogen Korea (Seoul, South Korea). Here, mRNA purification was carried out using Sera-mag Magnetic Oligo (dT) Beads, followed by buffer fragmentation. Reverse transcription was followed by PCR amplification to prepare the samples for sequencing, in an Illumina Hiseq-4000 sequencer (Illumina, San Diego, USA), keeping the strand information. The resulting raw sequences are available at the NCBI Sequence Read Archive (SRA) as stated in the section titled “Availability of data and materials”.
Transcriptome analysis
Raw reads obtained from Macrogen were quality assessed using fastQC software [71] and were quantified with RSEM [72] using our de novo European eel transcriptome [73] as a template. The differentially expressed transcripts were annotated using the Trinotate functional annotation pipeline [74] and assigned GO terms by blasting them to the EggNOG gene family database [75]. Successfully annotated transcripts have been described as genes in the results and discussion sections. Fisher’s analysis of enrichment was performed on these GO terms [76], to assess significantly affected functions and processes.
Immunofluorescence
The experiment (explained above) was repeated with ten fish per treatment group and from the Control. Immunolabelling of European eel Lhβ and Fshβ proteins was carried out using the pituitaries of these fish. For this procedure the fixed pituitaries were dehydrated in an increasing gradient series of ethanol solutions and preserved in 100% methanol at − 20 °C until rehydration in decreasing concentrations of ethanol, embedding in 3% agarose, and then cutting into 60 μm thick sagittal sections with a vibratome (Leica VT 1000 S, Leica Biosystems GmbH, Nussloch, Germany). 5–10 sections per sample were divided into 2 sets and incubated with agitation for 1 h at room temperature in blocking solution (normal goat serum 4%, dimethyl sulfoxide 1%, and Triton X-100 (Sigma-Aldrich) 0.3%, in PBST) followed by overnight incubation at 4 °C, with agitation, with a rabbit antibody specific to European eel Fshβ or Lhβ (Rara Avis Biotec S.L., Valencia, Spain), in either set. Hereafter the sections were incubated for 4 h at room temperature with a goat anti-rabbit IgG coupled to Rb488 (Jackson Immuno Research Europe Ltd.) as the secondary antibody. Finally, sections were treated with DAPI (4,6-diamidino-2-phenylindole dihydrochloride, 1:1000, Sigma-Aldrich) for overnight nuclear counterstaining at 4 °C. The stained sections were mounted on slides using Vectashield H-1000 (Vector laboratories, Burlingame, CA). The results were evaluated with a fluorescence microscope and the fields were captured with the same image parameters. Signal intensity was evaluated on a scale of 1–5 in a blind test. The highest signal from each group is presented in the results section as pictures taken with a Zeiss LSM710 laser scanning confocal microscope equipped with a 10X Plan Neofluar objective lens (N.A. 0.3). The presented pictures were adjusted for brightness and contrast using FIJI/ImageJ software (Fig. 4). Control experiments were performed on tissue slices using the same protocol but without the primary antibody.
Statistics
Results are shown as the mean ± standard error of the mean (SEM) and differences were considered significant when P-values < 0.05, when not otherwise specified. We used R version 3.1.3 (R core team, 2015) to perform Pearson’s Chi-squared tests with simulated P-values to compare the distribution of fin colour between the 3 treatment groups (T10, T20, and Tvar) and the Control group. After establishing data normality using the asymmetry standard coefficient and Curtosis coefficient we also used R to run general linear models (GLM) to identify significant differences between the groups in the remaining biometric parameters as well as differences in steroid levels. We used a generalized linear mixed model with a negative binominal distribution, and with "field "as a random effect, to compare cell counts and proportions of the histological testis analysis. For these test the following command line was executed with R version 3.1.3: “glmmPQL (cell type ~ treatment, random = ~1|field, family= negative.binomial (theta = 1), data=our_data)”. We furthermore used R to perform a Principal Component Analysis (PCA) for all the quantified expression data from the RNA-sequencing results. Only principal component 1 and 2 were included in the results as they together account for > 98% of the variance in the data, from all tissues. Significantly differentially expressed transcripts were located with DEseg [46] with a threshold for a false discovery corrected P-value (FDR) of < 0.05. R was also used to run one-way ANOVA tests of the TPM of both gonadotropin beta-subunits from the pituitaries; in the case of lhb a log transformation of the data was performed to improve the homogeneity of variance across the groups. Finally, R was used to create heat maps and unsupervised hierarchical clusters using a Euclidean distribution of all significantly differentiated expressed genes from each tissue.
Availability of data and materials
The raw RNA-sequencing reads from forebrain, pituitary, and testis samples from European eel (Anguilla anguilla) are available in GenBank [77] under accession no. SRP126643.
References
Jacoby D, Gollock M. Anguilla anguilla. The IUCN Red List of Threatened Species: IUCN; 2014. Version 2014.3. http://www.iucnredlist.org
Peñaranda DS, Gallego V, Rozenfeld C, Herranz-Jusdado JG, Pérez L, Gómez A, et al. Using specific recombinant gonadotropins to induce spermatogenesis and spermiation in the European eel (Anguilla anguilla). Theriogenology. 2018;107:6–20.
Pérez L, Asturiano JF, Tomás A, Zegrari S, Barrera R, Espinós FJ, et al. Induction of maturation and spermiation in the male European eel: assessment of sperm quality throughout treatment. J Fish Biol. 2000;57(6):1488–504.
Butts IAE, Sørensen SR, Politis SN, Pitcher TE, Tomkiewicz J. Standardization of fertilization protocols for the European eel, Anguilla anguilla. Aquaculture. 2014;426–427:9–13.
Mylonas CC, Duncan NJ, Asturiano JF. Hormonal manipulations for the enhancement of sperm production in cultured fish and evaluation of sperm quality. Aquaculture. 2017;472:21–44.
van Ginneken V, Antonissen E, Müller UK, Booms R, Eding E, Verreth J, et al. Eel migration to the Sargasso: remarkably high swimming efficiency and low energy costs. J Exp Biol. 2005;208:1329–35.
Aarestrup K, Økland F, Hansen MM, Righton D, Gargan P, Castonguay M, et al. Oceanic spawning migration of the European eel (Anguilla anguilla). Science. 2009;325:12–3.
Righton D, Westerberg H, Feunteun E, Okland F, Gargan P, Amilhat E, et al. Empirical observations of the spawning migration of European eels: the long and dangerous road to the Sargasso Sea. Sci Adv. 2016;2(10):e1501694.
Boëtius I, Boëtius J. Studies in the European Eel, Anguilla anguilla (L.). Experimental induction of the male sexual cycle, its relation to temperature and other factors. Medd Dan Fish. 1967;ser.;(4):339–405.
Peñaranda DS, Pérez L, Gallego V, Jover M, Tveiten H, Baloche S, et al. Molecular and physiological study of the artificial maturation process in European eel males: from brain to testis. Gen Comp Endocrinol. 2010;166(1):160–71.
Asturiano JF, Marco-Jiménez F, Pérez L, Balasch S, Garzón DL, Peñaranda DS, et al. Effects of hCG as spermiation inducer on European eel semen quality. Theriogenology. 2006;66(4):1012–20.
Webb MAH, Van Eenennaam JP, Doroshov SI, Moberg GP, Van Eenennaam JP. Preliminary observations on the effects of holding temperature on reproductive performance of female white sturgeon, Acipenser transmontanus Richardson. Aquaculture. 1999;176(3–4):315–29.
Webb MAH, Van Eenennaam JP, Feist GW, Linares-Casenave J, Fitzpatrick MS, Schreck CB, et al. Effects of thermal regime on ovarian maturation and plasma sex steroids in farmed white sturgeon, Acipenser transmontanus. Aquaculture. 2001;201(1–2):137–51.
Clark RW, Henderson-Arzapalo A, Sullivan CV. Disparate effects of constant and annually-cycling daylength and water temperature on reproductive maturation of striped bass (Morone saxatilis). Aquaculture. 2005;249(1–4):497–513.
Hokanson KEF. Temperature requirements of some percids and adaptations to the seasonal temperature cycle. J Fish Res Board Canada. 1977;34(10):1524–50.
Wang N, Teletchea F, Kestemont P, Milla S, Fontaine P. Photothermal control of the reproductive cycle in temperate fishes. Rev Aquac. 2010;2(4):209–22.
Tveiten H, Johnsen HK. Temperature experienced during vitellogenesis influences ovarian maturation and the timing of ovulation in common wolffish. J Fish Biol. 1999;55(4):809–19.
Davie A, Porter MJ, Bromage NR, Migaud H. The role of seasonally altering photoperiod in regulating physiology in Atlantic cod (Gadus morhua). Part II. Somatic growth. Can J Fish Aquat Sci. 2007;64(1):98–112.
Hansen T, Karlsen Ø, Taranger GL, Hemre GI, Holm JC, Kjesbu OS. Growth, gonadal development and spawning time of Atlantic cod (Gadus morhua) reared under different photoperiods. Aquaculture. 2001;203(1–2):51–67.
Suquet M, Normant Y, Gaignon JL, Quéméner L, Fauvel C. Effect of water temperature on individual reproductive activity of pollack (Pollachius pollachius). Aquaculture. 2005;243(1–4):113–20.
Prat F, Zanuy S, Bromage N, Carrillo M. Effects of constant short and long photoperiod regimes on the spawning performance and sex steroid levels of female and male sea bass. J Fish Biol. 1999;54(1):125–37.
Mañanós EL, Zanuy S, Carrillo M, Mananos EL, Zanuy S, Carrillo M. Photoperiodic manipulations of the reproductive cycle of sea bass (Dicentrarchus labrax) and their effects on gonadal development, and plasma 17 beta-estradiol and vitellogenin levels. Fish Physiol Biochem. 1997;16(64):211–22.
Migaud H, Fontaine P, Sulistyo I, Kestemont P, Gardeur JN. Induction of out-of-season spawning in Eurasian perch Perca fluviatilis: effects of rates of cooling and cooling durations on female gametogenesis and spawning. Aquaculture. 2002;205(3–4):253–67.
Baeza R, Mazzeo I, Vílchez MC, Gallego V, Peñaranda DS, Pérez L, et al. Effect of thermal regime on fatty acid dynamics in male European eels (Anguilla anguilla) during hormonally-induced spermatogenesis. Aquaculture. 2014;430:86–97.
Pérez L, Peñaranda DS, Dufour S, Baloche S, Palstra AP, Van Den Thillart GEEJM, et al. Influence of temperature regime on endocrine parameters and vitellogenesis during experimental maturation of European eel (Anguilla anguilla) females. Gen Comp Endocrinol. 2011;174(1):51–9.
Peñaranda DS, Morini M, Tveiten H, Vílchez MC, Gallego V, Dirks RP, et al. Temperature modulates testis steroidogenesis in European eel. Comp Biochem Physiol A Mol Integr Physiol. 2016;197:58–67.
Mazzeo I, Peñaranda DS, Gallego V, Baloche S, Nourizadeh-Lillabadi R, Tveiten H, et al. Temperature modulates the progression of vitellogenesis in the European eel. Aquaculture. 2014;434:38–47.
Ahn H, Yamada Y, Okamura A, Horie N, Mikawa N, Tanaka S, et al. Effect of water temperature on embryonic development and hatching time of the Japanese eel Anguilla japonica. Aquaculture. 2012;330–333:100–5.
Gallego V, Mazzeo I, Vílchez MC, Peñaranda DS, Carneiro PCF, Pérez L, et al. Study of the effects of thermal regime and alternative hormonal treatments on the reproductive performance of European eel males (Anguilla anguilla) during induced sexual maturation. Aquaculture. 2012;354–355:7–16.
Li H, Durbin R. Fast and accurate long-read alignment with burrows-wheeler transform. Bioinformatics. 2010;26(5):589–95.
Schulz RW, Renato L, França D, Lareyre J, Legac F, Chiarini-garcia H, et al. Spermatogenesis in fish. Gen Comp Endocrinol. 2010;165(3):390–411.
Miura T, Miura CI. Molecular control mechanisms of fish spermatogenesis. Fish Physiol Biochem. 2003;28:181–6.
Morini M, Peñaranda DS, Vílchez MC, Nourizadeh-Lillabadi R, Lafont AG, Dufour S, et al. The expression of nuclear and membrane estrogen receptors in the European eel throughout spermatogenesis. Comp Biochem Physiol -Part A Mol Integr Physiol. 2017;207:79–92.
Baeza R, Peñaranda DS, Vílchez MC, Tveiten H, Pérez L, Asturiano JF. Exploring correlations between sex steroids and fatty acids and their potential roles in the induced maturation of the male European eel. Aquaculture. 2015;435:328–35.
Pankhurst NW. Relation of visual changes to the onset of sexual-maturation in the European eel Anguilla anguilla (L). J Fish Biol. 1982;21(2):127–40.
Durif CMF, Dufour S, Elie P. Impact of silvering stage, age, body size and condition on reproductive potential of the European eel. Mar Ecol Prog Ser. 2006;327:171–81.
Schmitz M, Aroua S, Vidal B, Le Belle N, Elie P, Dufour S. Differential regulation of luteinizing hormone and follicle-stimulating hormone expression during ovarian development and under sexual steroid feedback in the European eel. Neuroendocrinology. 2005;81(2):107–19.
Kazeto Y, Kohara M, Miura T, Miura C, Yamaguchi S, Trant JM, et al. Japanese eel follicle-stimulating hormone (Fsh) and luteinizing hormone (Lh): production of biologically active recombinant Fsh and Lh by Drosophila S2 cells and their differential actions on the reproductive biology. Biol Reprod. 2008;79(5):938–46.
Ohta T, Miyake H, Miura C, Kamei H, Aida K, Miura T. Follicle-stimulating hormone induces spermatogenesis mediated by androgen production in Japanese eel, Anguilla japonica. Biol Reprod. 2007;77(6):970–7.
Miura T, Yamauchi K, Takahashi H, Nagahama Y. Hormonal induction of all stages of spermatogenesis in vitro in the male Japanese eel (Anguilla japonica). Physiology/Pharmacology. 1991;88:5774–8.
Lokman PM, Damsteegt EL, Wallace J, Downes M, Goodwin SL, Facoory LJ, et al. Dose-responses of male silver eels, Anguilla australis, to human chorionic gonadotropin and 11-ketotestosterone in vivo. Aquaculture. 2016;463:97–105.
Levavi-Sivan B, Bogerd J, Mañanós EL, Gómez A, Lareyre JJ. Perspectives on fish gonadotropins and their receptors. Gen Comp Endocrinol. 2010;165(3):412–37.
García-López Á, Bogerd J, Granneman JCM, Van Dijk W, Trant JM, Taranger GL, et al. Leydig cells express follicle-stimulating hormone receptors in African catfish. Endocrinology. 2009;150(1):357–65.
Planas JV, Swanson P. Maturation-associated changes in the response of the salmon testis to the steroidogenic actions of gonadotropins (GTH I and GTH II) in vitro. Biol Reprod. 1995;52(3):697–704.
Huang YS, Schmitz M, Le Belle N, Chang CF, Quérat B, Dufour S. Androgens stimulate gonadotropin-II β-subunit in eel pituitary cells in vitro. Mol Cell Endocrinol. 1997;131(2):157–66.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(R106):1–12.
Vidal B, Pasqualini C, Le Belle N, Holland MCH, Sbaihi M, Vernier P, et al. Dopamine inhibits luteinizing hormone synthesis and release in the juvenile European eel: a neuroendocrine lock for the onset of puberty. Biol Reprod. 2004;71(5):1491–500.
Komal P, Nashmi R. T-cell receptors modify neuronal function in the central nervous system. Biochem Pharmacol. 2015;97(4):512–7.
Boulanger LM. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64(1):93–109.
Huising MO, Stet RJM, Savelkoul HFJ, Verburg-Van Kemenade BML. The molecular evolution of the interleukin-1 family of cytokines; IL-18 in teleost fish. Dev Comp Immunol. 2004;28(5):395–413.
Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21(3):505–20.
Shatz CJ. MHC Class I: An unexpected role in neuronal plasticity. Neuron. 2009;64(1):40–5.
Needleman LA, Liu XB, El-Sabeawy F, Jones EG, McAllister AK. MHC class I molecules are present both pre- and postsynaptically in the visual cortex during postnatal development and in adulthood. Proc Natl Acad Sci. 2010;107(39):16999–7004.
Syken J, Shatz CJ. Expression of T cell receptor beta locus in central nervous system neurons. Proc Natl Acad Sci U S A. 2003;100(22):13048–53.
Komal P, Gudavicius G, Nelson CJ, Nashmi R. T-cell receptor activation decreases excitability of cortical interneurons by inhibiting 7 nicotinic receptors. J Neurosci. 2014;34(1):22–35.
Lee H, Brott BK, Kirkby LA, Adelson JD, Cheng S, Feller MB, et al. Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature. 2014;509(7499):195–200.
Pankhurst NW, Lythgoe JN. Changes in vision and olfaction during sexual maturation in the European eel Anguilla anguilla (L.). J Fish Biol. 1983;23(2):229–40.
Churcher AM, Pujolar J, Milan M, Hubbard PC, Martins RS, Saraiva JL, et al. Changes in the gene expression profiles of the brains of male European eels (Anguilla anguilla) during sexual maturation. BMC Genomics. 2014;15(1):799.
Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80.
Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–5.
Berr A, Shafiq S, Shen WH. Histone modifications in transcriptional activation during plant development. Biochim Biophys Acta - Gene Regul Mech. 2011;1809(10):567–76.
Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26(14):3346–59.
López-Olmeda JF, Sánchez-Vázquez FJ. Zebrafish temperature selection and synchronization of locomotor activity circadian rhythm to ahemeral cycles of light and temperature. Chronobiol Int. 2009;26(2):200–18.
Lahiri K, Vallone D, Gondi SB, Santoriello C, Dickmeis T, Foulkes NS. Temperature regulates transcription in the zebrafish circadian clock. PLoS Biol. 2005;3(11):2005–16.
Rainey MA, George M, Ying G, Akakura R, Burgess DJ, Siefker E, et al. The endocytic recycling regulator EHD1 is essential for spermatogenesis and male fertility in mice. BMC Dev Biol. 2010;37(10):1–19.
Burgerhout E, Minegishi Y, Brittijn SA, de Wijze DL, Henkel CV, Jansen HJ, et al. Changes in ovarian gene expression profiles and plasma hormone levels in maturing European eel (Anguilla anguilla); biomarkers for broodstock selection. Gen Comp Endocrinol. 2016;225:185–96.
Schulz R. Measurement of five androgens in the blood of immature and maturing male rainbow trout, Salmo gairdneri (Richardson). Steroids. 1985;46(2–3):717–26.
Frantzen M, Arnesen AM, Damsgård B, Tveiten H, Johnsen HK. Effects of photoperiod on sex steroids and gonad maturation in Arctic charr. Aquaculture. 2004;240(1–4):561–74.
Johnsen H, Tveiten H, Torgersen JS, Andersen Ø. Divergent and sex-dimorphic expression of the paralogs of the Sox9-Amh-Cyp19a1 regulatory cascade in developing and adult Atlantic cod (Gadus morhua L.). Mol Reprod Dev. 2013;80(5):358–70.
Peña-Llopis S, Brugarolas J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat Protoc. 2013 Nov;8(11):2240–55.
Andrews S. FastQC: A quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323.
Rozenfeld C, Blanca J, Albiach VG, García-Carpintero V, Herranz-Jusdado JG, Pérez L, et al. Large scale gene duplication affected the European eel (Anguilla anguilla) after the 3R teleost duplication. bioRxiv. 2017; preprint:232918.
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512.
Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:286–93.
Alexa A, Rahnenfuhrer J. topGO: Enrichment analysis for gene ontology. 2016. R package version 2.29.0.
National Center for Biotechnology Information (NCBI) GenBank. Http://www.ncbi.nlm.nih.gov/genbank. Access no SRP126643. Accessed 09-09-2018.
Acknowledgements
Not applicable.
Funding
This work was subsidized by the Spanish Ministry of Science and Innovation (REPRO-TEMP project, AGL2013–41646-R), the Spanish Ministry of Innovation, Science and Universities (EELGONIA project, RTI2018-096413-B-I00), and by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement N° 642893 (IMPRESS), including the CR and JGHJ predoctoral contracts. JGHJ was granted a Short Term Scientific Mission by the COST Office (COST Action AQUAGAMETE: Assessing and improving the quality of aquatic animal gametes to enhance aquatic resources. The need to harmonize and standardize evolving methodologies, and improve transfer from academia to industry). The funding bodies had no role in the study, analysis, interpretation of data, or in writing the manuscript.
Author information
Authors and Affiliations
Contributions
JFA, DSP, FAW, and LP conceived this research. JFA, CR, VG, JGHJ, and LP, carried out animal care, treatment, and sampling for the experiments. CR, DSP, performed RNA extraction and handled the contact and shipment of samples for the sequencing service. JGHJ, HT, and HKJ performed steroid quantifications. CR and RF performed immunofluorescence labelling. LP and CR produced and analysed the histological data. VGC, JC, and CR performed transcriptomic data analyses. CR wrote the manuscript. JFA and DSP assembled the main manuscript. All the co-authors contributed equally to the revision of the final version before submission.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The fish used for this study were handled in accordance with the European Union regulations concerning the protection of experimental animals (Dir 86/609/EEC) and with the recommendations given in the Guide for the Care and Use of Laboratory Animals of the Spanish Royal Decree 53/2013 regarding the protection of animals used for scientific purposes (BOE 2013). The applied protocols were approved by the Experimental Animal Ethics Committee from the Universitat Politècnica de València (UPV) and final permission was given by the local government (Generalitat Valenciana, Permit Number: 2014/VSC/PEA/00147). All efforts were made to minimize fish suffering.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional file
Additional file 1:
Figure S1. Boxplot of lhb and fshb expression. Figure S2. Confocal images of Fshβ histochemical labeled of European eel male pituitaries. Figure S3. Heat map and 2D hierarchical clusters of all differentially expressed genes found between the testes samples. Figure S4. Heat map and 2D hierarchical clusters of all differentially expressed genes found between the pituitary samples. Figure S5. Heat map and 2D hierarchical clusters of all differentially expressed genes found between the brain samples. Figure S6. Group distribution of differentially expressed transcripts. Figure S7. Boxplot of testes expression of selected genes. Table S1. Biometric measurements. Table S2. Enriched GO terms from the differentially expressed genes found between T10 and Tvar, in the pituitary. Table S3. Enriched GO terms from the differentially expressed genes found between T10 and Control, in the brain. (DOCX 3120 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Rozenfeld, C., García-Carpintero, V., Pérez, L. et al. Cold seawater induces early sexual developmental stages in the BPG axis of European eel males. BMC Genomics 20, 597 (2019). https://doi.org/10.1186/s12864-019-5969-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-5969-6