Abstract
Background
Southern root-knot nematode Meloidogyne incognita (Kofoid and White, 1919), Chitwood, 1949 is a key pest of agricultural crops. Pasteuria penetrans is a hyperparasitic bacterium capable of suppressing the nematode reproduction, and represents a typical coevolved pathogen-hyperparasite system. Attachment of Pasteuria endospores to the cuticle of second-stage nematode juveniles is the first and pivotal step in the bacterial infection. RNA-Seq was used to understand the early transcriptional response of the root-knot nematode at 8 h post Pasteuria endospore attachment.
Results
A total of 52,485 transcripts were assembled from the high quality (HQ) reads, out of which 582 transcripts were found differentially expressed in the Pasteuria endospore encumbered J2 s, of which 229 were up-regulated and 353 were down-regulated. Pasteuria infection caused a suppression of the protein synthesis machinery of the nematode. Several of the differentially expressed transcripts were putatively involved in nematode innate immunity, signaling, stress responses, endospore attachment process and post-attachment behavioral modification of the juveniles. The expression profiles of fifteen selected transcripts were validated to be true by the qRT PCR. RNAi based silencing of transcripts coding for fructose bisphosphate aldolase and glucosyl transferase caused a reduction in endospore attachment as compared to the controls, whereas, silencing of aspartic protease and ubiquitin coding transcripts resulted in higher incidence of endospore attachment on the nematode cuticle.
Conclusions
Here we provide evidence of an early transcriptional response by the nematode upon infection by Pasteuria prior to root invasion. We found that adhesion of Pasteuria endospores to the cuticle induced a down-regulated protein response in the nematode. In addition, we show that fructose bisphosphate aldolase, glucosyl transferase, aspartic protease and ubiquitin coding transcripts are involved in modulating the endospore attachment on the nematode cuticle. Our results add new and significant information to the existing knowledge on early molecular interaction between M. incognita and P. penetrans.
Similar content being viewed by others
Background
Following the publication of the Caenorhabditis elegans (Maupas, 1900) Dougherty, 1955 genome [104], some laboratories have become focused on using this nematode as a model for studying innate immunity [61, 71, 96, 111]. Due to the short developmental time of this nematode, the studies have necessarily focused on the infection of adults and earlier developmental stages, and in particular the non-feeding dauer stage have been neglected. The life-cycle of plant-parasitic nematodes commences when an infective juvenile hatches from an egg as a second-stage juvenile, and migrate through the soil seeking a suitable host plant before feeding starts. This period of time offers the opportunity to study early responses of the nematode to bacterial infection [26].
Pasteuria penetrans (Thorne, 1940) Sayre and Starr, 1985, a Gram-positive soil bacterium of the Bacillus–Clostridium clade, is a hyperparasite of the root-knot nematodes (RKN), and represents a typical naturally coevolved pathogen – hyperparasite system [15, 86]. This is an excellent system to study the early stages of the nematode infection processes by bacterial parasites. The life-cycle and developmental stages of P. penetrans inside M. incognita are well documented and understood [28, 82]. The bacterium completes its life-cycle in three phases, [1] Attachment and germination, [2] Rhizoid production and exponential growth; and [3] Sporogenesis [28]. There is a high degree of genetically regulated host specificity involved in this interaction. Pasteuria is highly selective and specific to their host; one population of Pasteuria will not recognize and infect other species in the same genus, and not even all populations of the same species [24]. The surface of nematode cuticle plays a decisive role in facilitating the specificity of the adhesion [27, 99] and the attachment of P. penetrans endospores to an as of yet uncharacterized cuticle receptor is the primary and arguably the most crucial step of the bacterial infection [28]. After the RKN J2 s establish permanent feeding sites in their plant hosts, the endospores perceive some currently unknown cue(s) from the nematode and germinate [25, 93]. The bacterium proliferates inside the worm’s body, kills it, and converts the females into an “endospore sac” containing millions of endospores [25, 82].
The recent development of genomic tools and technologies for the plant-parasitic nematodes has enabled researchers to investigate in detail at the molecular level the nematode’s interactions with their hosts, symbionts and pathogens/hyperparasites. It is known that hosts respond to pathogen attack by altering their gene expression; in the infection of Daphnia by Pasteuria ramosa, it was observed that the incompatible/resistant hosts responded by an up-regulated gene response, whereas the down-regulated gene response was pronounced in the compatible/susceptible host [64]. However, in a recently published study that used RNA-Seq to understand the nematode’s response to Pasteuria infection at three days post attachment, when nematode appeared less mobile and moribund because of the bacterial infection, it was found that 91% of the 445 differentially expressed genes were up-regulated [117]. This was contrary to the general understanding that a down-regulated gene response is exhibited by the susceptible hosts, which is probably caused by subversion of host immunity by the parasite [6]. Therefore, this large up-regulation in gene expression by Daphnia against Pasteuria infection over time, as reported by McTaggart et al. [64] and again by Zou et al. [117] using a population of M. incognita susceptible to Pasteuria penetrans at three days post attachment warranted further investigation.
In order to understand the nematode genes involved in the very first and crucial stage of the M. incognita – P. penetrans interaction, i.e. the recognition and adhesion of endospores, here we investigated the early transcriptional response of M. incognita at eight hours after their initial encounter and before the infective J2 s have invaded the plant root. Additionally, we also identified the functional role of five important differentially expressed genes in M. incognita – Pasteuria interaction by using RNAi based gene silencing and measuring their effects on endospore adhesion.
Methods
Biological materials
The single egg mass culture of an Indian isolate of M. incognita race 1 was increased on tomato plant (Solanum lycopersicum L. cv. Pusa ruby) in a glasshouse at ICAR- Indian Agricultural Research Institute, New Delhi, India. Nematode infected tomato roots were washed free of soil, egg mass were hand-picked and kept for hatching on a modified Baermann’s funnel assembly [110]. The freshly hatched J2 s were used for all of the experiments, viz., endospore attachment, RNA isolation, dsRNA treatment etc. The unused/left over J2 s were autoclaved and discarded.
Endospore attachment
Pasteuria penetrans (Strain AII-329: Pasteuria collection, ICAR-IARI, New Delhi, India) endospores were produced on M. incognita cultured on adzuki bean (Vigna angularis (Willd.) Ohwi and Ohashi) in CYG growth pouches (Mega International, St Paul, MN, USA) as described by Rao et al. [88]. Transfer of germinated seeds in growth pouches, setting up of root infection and post infection maintenance of plants were conducted as described earlier [84].
The freshly hatched ca. 20,000 J2 s were collected in a 1.5 ml microcentrifuge tube and mixed with 200 μl of endospore suspension (2.5 × 103 endospores ml− 1). The attachment of endospores was pursued by centrifugation method [41] and resulted in attachment of approximately 30–35 endospores on the cuticle surface of each juvenile (n = 200). The attachment of endospores onto the juveniles was confirmed microscopically. Following endospore attachment, the J2 s were washed thrice in M9 buffer (1 mM MgSO4, 22 mM KH2PO4, 42.3 mM Na2HPO4 and 85.6 mM NaCl; pH 7.0) to remove the free endospores. The endospore encumbered J2 s were incubated in fresh M9 buffer at room temperature (28 °C) for 8 h on a slowly moving rotator. The adherence of the endospores onto J2 surface was again confirmed microscopically after 8 h, and the juveniles were found to have similar numbers of attached spores as seen earlier. Following attachment, the J2 s were incubated at room temperature (28 °C) for 8 h on a slowly rotating incubator. The freshly hatched juveniles incubated in M9 buffer for 8 h without Pasteuria endospores served as a control.
RNA extraction
Total RNA was extracted from about 20,000 M. incognita non-encumbered and endospore encumbered J2 s with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer’s protocol. The RNA was treated with RQ1 RNase-Free DNase (Promega, Madison, WI, USA) to remove any genomic DNA contamination. The integrity of the isolated RNA was tested on a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The quality and concentration was determined by 1% agarose gel and NanoDrop-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The process was replicated twice.
cDNA synthesis, library preparation, RNA-sequencing
The total RNA was subjected to downstream processing for cDNA synthesis and library preparation. The extracted RNA was assessed for quality using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and RNA with an RNA integrity number (RIN) of 8.0 was used for mRNA purification. The mRNA (messenger RNA) was purified from approximately 5 μg of intact total RNA using oligodT beads (Illumina® TruSeq® RNA Sample Preparation Kit v2). The purified mRNA was fragmented in the presence of bivalent cations and first strand cDNA was synthesized using Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and random hexamer primers (Invitrogen, Carlsbad, CA, USA). Second strand cDNA was synthesized in the presence of DNA polymerase I and RNaseH following standard protocol (Illumina). The cDNA was cleaned using Agencourt AMPure XP purification kit (Beckman-Coulter, Brea, CA, USA), amplified, quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and checked for quality with a Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). In total, 4 libraries were prepared for non-encumbered and Pasteuria encumbered samples (2 each) as per the Illumina protocols. The cDNA libraries were then sequenced on the Illumina HiSeq platform by outsourcing to Bionivid Technologies Pvt. Ltd., Bangalore, India.
Transcriptome assembly, quantitation and identification of differentially expressed transcripts
All the Paired End fastq files were subjected to standard quality control using NGS QC toolkit, v2.3.3 [77]. Reads with adapter contamination were removed along with their mate pair. High Quality (HQ) reads from all the samples were merged together to generate a Primary Assembly using Trinity Assembler (Trinity RNA-Seq-v2.0.6) [36] with default k-mer length 25, minimum contig length 200 bp and minimum k-mer coverage as 5. Further amelioration of the transcripts was done by filtering on the basis of average depth (≥ 5) and coverage (≥ 70%) in the individual samples [5]. The ameliorated transcripts were then subjected to clustering using CD_HIT_EST (v4.6.1) to make the secondary and final assembly with sequence identity threshold as 0.8 and length difference cut off as 0.9. The redundant transcripts were removed by CD_HIT_EST to make the secondary assembly. We observed a higher percentage (~ 45%) of small transcripts (< 500 bp). These were further fileterd based on annotation obtained against NCBI NRDB protein database. Sequences < 500 bp, which remained un-annotated, were discarded and a final transcriptome assembly was generated.
The final assembly was used for quantitation of transcripts in each of the individual libraries by using RSEM method [58]. The assembly validated .bam (Binary Sequence Alignment/Map) file was processed using bedtools [87] and samtools [59] for quantitation (read count estimation) for each transcript in a library and also to calculate the total coverage and average depth of the transcriptome in each library. The differentially expressed transcripts were identified using DESeq R package [3] between the treatment (endospore encumbered J2 s) and control (non-encumbered J2 s) groups in replicate. The differential expression of transcripts was determined with log2fold change ≥2 & P value ≤0.05 obtained by DESeq analysis.
Transcript annotation
Homology based annotation for the final transcriptome was done against National Center for Biotechnology Information (NCBI) non-redundant (nr) protein database [2]. The filtration criteria used for blastx were: Evalue ≤0.001, Query Coverage ≥60 and Percentage Identity ≥40. The results were subjected to Gene Ontology (GO) and Pathway analysis using Blast2GO [22] and KAAS [69]. Additional analysis were performed to find the secreted peptides, neuropeptides and RNAi pathway genes present in the differentially expressed transcripts by using SignalP v4.1 [72], and by blast search against the local database of C. elegans neuropeptide sequences (Li and Kim, 2008) and M. incognita RNAi genes [1] at E value ≤0.001 and query coverage ≥60, respectively.
Validation of RNA-Seq gene expression data by qRT PCR
Quantitative real time PCR (qRT PCR) was carried out to confirm the expression pattern of fifteen selected transcripts differentially expressing in the range of + 2.01 fold to − 5.06 fold in the RNA-Seq experiment. These 15 transcripts (Table 1) were chosen based on their predicted role in nematode – Pasteuria interaction, and included ten down-regulated transcripts and five up-regulated transcripts. cDNA was prepared from the same RNA samples that were used for RNA-Seq. Approximately 500 ng of RNA was reverse transcribed using cDNA synthesis kit (Superscript VILO, Invitrogen, Carlsbad, CA, USA) and qRT PCR was performed in a realplex2 thermal cycler (Eppendorf, Hamburg, Germany) using SYBR Green Supermix Kit (Eurogentec, Liege, Belgium). Each reaction mixture contained a final volume of 10 μl, comprised of 5 μl of SYBR Green PCR Master mix (Eurogentec, Liege, Belgium), 750 nM of each primer and 1.5 ng of cDNA. To normalize the gene expression level 18S rRNA (Genbank accession: HE667742), a constitutively expressed gene was used as internal reference. Three biological and three technical replicates were maintained for each sample. The data were analyzed by ΔΔCt method [60] and results were expressed as log2-transformed fold change values and Dunnet’s multiple comparison was performed for determining the statistical significance of the expression data. The primer details for qRT PCR are provided in Table 1.
Functional validation of role of transcripts in Meloidogyne – Pasteuria interaction
To determine the role of up- and down-regulated transcripts in the Meloidogyne – Pasteuria interaction, five out of the 15 transcripts, i.e., TR10010, TR14793, TR26363, TR16177 and TR10990 were selected and silenced by RNAi. Conserved domains in the transcripts were analyzed by NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and specific primers were designed to amplify them (Table 1). The dsRNA for each of the five transcripts was prepared as described earlier [84]. The effect of dsRNA treatment on the attachment of endospores on nematode cuticle was tested by soaking the J2 s in dsRNA solution at 28 °C for 18 h [105], followed by mixing 100 μl of endospore suspension (2.5 × 103 ml− 1) with approximately 200 dsRNA treated J2 s as described earlier [41]. The level of transcript suppression after dsRNA feeding was quantified and analyzed by qRT PCR [84]. Freshly hatched J2 s and J2 s soaked in dsGFP were used as controls. The endospore attachment was quantified by observing ~ 30 nematodes with a Zeiss Axiocam compound microscope (Carl Zeiss, Oberkochen, Germany) and photographed. The assays were performed in triplicate.
Results
Transcriptome sequencing and differentially expressed genes
The RNA sequencing of the Pasteuria endospore encumbered M. incognita J2 s and non-encumbered J2 s generated 32 to 39 million reads per sample (Table 2). The number of high quality reads in the raw sequence data was more than 99% for each of the samples (Table 2). Quality filtering of the raw sequence data using NGS QC resulted in 32.69, 34.62, 39.38 and 37.29 million reads for each of the two samples of non-encumbered and endospore encumbered nematode J2 s, respectively (Table 2). The quality filtered HQ sequence data from all the samples were used to generate a common reference assembly using the Trinity assembler resulting in a total of 161,705 transcripts. The minimum and maximum transcript lengths were 224 and 8320 bp, respectively with the N50 transcript length 1004 bp (Table 3). Improving the assembly by amelioration, removing the duplicates by CD_HIT_EST and filtering un-annotated < 500 bp transcripts resulted in a final merged assembly of 52,485 transcripts (minimum length: 224 bp, maximum length: 8320 bp) with N50 transcript length 1159 bp & GC% of 33.99 (Table 3).
The sequence data from individual samples were analyzed for differentially expressed transcripts between the endospore encumbered and non-encumbered J2 s. Eight hours post attachment of Pasteuria endospores, a total of 582 transcripts (1.10% of total transcripts) were found to be differentially expressed, which included 353 down-regulated and 229 up-regulated transcripts (Table 4, Fig. 1, Additional file 1).
A heat map showing the clustering of the differentially expressed transcripts between non-encumbered and Pasteuria encumbered juveniles. The map was generated by using the Log2 fold change values derived from transcriptome data by using DESeq R package between the treatment (endospore encumbered J2 s) and control (Non-encumbered J2 s) groups. The differential expression of transcripts was determined at FPKM > 1(Fragments Per Kilobase of transcript per Million mapped reads) in either of the pair of samples with log2fold change ≥2 with a t-test P value ≤0.05
Characterization of differentially expressed transcripts
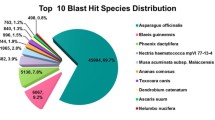
The annotation of 582 differentially expressed transcripts was done by blast search against NCBI nr database. Out of 582 transcripts, 246 transcripts showed nematode genes as the top hits (Fig. 2a). The three most frequent animal-parasitic nematodes found in the blast search were Ascaris suum Goeze, 1782 (42 hits), Strongyloides spp. (29 hits) and Ancylostoma spp. (26 hits), whereas M. incognita (12 hits), Bursaphelenchus xylophilus (Steiner, 1934) Nickle 1970 (6 hits) and M. hapla Chitwood, 1949 (5 hits) were top the top three plant-parasitic nematodes. Nineteen transcripts matched to Caenorhabditis spp., whereas one transcript showed the trematode species Schistosoma mansoni Sambon, 1907 as its topmost match. The differentially expressed transcripts were further functionally characterized into GO (Gene Ontology) categories of molecular function, biological processes and cellular components. The top ten GO enriched terms under each category are represented in Fig. 2b.
Donut charts showing the annotation of differentially expressed transcripts between the treatment (endospore encumbered J2 s) and control (Non-encumbered J2 s) groups. a. Out of 582 differentially expressed transcripts, 246 transcripts showed nematode genes as the top hits. Most of the transcripts matched to those of the animal parasitic nematodes. b. The Gene Ontology (GO) of the enriched transcripts. The top ten GO enriched terms under each category of cellular component, biological process and molecular function are represented. APN- animal parasitic nematodes, PPN- plant parasitic nematodes, FLN - free living nematodes
The transcripts showing highest down-regulation in the RNA-Seq experiment, TR38275 (− 7.0 folds) and TR22780 (− 6.9 folds) were annotated as heat shock protein 90 (HSP 90) and papain family cysteine protease, respectively (Additional file 1). The biggest group of down-regulated proteins were different ribosomal proteins (79 transcripts), whereas, hypotheticals represented the second largest group of the down-regulated proteins (49 transcripts). The heat shock proteins represented the third biggest group of down-regulated proteins (23 down-regulated transcripts) and included proteins such as HSP 70, HSP 20 and HSP 12.6. Eleven protease/proteinase (including two cysteine proteinases) and six kinases/phosphatases also showed down-regulation. Some other interesting proteins showing down-regulation were plectin (2 transcripts) and secretory proteins (3 transcripts), two fructose bisphosphate aldolase class-I family, one fatty acid and retinol binding protein, one signal transduction protein possibly involved in cell surface receptor signaling pathway (GO:0007166), three ubiquitin related proteins, three homologues of phospholipase B like 2 and one of phospholipase A2. In addition, M. incognita genes showing homology to tropomyosin (2 transcripts), actin-2 and tubulin (1 transcript each) were also found to be substantially down-regulated (Additional file 1).
The top three highly up-regulated transcripts in Pasteuria encumbered J2 s were TR25864, TR1903 and TR25239 that showed 9.4, 9.3 and 8.9 fold up-regulation in the RNA-Seq experiment. All of these three transcripts were uncharacterized or hypothetical proteins. In fact, hypotheticals, uncharacterized and unknown proteins were the dominant group under the up-regulated transcripts with 96 out of total 229 up-regulated transcripts falling into these categories. Cytochrome oxidase and major sperm protein domain containing proteins were the second most enriched transcript groups with 13 transcripts each. Blast based annotation showed that five transcripts matched to hormone receptor-like in 38 genes, four to transformation transcription domain-associated protein, two to venom allergen-like protein and one transcript matched to TK/FER protein kinase, UDP-glucosyl transferase and glutathione-S-transferase each (Additional file 1).
Analysis of the pathways represented by transcripts differentially expressed in the encumbered J2 s was performed by KAAS, an automatic genome annotation and pathway reconstruction tool. Annotation of the differentially expressed transcripts by KAAS showed that out of 582 transcripts, 148 could be mapped to 153 different pathways (Additional file 2). Ribosome (KO03010) was the most highly active pathway with 62 active transcripts, followed by thermogenesis (KO04714, 8 transcripts). A list of top 20 pathways and number of transcripts mapped to those pathways are shown in Table 5. A number of signaling pathways were active that are known to be involved in development and diverse functions in the encumbered J2 s such as ErbB signaling pathway (KO04012), Wnt signaling pathway (KO04310), Hedgehog signaling pathway (KO04341), TGF-beta signaling pathway (KO04350), Hippo signaling pathway (KO04390, KO04391), Apelin signaling pathway (KO04371), FoxO signaling pathway (KO04068), Rap1 signaling pathway (KO04015), PI3K-Akt signaling pathway (KO04151), AMPK signaling pathway (KO04152), cAMP signaling pathway (KO04024), cGMP-PKG signaling pathway (KO04022), MAPK signaling pathway (KO04010), mTOR signaling pathway (KO04150), HIF-1 signaling pathway (KO04066), calcium signaling pathway (KO04020), and sphingolipid signaling pathway (KO04071). In addition, pathways involved in recycling of cellular contents such as endocytosis (KO04144), phagosome (KO04145), lysosome (KO04142) and autophagy-animal (KO04140) were also found to be active in the encumbered J2 s.
The search for differentially regulated secreted peptides revealed that a total of 376 differentially expressed transcripts contained secretion signal, out of which 90 transcripts showed up-regulation, whereas 286 showed down-regulation. Of these 376 transcripts, only 320 could be annotated (Table 6, Additional file 2).
Validation of differentially expressed transcripts by qRT PCR
A total of fifteen transcripts, identified as differentially expressed in the RNA-Seq data, were taken up for validation by qRT PCR. Expression of 12 out of 15 transcripts was validated as significantly up- or down- regulated at 8 h post endospore encumbrance (Table 7). Unlike the RNA-Seq data, the fold change expression of the transcripts encoding heat shock protein 20, glutathione S-transferase and tropomyosin was not found to be significant. Data revealed that the selenium binding protein coding transcript (TR31579) showed maximum up-regulation by 6.89 fold, followed by the transcripts coding for glucosyl transferase (TR14793; 4.89 fold) and major sperm protein (TR26363; 2.92 fold), as compared to control. The transcripts showing highest down-regulation were TR14120 (phospholipase A2; − 7.98 fold), TR10010 (fructose bisphosphate aldolase; − 7.63fold) and TR11544 (fatty acid and retinol binding protein; − 6.09 fold), as compared to control. A comparison of the fold changes as detected by RNA-Seq and qRT PCR is provided in Table 7.
Functional evaluation of role of M. incognita genes on Pasteuria encumbrance by RNAi
Five transcripts were selected for functional validation based on their fold expression and predicted role in Meloidogyne – Pasteuria interaction. These included three down-regulated transcripts, viz., TR10010 (fructose bisphosphate aldolase), TR16177 (aspartic protease) and TR10990 (ubiquitin); and two up-regulated transcripts TR14793 (glucosyl transferase) and TR26363 (major sperm protein). The qRT PCR result showed that dsRNA treatment caused significant down-regulation of TR10010, TR16177, TR10990, TR14793 and TR26363 expression by 3.97 ± 0.26, 4.21 ± 0.37, 4.68 ± 0.12, 3.45 ± 0.33 and 2.95 ± 0.40 folds, respectively, as compared to the control. The dsRNA induced silencing of fructose bisphosphate aldolase (5 ± 3 endospores/J2) and glucosyl transferase (6 ± 4 endospores/J2) resulted in approximately six times lower endospore attachment as compared to the controls (water control: 32 ± 6 and dsGFP control: 31 ± 8 endospores/J2). On the other hand, silencing of aspartic protease (118 ± 12 endospores/J2) and ubiquitin (101 ± 8 endospores/J2) resulted in approximately three fold higher incidence of endospore attachment. However, there was no change on endospore attachment in the major sperm protein silenced J2 s (31 ± 6 endospores/J2), as compared to the controls (Fig. 3).
Effect of RNAi induced gene silencing of five transcripts on the attachment of Pasteuria penetrans endospores (seen as hemispherical structures) sticking onto Meloidogyne incognita J2 s. (a). non-silenced freshwater control J2 s, (b). dsGFP treated control J2 s, (c to g) – J2 s in which specific transcripts were RNAi silenced. The transcript ID and the average number of attached endospores are provided along with the name of the silenced genes. Silencing of TR10010 and TR14793 caused significantly reduced endospore attachment as compared to controls, whereas silencing of TR16117 and TR10990 caused a significant increase in the endospore attachment. Silencing of TR26363 did not cause any change in endospore attachment as compared to controls. (scale bar: 20 μm)
Discussion
Present RNA-Seq experiment provides a snapshot of the early molecular response by the M. incognita J2 s after exposure to P. penetrans endospores. Our results show that 582 nematode transcripts were differentially expressed at 8 h post Pasteuria endospore attachment of which 229 were up-regulated, and 353 were down-regulated. The expression patterns of 12 out of 15 differentially expressed (ten down- and five up-regulated) transcripts identified by the RNA-Seq were significantly validated to be true by qRT PCR. The functional role of five transcripts predicted to be important in Meloidogyne – Pasteuria interaction were validated by RNAi. The silencing of transcripts coding for fructose bisphosphate aldolase (TR10010) and glucosyl transferase (TR14793) resulted in approximately six times lower endospore attachment as compared to the controls, whereas, silencing of aspartic protease (TR16177) and ubiquitin (TR10990) coding transcripts resulted in approximately three fold higher incidence of endospore attachment. No change on endospore attachment was detected in major sperm protein (TR26363) silenced J2 s.
As established in several natural host-parasite interactions, viz., Daphnia – Pasteuria, Anopheles – Plasmodium and Bumblebee – Trypanosoma, a successful infection results from a compatible molecular interaction between the host and parasite genotypes [6, 55, 56, 64]. Daphnia magna Straus, 1820 responds to P. ramosa Metchnikoff, 1888 infection by rapid gene expression at 4 h, and the response tapers off with the passage of time at 8 and 12 h [64]. However, unlike Daphnia, which becomes infected after Pasteuria is orally ingested and adheres to the esophageal region, Meloidogyne becomes infected when Pasteuria spores adhere to the juvenile body surface and germinate when the nematode starts to feed on plants. In a study on Meloidogyne – Pasteuria interaction at three days post endospore adherence, Zou et al. [117] found that nematodes respond by an up-regulated gene expression, and identified 445 differentially expressed genes out of which 406 were up-regulated while 39 got down-regulated. Thirty seven immune responsive genes encoding collagens, cytochrome P450, lysozymes were among the other identified active proteins. Interestingly, Zou et al. [117] found that biosynthesis of unsaturated fatty acid pathways was up-regulated, while cytochrome P450 related genes were down-regulated. However, the findings from the Daphnia – P. ramosa system suggest that there may be an even earlier transcriptional response; hence, we focused on understanding the early transcriptional response by the nematode at eight hours after endospore attachment.
Our study showed that 153 pathways were active in Pasteuria encumbered M. incognita J2 s at 8 h post infection. As compared to our finding, 98 pathways were enriched at 3 days post Pasteuria attachment [117], of which 58 pathways were found to be active at both the time points (Additional file 2). ErbB signaling, Rap1 signaling, FoxO signaling, Hedgehog signaling, thermogenesis, longevity regulating pathways were some of the pathways active only at 8 h post endospore attachment; whereas pathways like cytochrome P450 (drug and xenobiotic metabolism), MAPK signaling, calcium signaling, HIF-1 signaling, mTOR signaling, PI3K-Akt signaling, Wnt signaling, TGF-beta signaling, Hippo signaling and regulation of actin cytoskeleton were active at both the time points. Some of the pathways active at 8 h assist cell-cell interaction and cellular adhesion in model organisms including C. elegans, for example, ErbB signaling [113], TGF-beta signaling [39], Hippo kinase cascade [114] and Rap1 signaling [8]. Other pathways like Wnt signaling [97], FoxO signaling [40], PI3K-Akt signaling [33], AMPK signaling [68], MAPK signaling [47], mTOR signaling [43] and HIF-1 signaling [116] operate in bacterial pathogenesis and stress responses. Taken together with the findings of Zou et al. [117], it appears that M. incognita immune pathways like cytochrome P450, MAPK signaling, HIF-1 signaling, mTOR signaling, PI3K-Akt signaling and TGF-beta signaling pathways were active upon infection by Pasteuria from 8 h to 3 days, in addition to the regulation of actin cytoskeleton pathway (Additional file 2).
Our study indicates that the nematode’s transcriptional responses after Pasteuria infection can be broadly understood as: (a) transcripts involved in nematode immunity, (b) transcripts involved in altering the cuticular surface coat property and thereby affecting endospore attachment, (c) transcripts involved in modulating the behaviour of the endospore encumbered juveniles.
Nematode immune responses triggered by Pasteuria
In addition to the immune pathways discussed above, at 8 h post endospore encumbrance, the ribosomal pathway (KO03010) was identified as the most affected with 62 differentially expressed transcripts. Besides, the ribosomal proteins represented the largest down-regulated group (79 transcripts) along with three ubiquitin related transcripts and one ZIP or ZRT/IRT-like protein (TR38155). The suppression of these transcripts suggest a reduction in the mRNA translational activity in Pasteuria encumbered M. incognita J2 s. This is consistent with the earlier reports of “effector-triggered” or “surveillance immunity”, where a down-regulation of host mRNA translation upon attack by bacterial pathogens has been reported in plants and in C. elegans [20, 29, 63, 67]. The bacterial pathogens disable the process of host mRNA translation, thereby preventing the production of anti-microbial molecules, and improve the chances of the infection [20]. It appears that a similar strategy is being used by Pasteuria while infecting M. incognita. The RNAi mediated knockdown of nematode ubiquitin also increased the endospore attachment on cuticular surface. Several bacterial pathogen effectors interact with eukaryotic ubiquitination pathways to exploit host functions [80]. The involvement of ubiquitin proteasome system, targeting proteins for degradation, has been established in C. elegans as an inducible response to infection [4, 20, 65]. Silencing of ubiquitin may lead to anomalous immune response where the cells fail to mount a sufficient immune response to remove the pathogen.
Approximately 13 autophagy related transcripts (e.g. endocytosis, KO04144; phagosome, KO04145; lysosome, KO04142 and autophagy-animal, KO04140) were down-regulated in the endospore encumbered M. incognita J2 s. It is well established that autophagy plays key role in pathogen defense [20]. Further, a down-regulation of 23 heat shock protein coding transcripts, for example, HSP70, HSP20 and HSP12.6 was observed. The HSPs are highly conserved group of proteins and are involved in protection against biotic and abiotic stress [19, 32, 44, 81, 91, 95, 115]. Similarly, HSP70s and HSP20 are known to play crucial role during disease stress response and serves as an endogenous danger signal [10, 23, 98]. Although down-regulation of HSPs during biotic stress is rare [78]; small HSPs (e.g. HSP17, HSP21 etc.) and HSP70 were found to be down-regulated in Arabidopsis when challenged by Pseudomonas syringae van Hall, resulting in suppression of host defense responses [10]. The down-regulation of autophagy related genes and HSPs by P. penetrans indicates pathogen induced host defense suppression.
In addition to the above mentioned pathways, several differentially expressed transcripts related to nematode immunity against bacterial pathogens were identified, such as, aspartic protease (TR16177), phospholipase A2 (TR14120), glutathione S-transferase (TR40461), selenium binding proteins (TR31579) and hormone receptor-like in 38 (Hr38) (TR5128, TR39260). Aspartic proteases function in the intracellular and extracellular degradation of proteins including processing of peptide hormones, antigens and immunoglobulins in parasitic nematodes [51, 100, 101]. In the present study, the RNAi induced inhibition of aspartic protease led to an increase in Pasteuria endospore adhesion on nematode cuticle indicating the defensive role of aspartic protease against bacterial pathogens. The inhibition of nematode phospholipase A2 by Pasteuria possibly prevents the bacterial infection structures from degradation, thereby allowing further infection [89]. Glutathione S-transferase (GST) was up-regulated in Pasteuria infected J2 s. In C. elegans and parasitic helminthes, GST is associated with an immune response and xenobiotic metabolism [11, 12, 18, 46, 62, 107, 108]. The up-regulation of GST may indicate nematode’s efforts to detoxify the potential Pasteuria effectors [86]. The RNAi mediated knockdown of M. incognita selenium binding protein (SeBP) significantly increased P. penetrans endospore attachment possibly through altering the cuticular surface coat property [83]. Lastly, the nuclear receptors (NRs) are ligand-dependent transcription factors that play pivotal roles in cell growth, differentiation, metabolism, reproduction and morphogenesis and immunity [13, 34]. Up-regulation of hormone receptor-like in 38 (Hr38) transcript, the Drosophila ortholog of the mammalian NGFI-B subfamily of orphan nuclear receptors [16] in M. incognita may be an indication of detection of the foreign nucleic acid particle (of Pasteuria), thereby triggering an immune response in M. incognita.
Modification of nematode cuticle biochemistry by Pasteuria
Mutations in genes involved in the building of complex cuticular surface components in C. elegans are known to affect bacterial adhesion [37]. Several differentially expressed transcripts identified in this study are predicted to interfere with attachment of endospores on the nematodes by altering the cuticle surface structure. These include TR10010 (fructose bisphosphate aldolase), TR14793 (glucosyl transferase), TR11544 (fatty acid and retinol binding (FAR) protein), and TR24724 (TK/FER kinase).
The fructose bisphosphate aldolase is a key enzyme of the glycolytic pathway [79] and its inhibition may contribute to accumulation of sugar molecules on the cuticle surface [30]. Similarly, glucosyl transferase catalyzes the transfer of sugar moieties to a wide range of acceptor molecules [73, 112]. RNAi induced in vitro inhibition of both these transcripts reduced the endospore attachment on nematode surface. Silencing fructose bisphosphate aldolase might have resulted in accumulation of sugar on the specific carbohydrate recognition sites on nematode mucin-like glycoprotein that binds to Pasteuria endospores [25, 85], thereby leading to reduction in attachment of endospores. Additionally, inhibition of glucosyl transferase may also result in mis-folding of the native glycoprotein molecules, thus resulting in decreased endospore attachment. Our findings are in concurrence with earlier observations on C. elegans, where the bus-8 mutant worms, defective in expressing glucosyl transferase were resistant to infection by Microbacterium nematophilum Hodgkin, Kuwabara and Corneliussen, 2000, due to failure of the bacterium to bind to the host surface [76]. The fatty acid and retinol binding (FAR) protein, uniquely present in nematodes, has been found to inhibit bacterial attachment onto nematode body surface by sequestering pharmacologically active lipids [45]. It has been demonstrated that the FAR protein is involved in the protection of M. incognita from attachment of P. penetrans endospores [84]. The TK/FER kinase regulates cadherin and integrin dependent cellular adhesion [9] and is also involved in nascent cell-cell adhesion via phosphorylation pathway [50]; and our results suggest it may also be involved in M. incognita and P. penetrans infection processes.
Alteration of nematode behavior and locomotion by Pasteuria
The endospore encumbered stressed nematode juveniles are slower in movement and finding their hosts [25, 84, 106]. The role of tropomyosin, actin and tubulin in regulation of muscle contraction has been established for several plant-parasitic, animal-parasitic and free living nematodes [38, 54]. As observed in our RNA-Seq data, down-regulation of these muscle-associated proteins upon endospore attachment may result in perturbed locomotion of the nematodes. The RNA-Seq data showed perturbation in a large number of neuropeptides in the Pasteuria encumbered juveniles in our study (Additional file 2). The neuropeptides of major groups, viz., FLPs, NLPs and ILPs affect numerous behavioral responses via various signaling pathways [14, 17, 21, 52, 109]. The association of FLPs and NLPs with nematode locomotion has been observed in C. elegans [48, 70, 90] and ILPs in the fruit fly movement [31]. Disruption of flp-18 in M. incognita by RNAi is known to reduce the plant parasitism [75]. The nematode behavioral alteration could also result from the effect of Pasteuria on the neuropeptides, as suggested by our results.
Apart from the above mentioned pathways and transcripts, two interesting observations from our study need mention. Firstly, we found that a major sperm protein (MSP) was up-regulated in infected juveniles, but RNAi mediated silencing of MSP coding transcript (TR26363) did not affect the adhesion of Pasteuria endospores onto nematode cuticle surface. MSP is involved in motility machinery and crawling movement of nematode sperms [92, 94], and is regulated at the onset of sexual differentiation in nematodes [49]. In C. elegans, MSP promotes the oocyte maturation and MAPK activation [53, 66]. It is well known that Pasteuria is largely confined to the reproductive system of M. incognita leading to complete destruction of its reproductive ability [7]. The sexual differentiation and gonad development process in M. incognita starts in the second-stage juveniles (J2 s) [74] and the up-regulation of MSP by Pasteuria might indicate an early interference with the nematode’s reproductive system.
Secondly, it has been observed that Pasteuria infected M. incognita have an increased life span of 10–12 days [25, 82], which could be a consequence of reduced mRNA translation in microbial infection. This corroborates with other studies in which the down-regulation of protein synthesis has resulted in increased lifespan [35], possibly by two different mechanisms. Firstly, reduced mRNA translation decreases the synthesis of normal as well as damaged proteins, resulting in lower accumulation of toxic proteins [42]. Secondly, as protein synthesis is a high cellular energy-consuming process [57], reduction of mRNA translation might increase energy availability and allow diversion of critical resources towards cellular maintenance and repair, thus promoting longevity [102, 103]. Our study shows a major reduction in protein synthesis in the Pasteuria encumbered nematodes thereby possibly extending lifespan when compared to the healthy nematodes.
Lastly, the hypothetical proteins represented the second largest group of the down-regulated proteins with 49 transcripts. There could be several interesting candidates within this group that may be directly or indirectly involved in the RKN – Pasteuria interaction.
Conclusion
Here we presented a transcriptomic analysis of M. incognita and P. penetrans interaction at eight hour post initial encounter. The transcriptome profile revealed that Pasteuria infection causes a reduction of the protein synthesis machinery of the nematode. We identified several differentially expressed transcripts putatively involved in nematode innate immunity, endospore attachment process and post-attachment behavioral modification of the juveniles. RNAi based functional validation of fructose bisphosphate aldolase (TR10010), glucosyl transferase (TR14793), aspartic protease (TR16177) and ubiquitin (TR10990) coding transcripts resulted in altered incidence of endospore attachment, whereas silencing of major sperm protein (TR26363) did not result in any alteration in Pasteuria attachment. Our results add new and significant information to the existing knowledge on early molecular interaction of M. incognita and P. penetrans.
Abbreviations
- GO:
-
gene ontology
- J2:
-
second stage juvenile
- KAAS:
-
KEGG Automated Annotation Server
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- qRT PCR:
-
quantitative real time PCR
- RKN:
-
root-knot nematodes
- RNAi:
-
RNA interference
References
Abad P, Gouzy J, Aury JM, Sereno PC, Danchin EG, Deleury E, et al. Genome sequence of the metazoan plan-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–15.
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106.
Bakowski MA, Desjardins CA, Smelkinson MG, Dunbar TA, Lopez-Moyado IF, Rifkin SA, et al. Ubiquitin-mediated response to microsporidia and virus infection in C. elegans. PLoS Pathog. 2014;10:e1004200.
Bankar KG, Todur VN, Shukla RN, Vasudevan M. Ameliorated de novo transcriptome assembly using Illumina paired end sequence data with trinity assembler. Genom Data. 2015;5:352–9.
Barribeau SM, Sadd BM, du Plessis L, Schmid-Hempel P. Gene expression differences underlying genotype-by-genotype specificity in a host-parasite system. Proc Natl Acad Sci U S A. 2014;111:3496–501.
Bird AF. The influence of the actinomycete, Pasteuria penetrans, on the host-parasite relationship of the plant-parasitic nematode, Meloidogyne javanica. Parasitology. 1986;93:571–80.
Boettner B, Van Aelst L. Control of cell adhesion dynamics by Rap1 signaling. Curr Opin Cell Biol. 2009;21:684–93.
Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, et al. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell–cell contacts upon externally applied stretch. Proc Nat Aca Sci U S A. 2012;109:12568–73.
Bricchi I, Bertea CM, Occhipinti A, Paponov IA, Maffei ME. Dynamics of membrane potential variation and gene expression induced by Spodoptera littoralis, Myzus persicae, and Pseudomonas syringae in Arabidopsis. PLoS One. 2012;(10):e46673.
Brophy PM, Crowley P, Barrett J. Detoxification reactions of Fasciola hepatica cytosolic glutathione transferases. Mol Biochem Parasitol. 1990;39:155–62.
Brophy PM, Pritchard DI. Parasitic helminth glutathione S-transferase: an update on their potential as targets for immuno and chemotherapy. Exp Parasitol. 1994;79:89–99.
Calgaro MR, Neto MDO, Figueira ACM, Santos MA, Portugal RV, Guzzi CA, et al. Orphan nuclear receptor NGFI-B forms dimers with nonclassical interface. Protein Sci. 2007;16:1762–72.
Chang SE, Han SS, Jung HJ, Choi JH. Neuropeptides and their receptors in psoriatic skin in relation to pruritus. Br J Dermatol. 2007;156:1272–7.
Charles L, Carbonne I, Davies KG, Bird D, Burke M, Kerry BR, et al. Phylogenetic analysis of Pasteuria penetrans using multiple genetic loci. J Bacteriol. 2005;187:5700–8.
Chen X, Rahman R, Guo F, Rosbash M. Genome-wide identification of neuronal activity-regulated genes in Drosophila. elife. 2016;5:e19942.
Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron. 2013;78:869–80.
Clark A. The comparative enzymology of the glutathione S-transferase from non-vertebrate organism. Comp Biochem Physiol. 1989;92:419–46.
Cohen IR, Young DB. Autoimmunity, microbial immunity and the immunological homuniculus. Immunol Today. 1991;12:105–10.
Cohen LB, Troemel ER. Microbial pathogenesis and host defense in the nematode C. elegans. Curr Opin Microbiol. 2015;23:94–101.
Cohen M, Reale V, Olofsson B, Knights A, Evans P, de Bono M. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. 2009;9:375–85.
Conesa A, Gotz S. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008;2008.
Das S, Mohapatra A, Sahoo PK. Expression analysis of heat shock protein genes during Aeromonas hydrophila infection in rohu, Labeo rohita, with special reference to molecular characterization of Grp78. Cell Stress Chaperones. 2015;20:73–84.
Davies KG, Danks C. Interspecific differences in the nematode surface coat between Meloidogyne incognita and M. arenaria related to the adhesion of the bacterium Pasteuria penetrans. Parasitology. 1992;105:475–80.
Davies KG, Kerry BR, Flynn CA. Observations on the pathogenicity of Pasteuria penetrans, a parasite of root-knot nematodes. Ann Appl Biol. 1988;112:1491–1501.
Davies KG, Laird V, Kerry BR. The motility, development and infection of Meloidogyne incognita encumbered with spores of the obligate hyperparasite Pasteuria penetrans. Revue de Nematologie. 1991;14:611–8.
Davies KG, Opperman CH. A potential role for collagen in the attachment of Pasteuria penetrans to nematode cuticle. IOBC/WPRS Bulletin. 2006;29:11–5.
Davies KG, Rowe J, Manzanilla-López R, Opperman CH. Re-evaluation of the life-cycle of the nematode-parasitic bacterium Pasteuria penetrans in root-knot nematodes, Meloidogyne spp. Nematology. 2011;13:825–35.
Dunbar TL, Yan Z, Balla KM, Smelkinson MG, Troemel ER. C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe. 2012;11:375–86.
Echevarria-Zomeno S, Ariza D, Jorge I, Lenz C, Del Campo A, Jorrin JV, et al. Changes in the protein profile of Quercus ilex leaves in response to drought stress and recovery. J Plant Physiol. 2009;166:233–45.
Erion R, Sehgal A. Regulation of insect behavior via the insulin-signaling pathway. Front Physiol. 2013;4:353.
Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82.
Gami MS, Wolkow CA. Studies of Caenorhabditis elegans DAF-2/insulin signaling reveal targets for pharmacological manipulation of lifespan. Aging Cell. 2006;5:31–7.
Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725.
Gonskikh Y, Polacek N. Alterations of the translation apparatus during aging and stress response. Mech Ageing Dev. 2017;168:30–6.
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–52.
Gravato-Nobre MJ, Hodgkin J. Microbial interactions with Caenorhabditis elegans: lessons from a model organism. In: Davies KG, Spiegel Y, editors. Biological control of plant-parasitic nematodes. Dordrecht: Springer; 2011. p. 65–90.
Guimaraes PM, Leal-Bertiolli SCM, Curtis RH, Davis EL, Bertioli DJ. Isolation of two cDNA encoding a tropomyosin and intermediate filament protein from soybean cyst nematde H glycines. Nematropica. 2001;33:87–95.
Heino J, Massague J. Cell adhesion to collagen and decreased myogenic gene expression implicated in the control of myogenesis by transforming growth factor beta. J Biol Chem. 1990;265:10181–4.
Hesp K, Smant G, Kammenga JE. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Exp Gerontol. 2015;72:1–7.
Hewlett TE, Dickson DW. A centrifugation method for attaching endospores of Pasteuria spp. to nematodes. Supplement to the. J Nematol. 1993;25:785–8.
Hipkiss AR. On why decreasing protein synthesis can increase lifespan. Mech Ageing Dev. 2007;128:412–4.
Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–45.
Kaufmann SHE, Schoel B. Heat shock proteins as antigens in immunity against infection and self. In: Morimoto RI, Tissieres A, Georgopoulos C, editors. The biology of heat shock proteins and molecular chaperones. Cold Spring Harbor laboratory press, cold Spring Harbor, NY; 1994. p. 495–531.
Kennedy MW, Garside LH, Goodrick LE, McDermott L, Brass A, Price NC, et al. The Ov20 protein of the parasitic nematode Onchocerca volvulus. A structurally novel class of small helix-rich retinol-binding proteins. J Biol Chem. 1997;(47):29442–8.
Ketterer B. Detoxication reactions of glutathione and glutathione transferases. Xenobiotica. 1986;16:957–73.
Kim ES, Kim MS, Moon A. TGF-β-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004;25:1375–82.
Kim K, Li C. Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol. 2004;475:540–50.
Klass M, Dow BA, Herndon M. Cell-specific transcriptional regulation of the major sperm protein in Caenorhabditis elegans. Dev Biol. 1982;93:152–64.
Kogata N, Masuda M, Kamioka Y, Yamagishi A, Endo A, Okada M, et al. Identification of Fer tyrosine kinase localized on microtubules as a platelet endothelial cell adhesion molecule-1 phosphorylating kinase in vascular endothelial cells. Mol Biol Cell. 2003;14:3553–64.
Krieger TJ, Hook VY. Purification and characterization of a cathepsin D protease from bovine chromaffin granules. Biochemistry. 1992;31:4223–31.
Kubiak TM, Larsen MJ, Zantello MR, Bowman JW, Nulf SC, Lowery DE. Functional annotation of the putative orphan Caenorhabditis elegans G protein-coupled receptor C10C6.2 as a FLP15 peptide receptor. J Biol Chem. 2003;278:42115–20.
Kuwabara PE. The multifaceted C. elegans major sperm protein: an ephrin signaling antagonist in oocyte maturation. Genes Dev. 2003;17:155–61.
Kwa MS, Veenstra JG, Van Dijk M, Roos MH. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J Mol Biol. 1995;246:500–10.
Lambrechts L, Halbert J, Durand P, Gouagna LC, Koella JC. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malar J. 2005;4:3.
Lambrechts L. Dissecting the genetic architecture of host-pathogen specificity. PLoS Pathog. 2010;6:e1001019.
Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–34.
Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCt method. Methods. 2001;25:402–8.
Mahajan-Miklos S, Tan MW, Rahme LG, Ausubel FM. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96:47–56.
Mannervik B. The isoenzymes of glutathione transferase. Adv Enzymol. 1985;57:357–417.
McEwan DL, Kirienko NV, Ausubel FM. Host translational inhibition by Pseudomonas aeruginosa exotoxin a triggers an immune response in Caenorhabditis elegans. Cell Host Microbe. 2012;11:364–74.
McTaggart SJ, Cezard T, Garbutt JS, Wilson PJ, Little TJ. Transcriptome profiling during a natural host-parasite interaction. BMC Genomics. 2015;16:643.
Melo JA, Ruvkun G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell. 2012;149:452–66.
Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans. Genes Dev. 2003;17:187–200.
Mohr I, Sonenberg N. Host translation at the nexus of infection and immunity. Cell Host Microbe. 2012;12:470–83.
Moreno-Arriola E, Hafidi ME, Ortega-Cuéllar D, Carvajal K. AMP-activated protein kinase regulates oxidative metabolism in Caenorhabditis elegans through the NHR-49 and MDT-15 transcriptional regulators. PLoS One. 2016;11:e0148089.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–5.
Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc Natl Acad Sci U S A. 2001;98:14000–5.
Nicholas HR, Hodgkin J. Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol Immunol. 2004;41:479–93.
Nielsen H. Predicting secretory proteins with SignalP. In: Kihara D, editor. Protein function prediction (methods in molecular biology vol. 1611): Springer; 2017. p. 59–73.
Nomura KH, Murata D, Hayashi Y, Dejima K, Mizuguchi S, Kage-Nakadai E, et al. Ceramide glucosyltransferase of the nematode Caenorhabditis elegans is involved in oocyte formation and in early embryonic cell division. Glycobiology. 2011;21:834–48.
Papadopoulou J, Traintaphyllou AC. Sex differentiation in Meloidogyne incognita and anatomical evidence of sex reversal. J Nematol. 1982;14:549–66.
Papolu PK, Gantasala NP, Kamaraju D, Banakar P, Sreevathsa R, Rao U. Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode, Meloidogyne incognita. PLoS One. 2013;8:e80603.
Partridge FA, Tearle AW, Gravato-Nobre MJ, Schafer WR, Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 2008;317:549–59.
Patel RK, Jain M. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 2012;7:e30619.
Pavlova EL, Rikhvanov EG, Tauson EL, Varakina NN, Gamburg KZ, Rusaleva TM, et al. Effect of salicylic acid on the development of induced thermotolerance and induction of heat shock protein synthesis in the Arabidopsis thaliana cell culture. Russ J Plant Physiol. 2009;56:68–73.
Penhoet EE, Rutter WJ. Catalytic and immunochemical properties of homomeric and heteromeric combinations of aldolase subunits. J Biol Chem. 1971;246:318–23.
Perrett CA, Lin DYW, Zhou D. Interactions of bacterial proteins with host eukaryotic ubiquitin pathways. Front Microbiol. 2011;2:143.
Peter E, Candido M. The small heat shock proteins of the nematode Caenorhabditis elegans: structure, regulation and biology. In: Arrigo AP, Müller WEG, editors. Progress in molecular and subcellular biology. Berlin, Heidelberg: Springer; 2002. p. 61–78.
Phani V, Rao U. Revisiting the life-cycle of Pasteuria penetrans infecting Meloidogyne incognita under soil-less medium, and effect of streptomycin sulfate on its development. J Nematol. 2018;50:91–8.
Phani V, Somvanshi VS, Rao U. Silencing of a Meloidogyne incognita selenium-binding protein alters the cuticular adhesion of Pasteuria penetrans endospores. Gene. 2018b;677:289–98.
Phani V, Shivakumara TN, Davies KG, Rao U. Meloidogyne incognita fatty acid- and retinol- binding protein (mi-FAR-1) affects nematode infection of plant roots and the attachment of Pasteuria penetrans endospores. Front Microbiol. 2017;8:2122.
Phani V, Shivakumara TN, Davies KG, Rao U. Knockdown of a mucin-like gene in Meloidogyne incognita (Nematoda) decreases attachment of endospores of Pasteuria penetrans to the infective juveniles and reduces nematode fecundity. Mol Plant Pathol. 2018a;19:2370–83.
Preston JF, Dickson DW, Maruniak JE, Nong G, Brito JA, Schmidt LM, et al. Pasteuria spp.: systematics and phylogeny of these bacterial parasites of phytopathogenic nematodes. J Nematol. 2003;35:198–207.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
Rao U, Mauchline TH, Davies KG. The 16S rRNA gene of Pasteuria penetrans provides an early diagnostic of infection of root-knot nematodes (Meloidogyne spp.). Nematology. 2012;14:799–804.
Raymond B, Leduc D, Ravaux L, Goffic RL, Candela T, Raymondjean M, et al. Edema toxin impairs Anthracidal phospholipase A2 expression by alveolar macrophages. PLoS Pathog. 2007;3:e187.
Reinitz CA, Herfel HG, Messinger LA, Stretton AO. Changes in locomotory behavior and cAMP produced in Ascaris suum by neuropeptides from Ascaris suum or Caenorhabditis elegans. Mol Biochem Parasitol. 2000;111:185–97.
Roberts R, Agius C, Saliba C, Bossier P, Sung Y. Heat shock proteins (chaperones) in fish and shellfish and their potential role in relation to fish health: a review. J Fish Dis. 2010;33:789–801.
Roberts TM, Stewart M. Role of major sperm protein (MSP) in the protrusion and retraction of Ascaris sperm. Int Rev Cell Mol Biol. 2012;297:265–93.
Sayre RM, Wergin WP. Bacterial parasite of a plant nematode: morphology and ultrastructure. J Bacteriol. 1977;129:1091–101.
Schormann N, Symersky J, Luo M. Structure of sperm-specific protein SSP-19 from Caenorhabditis elegans. Acta Crystallogr D Struct Biol. 2004;60:1840–5.
Shinnick TM. Heat shock proteins as antigens of bacterial and parasitic pathogens. Curr Top Microbiol Immunol. 1991;167:145–60.
Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect Immun. 2003;71:2208–17.
Silva-García O, Valdez-Alarcón JJ, Baizabal-Aguirre VM. The Wnt/β-catenin signaling pathway controls the inflammatory response in infections caused by pathogenic bacteria. Mediat Inflamm. 2014;2014:310183.
Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, et al. Dissection of the fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell. 1998;10:1055–68.
Spiegel Y, McClure MA. The surface coat of plant-parasitic nematodes: chemical composition, origin, and biological role- a review. J Nematol. 1995;27:127–34.
Szecsi PB. The aspartic proteases. Scand J Clin Lab Invest. 1992:52:5–22.
Tang J, Wong RN. Evolution in the structure and function of aspartic proteases. J Cell Biochem. 1987;33:53–63.
Tavernarakis N. Protein synthesis and aging: eIF4E and the soma vs. germline distinction. Cell Cycle. 2007;6:1168–71.
Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18:228–35.
The C. elegans sequencing consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8.
Urwin PE, Lilley CJ, Atkinson HJ. Ingestion of double-stranded RNA by pre-parasitic juvenile cyst nematodes leads to RNA interference. Mol Plant Microb Interact. 2002;15:747–52.
Vagelas IK, Dennett MD, Pembroke B, Gowen SR. Adhering Pasteuria penetrans endospores affect movements of rootknot nematode juveniles. Phytopathol Mediterr. 2012;51:618–24.
van Rossum AJ, Brophy PM, Tait A, Barrett J, Jefferies JR. Proteomic identification of glutathione S-transferases from the model nematode Caenorhabditis elegans. Proteomics. 2001;1:1463–8.
Vibanco-P’erez N, Landa A. Glutathione S-transferase in helminth parasites. Rev Lat Microbiol. 1998;40:73–85.
Waggoner LE, Hardaker LA, Golik S, Schafer WR. Effect of a neuropeptide gene on behavioral states in Caenorhabditis elegans egg-laying. Genetics. 2000;154:1181–92.
Whitehead AG, Hemming JR. A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann Appl Biol. 1965;55:25–38.
Wong D, Bazopoulou D, Pujol N, Tavernarakis N, Ewbank JJ. Genome-wide investigation reveals pathogen-specific and shared signatures in the response of Caenorhabditis elegans to infection. Genome Biol. 2007;8:R194.
Yan JP, He H, Zhang JJ, Liu Z, Wang JM, Li XF, et al. Overexpression of UDPGlucosyltransferase 71C5 increases glucose tolerance in Arabidopsis thaliana. Russ J Plant Physiol. 2010;(6):875–81.
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–37.
Yu FX, Guan KL. The hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–71.
Zevian SC, Yanowitz JL. Methodological considerations for heat shock of the nematode Caenorhabditis elegans. Methods. 2014;68:450–7.
Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA. The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS One. 2009;4:e6348.
Zou X, Lu Y, Sun Q, Huang H, Liu M, Bao S. Transcriptome analysis of Meloidogyne incognita encumbered by Pasteuria penetrans endospores provides new insights into bacteria and nematode interaction. Nematology. 2017;19:839–51.
Acknowledgements
VP acknowledges PG School, ICAR-IARI for support during the study. VSS and UR acknowledge the Director, ICAR-IARI and the Joint Director (Research), ICAR-IARI for support. We thank Ms. Roli Budhwar, Bionivid Technology Pvt. Ltd. for technical coordination and help.
Funding
Authors acknowledge funding from British Council UKIERI project grant DST 59FX34 2014, for designing of the study and collection of data. The role of funding body does not include data analysis, interpretation and manuscript writing. Ph.D. student VP acknowledges the PG School, ICAR-IARI for research fellowship.
Availability of data and materials
The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject no. PRJNA479713, SRA accession no. SRP152065.
Author information
Authors and Affiliations
Contributions
VP and UR conceived and designed the study. VP and VSS performed the experiments and analyzed the data. RNS assisted with the bioinformatic analysis. VP, VSS, RNS, KGD and UR wrote and revised the manuscript, and approved the final manuscript.
Corresponding authors
Ethics declarations
Authors’ information
All authors have substantially contributed in the present study and approved the final draft for submission.
Ethics approval and consent to participate
Not applicable on the manuscript.
Consent for publication
Not applicable on the manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
A list of all, and the differentially expressed transcripts between endospore encumbered and non-encumbered Meloidogyne incognita juveniles at 8 hours post-Pasteuria attachment. (XLSX 7442 kb)
Additional file 2:
KAAS pathway mapping, comparison of pathways active at 8 hours and 3 days, and annotation of neuropeptides and secreted peptides in the differentially expressed transcripts. (XLSX 534 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Phani, V., Somvanshi, V.S., Shukla, R.N. et al. A transcriptomic snapshot of early molecular communication between Pasteuria penetrans and Meloidogyne incognita. BMC Genomics 19, 850 (2018). https://doi.org/10.1186/s12864-018-5230-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-5230-8