Abstract
Background
Livestock production aims to provide meats of high and consistent eating quality. Insufficient intramuscular (IM) fat and excessive subcutaneous (SC) fat are paramount pork quality challenges. IM fat and SC fat, which are modulated by the adipogenesis of IM and SC adipocytes, play key roles in pork quality. Galectin-12 (LGALS12) was proven to be an important regulator of fat deposition in porcine. However, the current knowledge of the transcriptome-wide role of LGALS12 in adipocytes is still limited. This study was aimed to discover the different regulatory mechanisms of LGALS12 in porcine IM and SC adipocyte.
Results
The siRNA-mediated knockdown of the expression of LGALS12 identified 1075 and 3016 differentially expressed genes (DEGs) in IM and SC adipocytes, respectively. Among these, 585 were up- and 490 were downregulated in the IM adipocytes, while 2186 were up- and 830 were downregulated in the SC adipocytes. Moreover, 418 DGEs were observed only in the IM adipocytes, 2359 DGEs only in the SC adipocytes, and 657 DGEs in both types of adipocytes. According to Gene Ontology (GO) analysis, DEGs in both IM and SC adipocytes were mainly enriched in categories related to lipids or fat cell differentiation. Pathway analysis of the DEGs revealed 88 changed signaling pathways in the IM adipocytes and 86 in the SC adipocytes. The signaling pathways present in only one type of adipocyte were identified from among the top 50 signaling pathways in each type of adipocyte. Four signaling pathways, encompassing PI3K-AKT, cardiac muscle contraction, fatty acid metabolism and Ras, were significantly enriched in the IM adipocytes. On the other hand, four different signaling pathways, encompassing TNF, WNT, cGMP-PKG and NF-kappa B, were greatly enriched in the SC ones. The pathway changes were confirmed by chemical inhibition assays.
Conclusions
Our data reveals that LGALS12 knockdown alters the expression of numerous genes involved in key biological processes in the development of adipocytes. These observations provide a global view of the role of LGALS12 in porcine IM and SC adipocytes; thus, improving our understanding of the regulatory mechanisms by which this gene acts in fat development.
Similar content being viewed by others
Background
The intramuscular (IM) fat content is considered a crucial indicator of porcine meat quality, while subcutaneous (SC) fat affects the lean meat percentage of the carcass [1]. To meet the consumers’ increasing demands for high-quality pork, a main goal of breeding is to improve IM fat and to decrease the SC fat content [2]. Although IM fat and SC fat are mainly composed of adipocytes, their properties differ in many aspects. A wider characterization of isolated adipocytes from IM fat and SC fat revealed that not only lipogenesis, but also indicators of lipolysis, fatty-acid oxidation and basal-energy metabolism, are lower in abundance in adipocytes isolated from pig muscle than in fat cells isolated from other body fat depots [3, 4]. Our previous study also indicated that the intracellular triglyceride content of SC preadipocytes increased more dramatically than that of IM preadipocytes during cell differentiation, which was accompanied by lower expression levels of genes related to lipid metabolism such as PPARγ, C/EBPα, ATGL, HSL, LPL, aP2 and FAS, in IM adipocytes [5]. However, the expression of genes and proteins that participate in cell growth, such as insulin-like growth factor II (IGF-II) and prohibitin-1, was higher in IM than in SC adipocytes [6].
Galectins, a family of carbohydrate-binding proteins with 17 members, share the same gene sequence in the carbohydrate-recognition domains (CRDs) that have an affinity for β-galactosides [7]. Galectins 1, 2, 5, 7, 10, 11, and 13 through 17 are known as one-CRD-type galectins [8]. By contrast, galectins 4, 6, 8, 9, and 12 are two-CRD-type galectins, containing two homologous CRDs in a single polypeptide chain [8]. Galectin-3, the only chimeric galectin, contains one CRD and a non-lectin region comprising proline- and glycine-rich short tandem repeats [9]. Some galectins are widely distributed among different tissues, while others are tissue-specific. Galectin-12 (LGALS12) is preferentially expressed in adipocytes and proven to be an important regulator of lipid metabolism in mouse models [10]. In addition, studies have reported that LGALS12 mainly acts as a regulator of prolipolytic signaling and not as a major structural protein that shields the lipid droplet core from the action of lipases [11, 12]. Existing data indicate that the initial up-regulation of LGALS12 expression when cells undergo growth arrest is required for the response of preadipocytes to adipogenic hormone stimulation [13]. Ablation of this protein in mice leads to increased lipolysis, decreased adiposity, and amelioration of insulin resistance associated with weight gain [10]. Our previous study with porcine adipocytes showed that the knockdown of LGALS12 decreased adipogenesis, which indicates that it has potential value in pig breeding [14]. However, the regulatory mechanisms of LGALS12 in porcine adipocytes remain unclear.
In this study, we conducted a comprehensive transcriptome analysis of primary porcine IM and SC adipocytes under LGALS12-silencing and compared the expression profiles with the negative control group (NC) using RNA sequencing technology (RNA-Seq). We also identified the different responses of IM and SC adipocytes to LGALS12-silencing. Gene ontology analysis showed that LGALS12-siRNA interference led to the differential expression of genes in intramuscular adipocytes involved in biological processes such as lipid droplet, whereas the differentially expressed genes (DEGs) of subcutaneous adipocytes were involved in fat cell differentiation and response to lipid. Finally, differences of signaling pathways between the two types of adipocytes were identified and verified.
Results
RNA-Seq investigation of the effects of silencing LGALS12 expression in adipocytes
The expression levels of LGALS12 determined by qRT-PCR in LGALS12-siRNA treated intramuscular (IM) and subcutaneous (SC) adipocytes were about 50% lower than those of cells with NC-siRNA treatment (n = 3, P < 0.01) (Additional file 1: Figure S1), which indicated that LGALS12-siRNA effectively interfered with the expression of LGALS12. In addition, at day 10 of lipogenesis induction, triglyceride (TG) content was also decreased by approximately 70 and 63%, respectively, in intramuscular and subcutaneous adipocytes as revealed by TG content analysis (Additional file 2: Figure S2).
To obtain a global view of the role of the LGALS12 gene in porcine IM and SC adipocytes, comparative transcriptome analyses between NC-siRNA and LGALS12-siRNA treatment groups was performed. Totally, 23.91 ± 0.006, 23.93 ± 0.031, 23.95 ± 0.035 and 23.84 ± 0.047 million high quality clean reads were acquired from the SC-NC-siRNA, SC-LGALS12-siRNA, IM-NC-siRNA and IM-LGALS12-siRNA samples, respectively. For these four samples, 86.2 ± 0.11%, 86.4 ± 0.12%, 86.3 ± 0.07% and 86.2 ± 0.36% unique reads could be mapped in the current version of the pig genome (Sscrofa 11.1), corresponding to 15,484 ± 64, 15,840 ± 123, 15,400 ± 85 and 15,411 ± 27 genes, respectively (Table 1).
Identification of DEGs between the NC-siRNA and LGALS12-siRNA groups
The siRNA-mediated knockdown of the expression of LGALS12 identified a total of 3434 differentially expressed genes (DEGs) in both IM and SC adipocytes, including 1075 DEGs in IM adipocytes and 3016 DEGs in SC adipocytes (Figs. 1a, b). Among the 3434 DEGs, 418 were specifically found in IM adipocytes, 2359 in SC adipocytes, and 657 DEGs in both types of adipocytes (Fig. 1c). In porcine IM adipocytes, 585 DEGs were up- and 490 were downregulated upon the treatment with LGALS12-siRNA (Fig. 1a). Similarly, in the SC adipocytes, 2186 DEGs were up- and 830 downregulated (Fig. 1b).
RNA-Seq analysis of genes expressed in porcine adipocytes treated with NC-siRNA and LGALS12-siRNA. a Venn diagram showing the differentially expressed genes (DEGs) in porcine adipocytes after LGALS12-siRNA treatment. The numbers of up- and downregulated DEGs upon LGALS12-silencing in intramuscular (b) and subcutaneous adipocytes (c) are shown
To confirm the results of RNA-Seq, 12 genes related to the lipid metabolism were chosen for real-time quantitative real-time PCR (qRT-PCR) analysis. Figure 2 shows the gene expression patterns derived from the RNA-Seq (Fig. 2a, c) and qRT-PCR experiments (Fig. 2b, d). According to the RNA-Seq data, six genes changed in the same direction in both IM and SC adipocytes (Fig. 2a). Compared with the NC-siRNA group, the expression of PTGS2 was higher and that of ADIPOQ, PLIN1, PLIN4, FABP3 and FABP5 mRNA was lower in the LGALS12-siRNA group. Another six genes, which changed only in one type of adipocytes, were listed in Fig. 2c. With the LGALS12-siRNA treatment, the expression of SLC27A2, RNASEL and FOXO1 was significantly altered in IM adipocytes and the expression of CMKLR1, ADRB1 and HTR2A was significantly altered in SC adipocytes. The qRT-PCR results were completely in line with the RNA-Seq analysis results (Fig. 2d). These data confirmed that the results of RNA-Seq analysis were indeed reliable indicators of overall changes in gene expression, indicating that the RNA-Seq data were reliable and accurate.
Validation of the RNA-Seq data by qRT-PCR analysis. a, c The expression patterns of PTGS2, ADIPOQ, PLIN1, PLIN4, FABP3, FABP5, SLC27A2, RNASEL, FOXO1, CMKLR1, ADRB1 and HTR2A genes derived from the RNA-Seq. b, d RT-qPCR data of genes in porcine adipocytes after treatment with LGALS12-siRNA showed similar trends in gene expression profile to those obtained by RNA-Seq. The data are expressed as the means ± SE from 3 measurements, *P < 0.05, **P < 0.01, compared with the NC group
Transcription factors of the DEGs
Transcription factors (TFs) play important roles in the regulation of adipocyte differentiation. We compared TF expression between IM and SC adipocytes after the treatment with LGALS12-siRNA. In both IM and SC adipocytes, some DEGs were predicted to encode TFs based on the homology of DNA-binding domains. There were 208 predicted TFs in IM adipocytes, and 652 in SC adipocytes, which can be classified into 36 and 53 TF families, respectively. The distribution of the predicted TFs in the two types of adipocyte was very similar (Fig. 3a). The TF families found in the IM adipocytes were all present in SC adipocytes, among which zf-C2H2, Homeobox, bHLH, TF-bZIP were the largest four families (Fig. 3a).
Among these TFs, the expression of 101 was changed in both IM and SC adipocytes. These included C/EBPα, KLF15 and PPARγ, which are closely related to adipocyte differentiation (Fig. 3b). However, 660 TFs were found in only one of the types of adipocytes. For example, FOXO1, FOXO3 and KLF9 specifically decreased in IM adipocytes upon LGALS12-siRNA treatment (Fig. 3c), while PRDM1, NOTCH1, PRDM8 and E2F8 were specifically induced in in SC adipocytes (Fig. 3d).
Gene ontology (GO) analysis of the DEGs
GO analysis was preformed to further understand the biological functions of the DEGs in IM and SC adipocytes. Significant GO categories with P < 0.05 were selected. The results showed that DEGs related to growth factor activity, regulation of the inflammatory response, regulation of biological quality and G-protein coupled receptor binding were significantly enriched in both IM and SC adipocytes (Fig. 4a). However, some functional categories were found to be specifically enriched in IM adipocytes, such as lipid droplet, insulin-like growth factor binding and cytokine activity (Fig. 4b). In SC adipocytes, the genes were specifically clustered into the following functional groups: fat cell differentiation, brown fat cell differentiation and regulation of cell differentiation, response to lipids (Fig. 4c). Furthermore, in SC and IM adipocytes, the largest number of DEGs were involved in processes related to the lipid droplet, response to lipid, fat cell differentiation and brown fat cell differentiation categories (Fig. 4d).
Gene ontology analysis for differentially expressed genes. The results are summarized in the following three main categories: biological process, molecular function, and cellular component. The y-axis indicates functional groups. The x-axis indicates –log (P-value). GO annotations of unigenes significantly enriched (a) in both IM and SC adipocytes, (b) only in IM adipocytes, and (c) only in SC adipocytes. (d) Heat map of DEGs enriched in the lipid droplet, response to lipids, fat cell differentiation and brown fat cell differentiation categories
Analysis of pathways related to the DEGs
Pathway enrichment results showed that the DEGs were mainly involved in the PPAR signaling pathway, ECM-receptor interaction and cAMP signaling pathway in both IM and SC adipocytes (Fig. 5a).
KEGG pathway enrichment analysis of DEGs in intramuscular and subcutaneous adipocytes. The significantly enriched pathways (a) in both IM and SC adipocytes, (b) specifically in IM adipocytes or SC adipocytes. The x-axis indicates functional pathways. The y-axis indicates -log (P-value). (c) Heat map of DEGs enriched in the PI3K-AKT signaling pathway, fatty acid metabolism, TNF signaling pathway and WNT signaling pathway categories
Notably, the KEGG pathway analysis showed that the PI3K-Akt signaling pathway and fatty acid metabolism were specifically significantly enriched in IM adipocytes upon LGALS12-siRNA treatment (Fig. 5b). In SC adipocytes, the significantly enriched pathways of the DEGs under LGALS12-silencing mainly included the TNF and WNT signaling pathways (Fig. 5b). As shown in Fig. 5c, most of the DEGs were involved in key pathways, including the PI3K-AKT signaling pathway, fatty acid metabolism, TNF signaling pathway and WNT signaling pathway, which were clustered in both types of adipocytes.
Verification of the pathway analysis
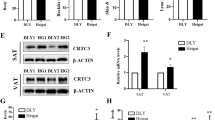
To further confirm the results of the pathway analysis, the specific WNT inhibitor Dickkopf-1 (DKK-1)/DKK and the PI3K-AKT inhibitor LY294002 were individually administrated to the two types of adipocytes at the later adipogenic induction stage. The induction of WNT5a (Fig. 6a) and β-catenin (Fig. 6b) by LGALS12-siRNA in SC adipocytes was completely abolished. By contrast, this phenomenon was not observed in IM adipocytes, because the WNT pathway was not induced by LGALS12-siRNA in these cells. Similarly, LY294002 was able to attenuate the function of the LGALS12 siRNA by rescuing the expression of AKT1 and AKT2 in IM adipocytes, but not in SC adipocyte (Fig. 6c, d).
The inhibition of adipogenesis by LGALS12 knockdown was attenuated by DKK and LY294002 through the WNT and PI3K-AKT signaling pathways, respectively. Adipocytes in the differentiation phase were treated with DKK or LY294002 for 2 days and the mRNA levels of WNT5a (a), β-catenin (b), AKT1 (c), AKT2 (d), FAS (f), HSL (g) and ATGL (h) were measured. e Analysis of triglyceride content in adipocytes. The results are expressed as the means ± SE of three independent experiments. *P < 0.05, **P < 0.01
In an earlier study, we demonstrated that a knockdown of LGALS12 could decrease the triglyceride (TG) content and alter the expression of FAS, HSL and ATGL in both IM and SC adipocytes [14]. In inhibitor experiments, the decrease of the TG content (Fig. 6e) and the changed expression of FAS (Fig. 6f), HSL (Fig. 6g) and ATGL (Fig. 6h) induced by LGALS12 knockdown were partially restored by 48 h of treatment with DKK in SC adipocytes, but not in IM adipocytes. Similarly, the treatment with LY294002 significantly blocked the effect of LGALS12 siRNA on the TG content (Fig. 6e) and the expression of genes associated with lipid metabolism such as FAS and HSL in IM adipocytes (Fig. 6f, g), but not in SC adipocytes. These results verified the pathway analysis of the DGEs. Taken together, the schema of the regulatory adipogenic mechanism of LGALS12 in in IM and SC adipocytes was proposed and shown in Fig. 7.
Discussion
The current study offers the first comprehensive insights into the role of the LGALS12 gene in porcine adipocytes using RNA-Seq technology. The number of total reads that were mapped to the reference genome met the high-quality criterion of the RNA-seq technology. Our data showed that the RNA-based silencing of LGALS12 respectively changed the expression levels of 1075 and 3016 genes in IM and SC adipocytes, according to pooled data of a total of three 3-day-old piglets from three independent litters. These DEGs were mainly associated with the lipid droplet, response to lipid, fat cell differentiation, PI3K-AKT signaling pathway, fatty acid metabolism, and TNF signaling pathway categories, suggesting that LGALS12 plays a crucial role in the process of fat deposition.
In previous studies, there was no information about LGALS12 affecting the expression of other LGALS family members. In this study, LGALS5, LGALS6, LGALS10 and LGALS11 were not detected, while the expression of LGALS2, LGALS4, LGALS7 and LGALS13 showed no significant changes upon LGALS12-silencing, suggesting that these genes might not be involved in LGALS12-mediated pathways. Interestingly, LGALS8 was significantly downregulated, while LGALS1 and LGALS3 were significantly upregulated by the silencing of LGALS12 in both IM and SC adipocytes. In general, ablation of LGALS3 accelerates the obesity and diabetes induced by a high-fat diet [15, 16]. By contrast, LGALS8 promotes proliferation and prevents apoptosis in cancerous cells [17, 18]. These findings suggest that LGALS12 might regulate lipid metabolism in concert with other members of the family.
Transcription factors play important roles in the regulation of adipocyte differentiation. Significant advances toward elucidating the regulatory mechanisms involved in adipocyte differentiation have been achieved mostly by identifying transcription factors that contribute to the adipogenic process. The transcription factor KLF15 is markedly up-regulated during the differentiation of adipocytes and controls the preadipocyte-to-adipocyte transition [19]. On the other hand, some genes are constitutively activated, such as the adipogenic transcription factors PPARγ and CEBPα, which are involved in regulating terminal differentiation [20]. In the present study, knockdown of LGALS12 was found to suppress the expression of KLF15, PPARγ and CEBPα in IM and SC adipocytes. The forkhead-type transcription factor family forkhead box class O (FOXO) is also thought to play a role in adipocyte differentiation [21]. Accordingly, the knockdown of FOXO1 expression by FOXO1-siRNA significantly suppressed adipocyte differentiation by downregulating the expression of PPARγ and C/EBPα [21]. KLF9 was found to activate the early phase of adipogenesis by enhancing the expression of the CEBPβ in 3 T3-L1 cells [22]. Our results showed that the expression of FOXO1 and KLF9 was specifically reduced by the silencing of LGALS12 in porcine IM adipocytes, while NOTCH1 and E2F8 were significantly induced in SC adipocytes, suggesting that different TFs respond to LGALS12 silencing in porcine IM and SC adipocytes.
Moreover, the interruption of LGALS12 impacted the fatty acid metabolism by regulating the levels of many key markers of adipogenesis. According to the current knowledge, ADIPOQ, PLINs and FABPs are central to adipogenesis [23, 24]. In this work, LGALS12 silencing downregulated the expression levels of ADIPOQ, PLIN1, PLIN4, FABP3, and FABP5, while it upregulated the levels of ACSL4, NR4A2, PTGS2, and UGT1A6 in IM and SC adipocytes. ADIPOQ is one of multiple adipocytokines secreted by adipose tissue, and it has been shown to modulate both glucose and lipid metabolism in vivo and in vitro [25]. Overexpression of ADIPOQ can enhance the proliferation of 3 T3-L1 fibroblasts, accelerate adipocyte differentiation, and, in fully differentiated adipocytes, augment both lipid accumulation and insulin-responsive glucose transport [25]. Members of the PLIN family of phosphoproteins are specifically located at the surface of intracellular lipid (triacylglycerol) droplets, the site of lipolysis [26, 27]. Overexpression of PLIN in fat-cell lines inhibits the tumor necrosis factor-α-mediated stimulation of lipolysis. PLIN ablation in mice leads to an increased lipolysis rate in fat cells [27]. In addition, fatty acid binding proteins (FABPs) play a crucial role in intracellular fatty acid transport by binding and properly targeting long-chain fatty acids to their correct metabolic sites [28, 29]. The FABP3 gene is also related to cytoplasmic hydrophobic ligand binding proteins and lipid metabolism by regulating the intracellular transport of long-chain fatty acids that bind to fatty acyl-CoA and acyl-L-carnitines [30]. The epithelial/epidermal-type FABP5 is also expressed in adipose tissue. The FABP4 and FABP5 genes are co-regulated in other mammals, and the FABP4:FABP5 ratio or the absolute FABP content in adipose tissue might be critical for the regulation of lipid metabolism [31]. The present study indicates that the expression levels of ADIPOQ, PLINs, and FABPs were downregulated upon LGALS12-silencing, providing strong evidence that silencing of LGALS12 might impact both IM and SC adipocytes by impairing the progression of adipogenesis.
However, adipocytes derived from porcine subcutaneous and intramuscular fat tissue have distinct adipogenic potentials, and the role of LGALS12 in the adipogenesis of SC and IM adipocytes exhibited some differences. Therefore, a specific pattern of gene expression was observed in the SC or IM adipocytes upon the LGALS12-siRNA treatment. In IM adipocytes, LGALS12 silencing regulated the expressions of lipid droplet-related genes including CIDEC, RSAD2 and DGAT2. In SC adipocytes, LGALS12 silencing had an effect on genes related to the brown fat cell differentiation process, such as ADRB1, ADIG and SLC2A4.
To understand the regulatory network of LGALS12 in SC and IM adipocytes, KEGG pathway analysis was used to explore the signaling pathways of the DEGs identified in SC and IM adipocytes. Several adipogenesis-related pathways, such as the PPAR signaling pathway, MAPK signaling pathway and ECM-receptor interaction pathway, were found to be affected by LGALS12 silencing in both SC and IM adipocytes. PPARs are one of the most important groups of receptors in adipocytes, where they are involved in the regulation of adipocyte differentiation and various metabolic activities [32]. The ECM-receptor interaction pathway can directly or indirectly influence cellular behaviors such as adhesion and migration [33]. The the shape change of fibroblastic preadipocytes to rounded, mature adipocytes is accompanied by changes of cytoskeletal organization and contacts with the ECM [34]. In addition, the ERK, p38 and JNK mitogen activated protein kinases (MAPKs) are involved in intracellular signaling pathways that play a pivotal role in many essential cellular processes such as proliferation and differentiation. MAPK pathways are able to regulate adipogenesis at each steps of the process, from stem cells to adipocytes [35, 36]. Notably, the KEGG pathway analysis showed that LGALS12 silencing significantly enriched the ECM-receptor interaction pathway and cAMP signaling pathway in both IM and SC adipocytes. Therefore, our data support the idea that the ECM-receptor interaction pathway and cAMP signaling pathway may participate in adipocyte differentiation. The intracellular PI3K-Akt signaling pathway is involved in the regulation of many cellular processes. In particular, several lines of evidence have implicated the PI3K-Akt signaling pathway as a positive regulator of adipocyte differentiation [37]. Disruption of PI3K function by pharmacological inhibitors or dominant negative mutations abolishes adipocyte differentiation from preadipocytes [37]. In this study, the KEGG pathway analysis showed that the PI3K-Akt signaling pathway was specifically significantly enriched in IM adipocytes after LGALS12-siRNA treatment, which indicates that LGALS12 regulates IM adipocyte differentiation via the PI3K-Akt signaling pathway. Previous studies demonstrated the involvement of WNT pathways in the regulation of white and brown adipocyte differentiation in vitro and in vivo. Disruption of WNT/β-catenin signaling leads to spontaneous adipogenesis, which indicates that endogenous WNT signals restrain preadipocyte differentiation [38, 39]. In differentiated brown adipocytes, the activation of WNT signaling suppresses the expression of uncoupling protein 1 (UCP1) through PGC1A repression without influencing common adipocyte markers [40]. In the present study, the WNT signaling pathway was specifically enriched in SC adipocytes treated with LGALS12-siRNA. Thus, we speculate that LGALS12 regulates SC adipocyte differentiation via the WNT signaling pathway.
Conclusions
Although it is known as a key regulator of multiple biological processes related to fat development, knowledge on the function of LGALS12 in porcine adipocytes is still limited. In this study, LGALS12 was knocked down in porcine IM and SC adipocytes and transcriptional profiles of the two types of adipocytes were compared. The LGALS12 knockdown altered the expression of numerous genes involved in key biological processes in the development of adipocytes. Our data provide a novel global view of the role of the LGALS12 gene in porcine IM and SC adipocytes, thus improving our understanding of the regulatory mechanisms by which LGALS12 influences porcine fat development.
Methods
Animals
Three 3-day-old piglets, selected from different litters (Jiaxing Black pig breed, male), were provided by Zhejiang Qinglian Food Limited by Share Ltd. (Jiaxing, Zhejiang Province, China). The piglets were housed in a temperature-controlled location with a 12 h light-dark cycle; they had free access to breast milk. To execute three 3-day-old piglets (1–2 kg) of euthanasia. Euthanasia procedure: Pigs were carried to the laboratory which provided isolation, thereby minimizing noise and distractions. To habituate pigs to the CO2 euthanasia box, the pig pair was placed in the box for 10 min to 60 min and then taken back to laboratory. The longissimus dorsi muscle and subcutaneous adipose tissues were collected for the primary culture of intramuscular (IM) and subcutaneous (SC) adipocytes, respectively. The experimental procedure was in accordance with the guidelines of the Jiaxing University Animal Care Committee.
Cell culture and adipocyte differentiation
Porcine longissimus dorsi muscle and subcutaneous adipose tissue were harvested from piglets (3d, male) under aseptic conditions. Isolated tissues were minced and digested with 1 mg/mL collagenase type I (Invitrogen, Carlsbad, CA, USA) at 37 °C for 60 min, followed by filtration through 212 μm and 75 μm nylon meshes. Adipose-derived stromal-vascular (SV) cells were collected by centrifugation at 1360×g for 7 min and grown in DMEM/F12 medium (Hyclone, USA) containing 1% antibiotic/antimycotic solution (SV30010; Hyclone, USA) and 10% fetal bovine serum (FBS; Gibco, USB) at 37 °C in a humidified atmosphere comprising 5% CO2. The cells were cultured to confluence (designated as experimental day 0) in the growth medium, and then induced to differentiate using a differentiation cocktail comprising DMEM/F12 supplemented with 10% FBS, 0.5 mM isobutyl methylxanthine (IBMX; Sigma, USA), 0.5 mM dexamethasone (Sigma, USA), and 20 nM insulin (Sigma, USA) for 2 days. The cells were then maintained in DMEM/F12 with 10% FBS and 20 nM insulin for another 4–6 days. During the differentiation process, the medium was changed every other day [41].
Transfection of adipocytes with siRNA
The sequence under the GenBank accession nr. NM_001142844.1 was used to design oligonucleotides for LGALS12 and NC-siRNA using the online software rnaidesigner (https://rnaidesigner.thermofisher.com/rnaiexpress/) and constructed as follows: LGALS12-siRNA 5′-GCGUGAAUGGACUCCACUUTT-3′, 5′-AAGUGGAGUCCAUUCACGCTT-3′; NC-siRNA 5′-UUCUCCGAACGUGUCACGUTT-3′, 5′-ACGUGACACGUUCGGAGAATT-3′. Cells were transfected using Lipofectamine 2000 (Invitrogen, USA) as described in the manufacturer’s protocol. Four hours before transfection, the medium was changed to Opti-MEM® Medium (Gibco, USA). The siRNA (20 μM) was incubated with 5 μl Lipofectamine® 2000 transfection reagent in Opti-MEM® Medium for 20 min at room temperature before transfection. Six hours later, the medium was switched to culture medium. Two days before induction, porcine adipocytes were transfected with control or LGALS12 siRNA [42]. Details are shown in Additional file 3: Figure S3.
RNA extraction, library preparation and RNA-seq
Total RNA was isolated from three different biological replicates for each sample using TRIzol Reagent (Invitrogen). The sequencing process was carried out by BGI (Beijing Genomics Institute). RNA quality was tested using an Agilent 2100 instrument (Agilent Technologies Inc., USA), a NanoDrop 2000 (Thermo Fisher Scientific Inc., USA) and agarose gel electrophoresis. Three replicate RNA samples were prepared for cDNA library construction. Specifically, mRNAs with poly A tails were enriched using Oligo (dT) magnetic beads. The target RNA was fragmented and reversely transcribed to double-strand cDNA (ds-cDNA) using N6 random primers. Ds-cDNA ends were repaired with phosphate at 5′ end and sticky ‘A’ at the 3′ end, and then ligated and an adaptor added with sticky ‘T’ at the 3′ end. Next, the ligation products were amplified using two specific primers. The PCR products were denatured by heat and the single strand DNA was cyclized using a splint oligo and DNA ligase. Finally, the cDNA library underwent transcriptome sequencing using BGISEQ-500 RNA-Seq [43]. The sequencing created 50-bp reads using single ends (SE50).
Read mapping and gene quantification
In this project, BGISEQ-500 platform was used to sequence samples. And then primary sequencing data (raw reads) was filtered to remove low quality reads. After reads filtering, HISAT [44] was used to map clean reads to reference genome (Sscrofa 11.1), and Bowtie2 [45] was used to map clean reads to reference transcripts. The alignment results were subjected to a comprehensive analysis; both randomness and gene coverage were evaluated. Gene expression levels were calculated using the Fragments Per Kilobase of exon model per Million mapped reads (FPKM) method provided by RSEM software [46].
Gene expression analysis
The FPKM value and DEGseq algorithm were used to measure the gene expression levels and filter the differentially expressed genes (DEGs), respectively. The criteria for filtering DEGs were as follows: Fold Change ≧ 2 and Adjusted P-value ≦ 0.001. Hierarchical clustering analysis of DEGs was performed using Cluster 3.0 with the average linkage and Euclidean distance metric, and the result was visualized using Java TreeView (version 1.1.6r2, Stanford University, Stanford, CA, USA).
Functional enrichment analysis
Gene ontology (GO) and pathway analysis of DEGs were conducted using the Gene Ontology (http://www.geneontology.org/) and KEGG (http://www.genome.jp/kegg/) servers, respectively. Fisher’s exact test was applied to identify the significant GO categories or pathways. Terms with corrected P-values of less than 0.05 were considered to be significantly enriched. GO functional enrichment was performed using phyper, a function of R.
Quantitative real-time RT-PCR
Gene-specific primers were designed using PRIMER 5.0 software (Additional file 4: Table S1). Primer quality and PCR amplification efficiency was confirmed by RT-PCR. The RNA samples were the same as those used for RNA-Seq. The first-strand cDNA was produced from 500 ng total RNA usng the PrimeScript™ RT reagent Kit with gDNA Eraser (Takara, Japan). The real-time PCR was conducted using One Step TB Green™ PrimeScript™ RT-PCR Kit (Takara, Japan) on a Bio-Rad system. All samples were assayed in triplicate wells and results of each treatment were presented as means±SE of three biological replicates. The temperature program was as follows: 95 °C for 1 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s. The relative mRNA abundance of each gene was normalized to the expression level of the housekeeping gene β-actin. 2-△△Ct method was used to calculate expression changes [47].
Statistical analysis
The experiment was set up in a completely randomized design with three biological replicates (piglets from independent litters) for each sample. The mean value for three technological replicates within each pig was calculated. The data are presented as the means ±standard error (SE). In each experiment, all determinations were performed at least in triplicate. SPSS 10.0 (IBM Corp., USA) was used for statistical analyses. Statistical differences between NC-siRNA and LGALS12-siRNA groups were examined using Student’s t-test. P-values of less than 0.05 and 0.01 were considered to indicate significant and very significant differences, respectively.
Availability of data and materials
The data sets supporting the results of this article are included within the manuscript and its additional files. The raw datasets generated during the current study are not publicly available due as analysis is still ongoing, but are available from the corresponding author on reasonable request.
Abbreviations
- ADIPOQ:
-
Adiponectin
- ADRB1:
-
β1-adrenoceptor
- aP2:
-
Fatty acid binding protein 4
- ATGL:
-
Adipose Triglyceride Lipase
- C/EBPα:
-
CCAAT/enhancer binding protein-α
- CMKLR1:
-
Chemokine-like receptor-1
- FABP3:
-
Fatty acid binding protein 4
- FABP5:
-
Fatty acid binding protein 5
- FAS:
-
Fatty acid synthetase
- FOXO1:
-
Forkhead box O1
- HSL:
-
Hormone-sensitive lipase
- HTR2A:
-
5HT receptor type 2A
- IM:
-
Intramuscular
- LGALS12:
-
Galectin-12
- LPL:
-
Lipoprotein Lipase
- PLIN1:
-
Perilipin 1
- PLIN4:
-
Perilipin 4
- PPARγ:
-
Peroxisome proliferator-activated receptor γ
- RNASEL:
-
Ribonuclease L
- SC:
-
Subcutaneous
- SLC27A2:
-
Solute carrier family 27 (fatty acid transporter) member 2
- UCP1:
-
Uncoupling protein 1
References
Grunert KG, Bredahl L, Brunsø K. Consumer perception of meat quality and implications for product development in the meat sector-a review. Meat Sci. 2004;66:259–72.
Fernandez X, Monin G, Talmant A, Mourot J, Lebret B. Influence of intramuscular fat content on the quality of pig meat-1. Composition of the lipid fraction and sensory characteristics of m. longissimus lumborum. Meat Sci. 1999;53:59–65.
Gondret F, Guitton N, Guillerm-Regost C, Louveau I. Regional differences in porcine adipocytes isolated from skeletal muscle and adipose tissues as identified by a proteomic approach. J Anim Sci. 2008;86:2115–25.
Kouba M, Bonneau M. Compared development of intermuscular and subcutaneous fat in carcass and primal cuts of growing pigs from 30 to 140 kg body weight. Meat Sci. 2009;81:270–4.
Wu W, Zhang J, Zhao C, Sun Y, Pang W, Yang G. CTRP6 regulates porcine adipocyte proliferation and differentiation by the AdipoR1/MAPK signaling pathway. J Agric Food Chem. 2017;65:5512–22.
Gardan D, Gondret F, Louveau I. Lipid metabolism and secretory function of porcine intramuscular adipocytes compared with subcutaneous and perirenal adipocytes. American Journal of Physiology-Endocrinology and Metabolism. 2006;291:372–80.
Wan L, Yang RY, Liu FT. Galectin-12 in cellular differentiation, apoptosis and polarization. Int J Mol Sci. 2018;19:176.
Viguier M, Advedissian T, Delacour D, Poirier F, Deshayes F. Galectins in epithelial functions. Tissue barriers. 2014;2:29103.
Itabashi T, Arima Y, Kamimura D, Higuchi K, Bando Y, Takahashi-Iwanagaa H, Murakamic M, Watanabeb M, Iwanagaa T, Nio-Kobayashia J. Cell-and stage-specific localization of galectin-3, a β-galactoside-binding lectin, in a mouse model of experimental autoimmune encephalomyelitis. Neurochem Int. 2018;118:176–84.
Yang RY, Yu L, Graham JL, Hsu DK, Lloyd KC, Havel PJ, Liu FT. Ablation of a galectin-preferentially expressed in adipocytes increases lipolysis, reduces adiposity, and improves insulin sensitivity in mice. Proc Natl Acad Sci. 2011;108:18696–701.
Yang RY, Havel P, Liu FT. Galectin-12: a protein associated with lipid droplets that regulates lipid metabolism and energy balance. Adipocyte. 2012;1:96–100.
Yang RY, Hsu DK, Yu L, Chen HY, Liu FT. Galectin-12 is required for adipogenic signaling and adipocyte differentiation. J Biol Chem. 2004;279:29761–6.
Yang RY, Xue H, Yu L, Velayosbaeza A, Monaco AP, Liu FT. Identification of VPS13C as a galectin-12-binding protein that regulates galectin-12 protein stability and adipogenesis. PLoS One. 2016;11:e0153534.
Wu WJ, Yin YJ, Xu K, Peng YJ, Zhang J. Knockdown of galectin-12 inhibits porcine adipocyte adipogenesis via PKA-Erk1/2 signalling pathway. Acta Biochim Biophys Sin. 2018;50:960–7.
Pejnovic N, Pantic J, Jovanovic I, Radosavljevic G, Milovanovic M, Nikolic I, Zdravkovic N, Djukic A, Arsenijevic N, Lukic M. Galectin-3 deficiency accelerates high-fat diet induced obesity and amplifies inflammation in adipose tissue and pancreatic islets. Diabetes. 2013;62:1932–44.
Baek JH, Kim SJ, Gu KH, Lee HW, Kim JH, Hwang KA, Song J, Chun KH. Galectin-3 activates PPARγ and supports white adipose tissue formation and high-fat diet-induced obesity. Endocrinology. 2015;156:147–56.
Nagy N, Bronckart Y, Camby I, Legendre H, Lahm H, Kaltner H, Hadari Y, Van HP, Yeaton P, Pector JC, Zick Y, Salmon I, Danguy A, Kiss R, Gabius HJ. Galectin-8 expression decreases in cancer compared with normal and dysplastic human colon tissue and acts significantly on human colon cancer cell migration as a suppressor. Gut. 2002;50:392–401.
Hadari YR, Arbel-Goren R, Levy Y, Amsterdam A, Alon R, Zakut R, Zick Y. Galectin-8 binding to integrins inhibits cell adhesion and induces apoptosis. J Cell Sci. 2000;113:2385–97.
Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, Kadowaki T, Matsuki Y, Ogawa W, Hiramatsu R, Kasuga M. Role of Krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–75.
White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318:10–4.
Munekata K, Sakamoto K. Forkhead transcription factor Foxo1 is essential for adipocyte differentiation. In Vitro Cellular and Developmental Biology-Animal. 2009;45:642.
Kimura H, Fujimori K. Activation of early phase of adipogenesis through Krüppel-like factor KLF9-mediated, enhanced expression of CCAAT/enhancer-binding protein β in 3T3-L1 cells. Gene. 2014;534:169–76.
Moreno-Navarrete JM, Fernández-Real JM. Adipocyte differentiation. Adipose tissue biology. Cham: Springer; 2017. p. 69–90.
Wang H, Yan WJ, Zhang JL, Zhang FY, Gao C, Wang YJ, Bondlaw W, Tao L. Adiponectin partially rescues high glucose/high fat-induced impairment of mitochondrial biogenesis and function in a PGC-1α dependent manner. Eur Rev Med Pharmacol Sci. 2017;21:590–9.
Fu Y, Luo N, Klein RL, Garvey WT. Adiponectin promotes adipocyte differentiation, insulin sensitivity, and lipid accumulation. J Lipid Res. 2005;46:1369–79.
Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intracellular lipid storage droplet proteins. J Lipid Res. 2010;51:468–71.
Mottagui-Tabar S, Ryden M, Löfgren P, Faulds G, Hoffstedt J. Evidence for an important role of perilipin in the regulation of human adipocyte lipolysis. Diabetologia. 2003;46:789–97.
Schaap FG, Binas B, Danneberg H, Van GJ, Glatz JF. Impaired long-chain fatty acid utilization by cardiac myocytes isolated from mice lacking the heart-type fatty acid binding protein gene. Circ Res. 1999;85:329–37.
Schroeder F, Petrescu AD, Huang H, Atshaves BP, Mcintosh AL. Role of fatty acid binding proteins and long chain fatty acids in modulating nuclear receptors and gene transcription. Lipids. 2008;43:1–17.
Cho KH, Kim MJ, Jeon GJ, Chung HY. Association of genetic variants for FABP3 gene with back fat thickness and intramuscular fat content in pig. Mol Biol Rep. 2011;38:2161–6.
Samulin J, Berget I, Lien S, Sundvold H. Differential gene expression of fatty acid binding proteins during porcine adipogenesis. Comp Biochem Physiol B Biochem Mol Biol. 2008;151:147–52.
Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–92.
Myers JP, Santiago-Medina M, Gomez TM. Regulation of axonal outgrowth and pathfinding by integrin-ECM interactions. Developmental neurobiology. 2011;71:901–23.
Huang G, Greenspan DS. ECM roles in the function of metabolic tissues. Trends Endocrinol Metab. 2012;23:16–22.
Bost F, Aouadi M, Caron L, Binétruy B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie. 2005;87:51–6.
Zhan Y, Kim S, Izumi Y, Izumiya Y, Nakao T, Miyazaki H, Iwao H. Role of JNK, p38, and ERK in platelet-derived growth factor-induced vascular proliferation, migration, and gene expression. Arterioscler Thromb Vasc Biol. 2003;23:795–801.
Li H, Li T, Fan J, Li T, Fan L. miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death Differ. 2015;22:1935.
Choi SY, Song MS, Ryu PD, Lam AT, Joo SW. Gold nanoparticles promote osteogenic differentiation in human adipose-derived mesenchymal stem cells through the Wnt/β-catenin signaling pathway. Int J Nanomedicine. 2015;10:4383.
Kim MB, Song Y, Kim C, Hwang JK. Kirenol inhibits adipogenesis through activation of the Wnt/β-catenin signaling pathway in 3T3-L1 adipocytes. Biochem Biophys Res Commun. 2014;445:433–8.
Kang S, Bajnok L, Longo KA, Petersen RK, Hansen JB. Effects of Wnt signaling on brown adipocyte differentiation and metabolism mediated by PGC-1α. Mol Cell Biol. 2005;25:1272–82.
Wu WJ, Mo DL, Zhao CZ, Zhao C, Chen YS, Pang WJ, Yang GS. Knockdown of CTRP6 inhibits adipogenesis via lipogenic marker genes and Erk1/2 signalling pathway. Cell Biol Int. 2015;39:554–62.
Wu W, Sun Y, Zhao C, Zhao C, Chen X, Wang G, Pang W, Yang G. Lipogenesis in myoblasts and its regulation of CTRP6 by AdipoR1/Erk/PPARγ signaling pathway. Acta Biochem Biophys Sin. 2016;48:509–19.
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–60.
Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25–34.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. Bmc Bioinformatics. 2011;12:323.
Wu W, Zhang J, Zhao C, Sun Y, Yin Y, Peng Y, Pang W, Yang G. Lentivirus-mediated CTRP6 silencing ameliorates diet-induced obesity in mice. Exp Cell Res. 2018;367:15–23.
Acknowledgements
The authors are grateful to Zhejiang Qinglian Food Limited by Share Ltd. and Jiaxing Dayunhe Ecological Animal Husbandry Co., Ltd. for providing experiment animals.
Funding
This work was supported by Zhejiang Natural Science Foundation (No. LQ18C170002 and LY17C170003), Initial Scientific Research Fund of Young Teachers in Jiaxing University (No.70516055), General Research Project of Zhejiang Provincial Department of Education (No.Y201738243), and The Key Science and Technology Innovation Team of Jiaxing (Group of Ecological Restoration and Ecological Engineering), which involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript. We are very grateful for the support provided by these funds.
Author information
Authors and Affiliations
Contributions
All authors have read and approved the manuscript. WW designed the experiments, performed cell culture, wrote the paper, and provided the experimental funding. WW, DZ, YY and JZ designed, performed, analyzed and interpreted the study. XH and KX constructed siRNA oligonucleotides and literature search. MJ performed cell culture and experimental statistics. JZ designed the experiments, revised the paper, and provided the experimental funding. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Jiaxing University Animal Care Committee provided official ethics board approval for this study. Tissues used in this study were collected from pigs maintained and used according to the approved protocols.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. Efficiency of LGALS12 siRNA in intramuscular and subcutaneous adipocytes. The knockdown efficiencies of LGALS12 siRNAs were measured by qPCR at day 1 after treatment with LGALS12-siRNA. The LGALS12 gene expression patterns were derived from qRT-PCR experiments. The data are expressed as means ± SEM n = 3, *P < 0.05, **P < 0.01, compared with the NC group. (PDF 37 kb)
Additional file 2:
Figure S2. LGALS12 knockdown inhibits lipid accumulation in adipocytes. Triglyceride content in intramuscular and subcutaneous adipocytes. Data expressed as mean ± SEM n = 3, *P < 0.05, **P < 0.01, compared with NC group. (PDF 217 kb)
Additional file 3:
Figure S3. Experimental design for assessing the effect of LGALS12 on the adipogenesis in IM and SC adipocytes (PDF 581 kb)
Additional file 4:
Table S1. Primers (S, sense; A, antisense) for real time PCR. (DOC 73 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wu, W., Zhang, D., Yin, Y. et al. Comprehensive transcriptomic view of the role of the LGALS12 gene in porcine subcutaneous and intramuscular adipocytes. BMC Genomics 20, 509 (2019). https://doi.org/10.1186/s12864-019-5891-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-5891-y