Abstract
Background
The FK506-binding proteins (FKBPs) play diverse roles in numerous critical processes for plant growth, development, and abiotic stress responses. However, the FKBP gene family in the important fruit crop apple (Malus × domestica Borkh.) has not been studied as thoroughly as in other species. Our research objective was to investigate the mechanisms by which apple FKBPs enable apple plants to tolerate the effects of abiotic stresses.
Results
Using bioinformatics-based methods, RT-PCR, and qRT-PCR technologies, we identified 38 FKBP genes and cloned 16 of them in the apple genome. The phylogenetic analysis revealed three major groups within that family. The results from sequence alignments, 3-D structures, phylogenetics, and analyses of conserved domains indicated that apple FKBPs are highly and structurally conserved. Furthermore, genomics structure analysis showed that those genes are also highly and structurally conserved in several other species. Comprehensive qRT-PCR analysis found various expression patterns for MdFKBPs in different tissues and in plant responses to water-deficit and salt stresses. Based on the results from interaction network and co-expression analyses, we determined that the pairing in the MdFKBP62a/MdFKBP65a/b-mediated network is involved in water-deficit and salt-stress signaling, both of which are uniformly up-regulated through interactions with heat shock proteins in apple.
Conclusions
These results provide new insight for further study of FKBP genes and their functions in abiotic stress response and multiple metabolic and physiological processes in apple.
Similar content being viewed by others
Background
Immunophilins are cellular receptor proteins for immunosuppressive drugs that can be combined with FK506, Cyclosporine-A (CsA), and rapamycin [1, 2]. As agents of immunosuppression, immunophilins are divided into two categories: cyclophilins, which can bind to CsA; and FK506-binding proteins (FKBPs), which can bind to FK506 and rapamycin [2,3,4]. The FKBPs belong to a superfamily for peptidyl-prolyl cis-transisomerase (PPIase, EC 5.1.2.8), which catalyzes the rotation of the peptide bond that immediately precedes a proline residue between the cis and trans configurations; family members are present in all organisms and almost all subcellular compartments, from yeasts to humans [1, 2, 5, 6].
All FKBPs contain at least one FK506-binding domain (FKBd) of approximately 110 amino acids that provides the active site for PPIase catalysis and the receptor site for proline and proline analogues [1, 2, 7]. In FKBPs, the formation of a well-conserved tertiary structure of FKBd primarily contains six anti-parallel beta sheets that form a concave surface and hydrophobic sidechains; a short alpha helix; and several solvent-exposed loops that interact with substrates bound at the FKBP active site [1, 2, 7]. Members of FKBP families in plants range significantly in size, from single-domain (SD) isoforms that comprise a single FKBd to multiple-domain (MD) proteins that contain up to three FKBds, along with a tetratricopeptide repeat (TPR), C-terminal calmodulin-binding domains, or a coiled–coil domain [1, 2, 6,7,8]. In plants, FKBPs have been isolated from the mitochondrion, endoplasmic reticulum, cytosol, and nucleus [9,10,11,12]. However, most FKBP subcellular locations are expressed in the chloroplast thylakoid [13,14,15].
Over the past 20 years, researchers have reported that plant FKBPs perform diverse functions in numerous critical processes for growth and development, including signal transduction, hormonal pathways, stress responses, photosynthesis, and gene transcription [1, 2, 5,6,7,8, 16,17,18]. Both AtFKBP12 from Arabidopis thaliana (hereafter Arabidopsis) and PwFKBP12 (from Picea wilsonii) are involved in cell cycle regulation and embryo development, and they also participate in controlling the direction of pollen tube growth through their relationship with FKBP12 interacting protein 37 kD (AtFIP37) and a putative CCAAT-binding transcription factor protein PwHAP5, respectively [19,20,21]. In the green algae Chlamydomonas reinhardtii and maize (Zea mays; Zm), FKBP12 and rapamycin form a complex that inhibits TOR kinase, an important regulator of plant germination and development [22, 23]. Overexpression of Polytrichastrum alpinum PaFKBP12 enhances tolerance to heat, salt, and drought stresses in Arabidopsis [24]. The FKBP62 (ROF1) and FKBP65 (ROF2) proteins from Arabidopsis and the closely related FKBPs, FKBP73, and FKBP77 from wheat (Triticum aestivum) are induced by malondialdehyde (MDA) treatment, wounding, and NaCl stress, all of which up-regulate the expression of genes involved in abiotic stress responses [1, 7, 25, 26]. Moreover, AtFKBP20–2 in the chloroplast thylakoid lumen is required for accumulation of the PSII supercomplex in Arabidopsis [16]. The AtFKBP72 (PAS1) protein helps control cell proliferation, and pas1 mutant plants show severe developmental defects [27]. Furthermore, AtFKBP42 (TWD1) modulates the activities of P-glycoproteins, influencing auxin efflux from the root apoplast into the cytoplasm by interacting with the B family of ABC transporter (ABCB), and also regulating brassinosteroid signaling through an interaction with the receptor kinase BRASSINOSTEROID-INSENSITIVE 1 (BRI1) [28,29,30,31,32]. Although twd1 mutant plants exhibit disoriented growth for all organs, they still develop fertile flowers and seeds [28]. Whereas members of the FKBP family have conserved some of their basic functions in higher eukaryotes, other biological functions are unique to each member, such that one member cannot completely compensate for the absence of another [33]. Therefore, researchers must characterize each member independently to uncover those unique functions.
In many plant species, increasing numbers of FKBP gene family members have been identified and characterized based on the highly conserved FK506-binding domain reported for Arabidopsis (At) [34], rice (Oryza sativa; Os) [8, 35, 36], maize [37], grapevine (Vitis vinifera; Vv) [3], strawberry (Fragaria ananassa; Fa) [5], and peach (Prunus persica; Pp) [6]. Although considerable details are known about this family in several species because it has been subjected to extensive genomic analysis; members in apple (Malus × domestica Borkh.) have not been as thoroughly investigated. Nevertheless, recent completion of the draft genome sequence for apple has enabled genome-wide analyses of FKBP genes in that species [38,39,40]. Here, this work identified the FKBP genes in apple and examined their FK506-binding domain, protein and gene structures, conserved domains, phylogenetic relationships, chromosomal locations, cis-acting elements, and expression patterns for MdFKBPs cloned from various tissues and in response to water deficits and salt stress. To our knowledge, this is the first comprehensive study of the FKBP gene family in apple. These results could facilitate future investigations into the functions of these genes in apple growth, developmental processes, and abiotic stress responses, and would lay a solid foundation for efforts to introduce improved apple cultivars.
Methods
Identification of apple FKBP genes
The database of the Arabidopsis FKBP family was downloaded from the TAIR website (http://www.arabidopsis.org/) [34]. As query sequences for BlastP (http://www.rosaceae.org/tools/ncbi_blast) against predicted apple proteins, we used 23 Arabidopsis FKBP proteins and the consensus protein sequences of the FK506-binding domain Hidden Markov Model (HMM) profile (FKBP_C, PF00254) from the Pfam database (http://pfam.xfam.org/family/PF00254). We then searched all of those FKBP sequences against the apple genome database (https://www.rosaceae.org/gb/gbrowse/malus_x_domestica/) with HMMER v3.0 and BlastP [41]. Confirming the reliability of those protein sequences ensured that the FK506-binding domain was present in each candidate MdFKBP protein. For this, we used the Pfam database (http://pfam.sanger.ac.uk/search) and NCBI-Conserved Domain Search (NCBI-CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) [41].
Sequence logo and structure modeling
Sequence logos for the FK506-binding domain in 22 FKBP12 genes and the TPR domain in 25 FKBP42 genes were generated by the application WebLogo (http://weblogo.threeplusone.com) [42]. The web server SWISS-MODEL (http://swissmodel.expasy.org/) [43] was used for modeling and predicting the homology of protein structures for MdFKBP12, MdFKBP42, MdFKBP62, and MdFKBP72. The proposed 3-D structure was modelled on the original NMR structure in PDB ID: 5HWB [44], 2IF4 [45], 1KT1 [46], and 3JYM [47], and RasTop 2.2 software (http://www.geneinfinity.org/rastop/) was used to present that model [41].
Sequence alignment and phylogenetic analysis
Multiple sequence alignments of 36 MdFKBP protein sequences were performed for using DNAMAN 6.0.3.99 with its default parameters. The phylogenetic tree for the MdFKBP gene family was constructed with MEGA 5.2 software (www.megasoftware.net) and the Neighbour-Joining (NJ) method, utilizing the amino acid sequences for those 36 proteins as well as FKBP12 proteins from other species. The following parameters were used in the NJ method: bootstrap (1000 replicates), complete deletion, and amino:p-distance. Protein–protein interactions were determined using STRING 10.0 (http://string-db.org/), setting the confidence score at > 0.9 [48, 49].
Analyses of intron-exon structure, genome distribution, and gene duplications
Genomic sequences (apple v1.0), gene distributions on chromosomes, and genome locations for the FKBP genes in apple were downloaded from the apple genome database (Additional file 1: Table S1). Data for intron-exon distribution of the FKBP12, FKBP42, FKBP62, and FKBP72 genes in various species were downloaded from the resource PLAZA 3.0 (http://bioinformatics.psb.ugent.be/plaza/) (Additional file 2: Table S2) [50]. The MdFKBP genes were mapped onto chromosomes by using MapInspect (www.plantbreeding.wur.nl/UK/software_mapinspect.html), a locational software for identifying chromosomal positions [41]. We followed the method of Hu and Liu [51] for investigating segmental and tandem duplication events.
Prediction of cis-acting elements in promoters
To examine the putative cis-acting elements in the promoters of apple FKBP genes, we isolated sequences that were 1500 bp upstream of the translational start codon, using the contig sequences of that genome and PCR amplification. Details for the promoters used here are listed in Table 2 and Additional file 3: Table S3. Possible cis-acting elements in those promoters were then predicted according to the Plant CARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Plant materials and treatments
For gene cloning and expression analysis, young roots, stems, and leaves, as well as flowers and mature fruit (70 mm, red peel, 150 d after bloom) were collected from apple plants that were five-years-old after bud-grafting. The scion was Malus domestica ‘Golden Delicious’ and the rootstock was M. hupehensis. Samples used for examining the effects of water deficits and NaCl stress were harvested from plants 3 months after bud-grafting was performed with ‘Golden Delicious’ scions and M. hupehensis rootstocks. These grafted plants were grown in pots (height, 320 mm; diameter, 300 mm) in a greenhouse and treatments began when the plants were approximately 500 mm tall. To induce a water deficit, irrigation was withheld from certain plants for up to 8 d while the designated control plants continued to receive normally scheduled irrigation [52]. For salt-stress treatment, plants were irrigated for 2 d with a 200 mM NaCl solution [52]. The sampling schedule involved harvesting mature leaves at the middle nodes on Days 0, 4, and 8 of the water deficit; or on Days 0, 1, and 2 during salt treatment. All tissues were frozen immediately in liquid N2 and stored at − 80 °C.
Cloning of MdFKBPs and qRT-PCR analysis of expression
Total RNA was extracted from previously frozen tissues according to the hot borate method [53]. Two micrograms of total RNA were used to synthesize first-strand cDNA. For cloning MdFKBP, complete open reading frames (ORFs) were obtained by RT-PCR, using specific primers listed in Additional file 4: Table S4. The 5’- and 3’-untranslated regions (UTRs) were obtained with a Rapid Amplification for cDNA Ends kit (TaKaRa, Dalian, China). For the qRT-PCR assays, reverse-transcription was performed with 1 μg of total RNA from each sample, followed by PCR-amplification of 1 μL of the product. We conducted the qRT-PCR assays in 20-μL reaction mixtures that contained 10 μL of SYBR® Premix Ex Taq™ (TaKaRa), and used an iQ5 instrument (Bio-Rad, Hercules, CA, USA) as described before [54]. Thermal-cycling included an initial 3 min at 95 °C; then 40 cycles of 10 s at 95 °C, 30 s at 58 °C, and 15 s at 72 °C; followed by 3 min at 72 °C and then 81 cycles of 7 s each, increasing by an increment of 0.5 °C from 55 °C to 95 °C. Three biological replicates were tested in each assay, and △Ct values were calculated by using MdMDH as our endogenous control [55]. Relative quantification values were calculated according to the 2-△△Ct method [56] and dissociation curve analysis was performed for determining the specificity of the amplifications. Heat maps of MdFKBP family members were constructed and hierarchical clustering was conducted with the MeV v4.8.1 software package [57].
Statistical analysis
All data were analyzed with IBM SPSS Statistics v20. One-way ANOVA and Tukey’s tests were used to compare the results from abiotic stress treatments against their respective controls. Values were considered significantly different at p < 0.05.
Results
Identification and annotation of apple FKBP genes
To identify the genes in the apple genome that encode FK506-binding proteins, we conducted a BlastP of the apple genome database and identified 42 putative FKBP family genes in that genome. We then used the Pfam and NCBI-CDD databases to verify that all of them were FKBP members by searching for the FK506-binding domain in the amino acid sequences encoded by all 42 genes. From this, we confirmed the identity of 36 typical apple FKBP genes in the original dataset (Table 1). In addition to those 36, we found four genes with partial FKBP ORFs in the apple genome (Additional file 5: Table S5). However, we were unable to analyze those genes further because of their incomplete ORFs. Another two were identified as TIG genes. However, unlike other species, the apple TIG genes do not contain the FKBP_C domain. We classified these 36 apple FKBP genes as either SD members with an FKBP catalytic domain, or MD members with a TPR domain. Among those 36, 29 genes were characterized as SD members, three genes (MDP0000205111, MDP0000141863, and MDP0000182579) were predicted to encode proteins containing two FKBP_C domains, and four (MDP0000175388, MDP0000292276, MDP0000858936, and MDP0000146478) contained three FKBP_C domains (Table 1).
For the nomenclature of the apple FKBP genes, we followed the previously published rules for Arabidopsis [34] and rice [36]. Thus, they were named for the FKBP506-binding protein and labeled according to their orthology with reported isoforms in Arabidopsis, based on their estimated molecular weights. For some genes in which more than two proteins showed an orthologous sequence with the same gene in Arabidopsis, lower-case extension letters were added to the name according to the order of similarity (Table 1). For example, the putative FKBPs from apple that shared the highest amino acid identity (75, 58, 62, 72, 74, and 67%) with AtFKBP42 were denoted as ‘MdFKBP42a, -42b, -42c, -42d, -42e, and -42f’, despite their respective predicted molecular weights being 42, 56, 38, 22, 29, and 27 kDa. In addition, FKBP15–3, FKBP17–2, and FKBP17–3 were not predicted in apple, while two new members (MdFKBPa and MdFKBPb) were found with the number of their FKBd being 2 and 1, respectively. We next cloned all of the full-length apple FKBP genes based on predicted nucleotide sequences in the apple genome and in the NCBI Nucleotide Database. As shown in Table 1, this revealed that the full-length cDNAs of MdFKBP12, − 15-2, − 16-2, − 16-3, − 17-1, − 18, − 19, − 20-1a, −42a, − 43, −53a, −62a, −65a, −65b, −72a, and MdTIGa had been isolated and confirmed by RT-PCR. Subsequently, the corresponding 5′- and 3’-UTRs of each gene were amplified.
Multiple sequence alignments and 3-D structures of MdFKBP12, MdFKBP42, MdFKBP62, and MdFKBP72 proteins
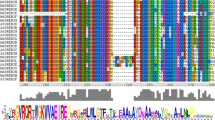
Multiple alignments demonstrated that the FK506-binding domains were conserved among the MdFKBP proteins (Fig. 1). To characterize the FK506-binding domain in the MdFKBP12 protein and the TPR domain in MdFKBP42 protein, we produced sequence logos, which enabled us to determine that the FK506 binding domain and the TPR domain from various species were highly conserved at each residue position (Fig. 2a, b; Additional file 6: Table S6). We then used the SWISS-MODEL web server for modeling and analysis of homology among protein structures. From this, we built homology models for MdFKBP12, MdFKBP42, MdFKBP62, and MdFKBP72 protein homology models and evaluated them with homologous templates 5HWB.pdb, 2IF4.pdb, 1KT1.pdb, and 3JYM.pdb, respectively. The results indicated that the MdFKBP12 structure most closely matched that of the fungal FKBP12s [44] (Fig. 2c; PDB ID: 5HWB.1.A; root-mean-square deviation, or RMSD, = 2.05 Å; and 52.43% sequence identity for residues 1–112), the MdFKBP42 structure most closely matched AtFKBP42 [45] (Fig. 2d; PDB ID: 2IF4.1.A; RMSD = 2.85 Å; and 79.01% sequence identity for residues 42–299), the MdFKBP62 structure most closely matched the squirrel monkey FKBP51 [46] (Fig. 2e; PDB ID: 1KT1.1.A; RMSD = 2.80 Å; and 41.03% sequence identity for residues 35–552), and the MdFKBP72 structure most closely matched wheat FKBP73 [47] (Fig. 2f; PDB ID: 3JYM.1.A; RMSD = 2.28 Å; and 32.19% sequence identity for residues 48–384).
The secondary and three-dimensional structure of FKBP genes. Sequence logos of FK506-binding domains (a) and TPR domains (b) in 25 FKBP12 and 25 FKBP42 genes, respectively. Heights of symbols within each stack indicate relative frequency of each amino acid at that position. Logos were obtained through multiple alignments of 25 FKBP12 and 25 FKBP42 protein sequences. The a-helix and b-sheet appear at top of corresponding amino acid sequences. Predicted three-dimensional tertiary structural modes of MdFKBP12 (c), MdFKBP42 (d), MdFKBP62 (e), and MdFKBP72 (f) proteins (PDB IDs: 5HWB.1.A, 2IF4.1.A, 1KT1.1.A, and 3JYM.1.A, respectively). Yellow, red, and green-blue indicate β-Sheets, α-helices, and strands, respectively. RasTop software was used to generate 3-D representation
Phylogenetic tree and analysis of conserved domains among apple FKBP genes
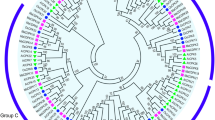
To examine the evolutionary relationships among apple FKBPs, the full-length amino acid sequences of 36 apple FKBPs were used, plus ZmFKBP12, OsFKBP12, VvFKBP12, PpFKBP12, and AtFKBP12, to generate a phylogenetic tree. These 41 total FKBPs were assigned to one of three groups (Fig. 3). In Group I, most of the 24 putative FKBPs each had a single FKBd. The exceptions in this group were MdFKBP42a, −b, −c, −e, and -f, each of which contained one FKBd and one TPR domain. The Group II proteins were MdFKBP13, − 16-3, − 19, − 20-2a, − 20-2b, − 20-2c, and -b, each of which contained only a single FKBd. In Group III, MdFKBP62a, −62b, −65a, −65b, −72a, −72b, and -a were multi-domain while MdFKBP65c, −65d, and -72c each contained only one FKBd (Fig. 4).
Phylogenetic tree of amino acid sequences from 36 MdFKBPs, ZmFKBP12, OsFKBP12, VvFKBP12, PpFKBP12, and AtFKBP12. Neighbor-Joining phylogenetic tree was constructed with MEGA 5 software, using 41 full-length amino acid sequences from 6 species. Three protein groups on tree are represented by Roman numerals
Distribution of functional domains in 38 apple FKBPs. Three conserved domains shown in colored boxes were identified in apple FKBP protein sequences using NCBI batch web Conserved Domains Search tool. Blue box, FKBP_C domain; red box, TPR domain; and purple box, Tigger_C domain. Order in which motifs appears corresponds with their positions in individual protein sequences
Genome distribution of apple FKBP genes
The genomic locations of apple FKBP genes were determined by mapping each to genes identified in the apple genome database based on their mapping coordinates. These MdFKBPs were unevenly distributed among the 13 chromosomes (Fig. 5), with Chromosome (Chr) 2 having the most, i.e., six, followed by Chr4 (5 genes), and Chr3 and Chr6, each containing only one MdKFBP.
This gene family is believed to have expanded during the process of genome-evolution for apple [58]. To uncover the mechanism underlying this expansion, we investigated gene-duplication events, including tandem and segmental duplications, and found that many MdFKBP genes (55.3%, 21/38) were present in two or more copies (Fig. 5). While 12 have undergone tandem duplication, 11 have undergone segment duplication. Whereas those tandem duplications produced FKBP gene clusters or hotspots, the segment duplications produced many homologs of FKBP genes on different chromosomes (see red and blue fonts in Fig. 5). It is thought that a relatively recent (60–65 million years ago) genome-wide duplication event resulted in the transition of nine ancestral chromosomes to 17 chromosomes within the Pyreae tribe [38]. Here, our results determined that multiple pairs were each linked to at least 12 potential chromosomal segmental duplications (Fig. 5, pairs of bars in dark-grey areas). These pairs included large sections of Chr2 and Chr7, Chr4 and Chr12, and Chr5 and Chr10.
Exon-intron structures of FKBP genes in various species
To gain insight into their structural diversity, we analyzed the exon-intron organization of coding sequences for FKBP genes from various species. As typical genes, we selected FKBP12s, containing one FKBd; FKBP42s, with one FKBd and one TPR domain; FKBP62s, with two FKBds and one TPR domain; and FKBP72s, with three FKBds and one TPR domain (Fig. 4). Among the species tested here, we found that these genes typically contained four (FKBP12), six (FKBP42), 12 (FKBP62), or 19 (FKBP72) introns (Fig. 6; Additional file 2: Table S2). We also compared gene lengths and noted that they were restricted in all exons of the FKBP12 genes (Fig. 6a). The lengths of the FKBP42 genes were most restricted in Exons 3 through 6, while Exons 1, 2, and 7 displayed comparatively more variation (Fig. 6b). For the FKBP62 genes, their lengths were most restricted in Exons 2 through 11, while Exons 1, 12, and 13 showed comparatively less variation (Fig. 6c); Finally, for the FKBP72 genes, their lengths were most restricted in Exons 2 through 19, and Exons 1 and 20 exhibited comparatively higher variation (Fig. 6d).
Exon length distribution analysis of FKBP12, FKBP42, FKBP62, and FKBP72 in various plant species. Analysis of exon length distributions for FKBP12 (a), FKBP42 (b), FKBP62 (c), and FKBP72 (d), based on Boxplot depictions in SigmaPlot 12.0 program. Each box represents exon size range in which 50% of values for a particular exon are grouped. Mean value is indicated by long red line
Promoter sequences of MdFKBP genes
To investigate putative cis-acting elements in their promoter regions, we isolated approximately 1500-bp genomic sequences upstream of the start codon from our MdFKBPs (Additional file 1: Table S1). In addition to some types of cis-elements involved in light-responsiveness (Additional file 3: Table S3), many types responsive to stresses and correlative hormones were found in those promoter regions. In total, 11 types of cis-elements were discovered in the 15 promoters. They were associated with responses to hypoxia, heat, chilling, drought, pathogens, wound, or correlative hormones such as salicylic acid (SA), methyl jasmonate (MeJA), abscisic acid (ABA), or ethylene (Table 2). The results concluded that these cis-elements play various important roles in plant stress responses.
Expression profiles for MdFKBP genes
Knowing the patterns of expression in various tissue types can help us understand gene functions. We isolated the full-length cDNA, 5’-UTR, and 3’-UTR sequences of 16 MdFKBP genes and used specific primers for our qRT-PCR assays (Additional file 4: Table S4). As shown in Fig. 7, MdFKBPs were constitutively expressed in the five tissues examined here, albeit at different levels of transcription. For example, MdFKBP12, − 16-2, − 16-3, − 17-1, − 18, − 19, − 20-1a, −53a, −65b, and MdTIGa were most highly expressed in the leaves, while expression of MdFKBP15–2, −42a, − 43, −62a, −65a, and -72a was highest in the roots. Under water-deficit conditions, expression of MdFKBP12, − 20-1a, −42a, − 43, −53a, −62a, −65a, −65b, −72a, and MdTIGa was significantly induced (p < 0.05) from that detected in the control plants while the expression of other MdFKBP genes remained constant (Fig. 8). In response to NaCl stress, transcripts of MdFKBP42a, − 43, −53a, −62a, −65a, −65b, and MdTIGa mRNAs were significantly accumulated (p < 0.05) when compared with the untreated controls (Fig. 9).
Expression heat map of MdFKBPs under water deficit stress. After qRT-PCR data were re-analyzed, relative expression was calculated with respect to control samples (i.e., Day 0). Heat maps were generated using TIGR MeV v4.8.1 software. Bar at bottom of each heat map presents relative expression values: 0, down-regulated; 1.0, expression unaltered; or 1.4, up-regulated
Expression heat map of MdFKBPs under NaCl stress. After qRT-PCR data were re-analyzed, relative expression was calculated with respect to control samples (i.e., Day 0). Heat maps were generated using TIGR MeV v4.8.1 software. Bar at bottom of each heat map presents relative expression values: 0, down-regulated; 1.0, expression unaltered; or 1.4, up-regulated
Analyses of MdFKBP62a and MdFKBP65a/b interaction network and co-expression
Although orthologous ROF1/ROF2 pairs are known to function in abiotic stress responses in some species [1, 2, 7, 27, 59,60,61], they have not been characterized in apple. Therefore, we chose the FKBP62/65 pair to investigate potential protein–protein interactions. An Arabidopsis ROF1/ROF2-mediated network, comprising ROF1, ROF2, AtFKBP18, and eight other interactive proteins, was generated using STRING 10.0, an online database that can, with high confidence, identify interactive proteins. We then determined the homologs of those proteins through reciprocal BlastP analyses of the apple genome (Fig. 10; Additional file 7: Table S7; Additional file 8: Table S8), and used the predicted nucleotide sequences to identify reciprocal 5’- or 3’-UTR expressed sequence tags (ESTs) in the >NCBI EST Database (Additional file 7: Table S7). Our qRT-PCR analysis indicated that, under water-deficit or NaCl treatments, 10 gene pairs (ROF1:MdFKBP62a-ROF2:MdFKBP65a/b; ROF1:MdFKBP62a-HSFA2:MdHSFA2; ROF1:MdFKBP62a-HSP60:MdHSP60; ROF1:MdFKBP62a-HSP90.1:MdHSP90.1; ROF1:MdFKBP62a-HSP81–2:MdHSP81–2; ROF1:MdFKBP62a-HSP81.4:MdHSP81.4; ROF1:MdFKBP62a-HSP70–15:MdHSP70–15; ROF1:MdFKBP62a-HSP89.1:MdHSP89.1; ROF2:MdFKBP65a/b- HSFA2:MdHSFA2; and ROF2:MdFKBP65a/b- HSP90.1:MdHSP90.1) were co-expressed and showed uniform upregulation (Figs. 8, 9, 10). All of these results demonstrated that genes within this apple MdFKBP62a:MdFKBP65a/b-mediated network have potentially important roles in water-deficit and NaCl-stress signaling.
Discussion
The proteins encoded by FKBP genes comprise large families and are broadly distributed in higher plants [1, 7, 18]. Apple is an economically important woody plant and the most widely cultivated fruit crop in the world. Sequencing of the apple genome has provided a good platform for genome-wide analyses of all putative gene families in apple, including the DREB [62], MYB [63], MADS-box [41], and WRKY [64, 65] families. However, genome-wide information about the FKBP gene family in apple has remained unknown while members of that family have been identified in other plant species [3, 5, 6, 8, 34,35,36,37, 66]. Moreover, the content of FKBP genes varies substantially among species. For example, 23, 29, 24, 23, 21, and 23 FKBP genes have been reported for Arabidopsis [34], rice [8, 35, 36], maize [37], grape [3], strawberry [5], and peach [6]. Here, we determined that the apple genome contains 38 FKBP genes, making this family much larger than in any other species.
When members are compared among species, it is apparent that some have disappeared or are repeated. For example, in a comparison with AtFKBP members, FKBP15–2, FKBP15–3, FKBP17–3, and FKBP53 are absent while FKBPa, FKBPb, and FKBPc are new members in grapevine [3]. In peach, FKBP15–2, FKBP15–3, FKBP17–3, and FKBP43 do not exist but two copies of FKBP42, FKBP53, and FKBP62 occur [6]. Similar trends have been noted with rice [36], maize [37], and strawberry [5]. We found two copies of FKBP15 in apple (versus three copies in Arabidopsis), one copy of FKBP17 (three in Arabidopsis), five copies of FKBP20 (two in Arabidopsis), six copies of FKBP42 (one in Arabidopsis), two copies of FKBP53 (one in Arabidopsis), two copies of FKBP62 (one in Arabidopsis), four copies of FKBP65 (one in Arabidopsis), and three copies of FKBP72 (one in Arabidopsis). Based on traditional FKBP nomenclature rules, we named MdFKBPa and MdFKBPb as new members of that family. We speculated that the presence of additional genes, or the disappearance/repeat of others in the apple genome, reflects the great need for these genes to drive the complicated enzymatic activity associated with these woody perennial plants. These apple FKBPs are highly and structurally conserved, based on analyses of sequence alignments, 3-D structures, phylogenetics, and conserved domains (Figs. 1, 2, 3, 4). Similar results have been reported for Arabidopsis [34], rice [36], maize [37], grape [3], strawberry [5], and peach [6]. In addition, FKBP12 shares the same exon-intron structure and the same exon length as found in other species, as well as in FKBP42, FKBP62 and FKBP72 (Fig. 6), again suggesting that genes in that family are highly conserved. Because some members have increased in number, disappeared, or been repeated in some species, we believe this phenomenon means that the content of these FKBP genes has varied during the evolution of this family.
Segmental, tandem, and whole-genome duplications are critical for both the diversification of gene function and the rearrangement and expansion of genomes [41, 62,63,64,65]. Whole-genome duplication events have occurred in apple [38], and tandem, segmental, and whole-genome duplications have caused some apple gene families to expand, including the MYB [63], MADS-box [41], and WRKY [64] families. We learned here that 11 MdFKBP genes have undergone segment duplication and 12 have undergone tandem duplication (Fig. 5). In addition, multiple pairs have each been linked to 12 potential chromosomal segmental duplications (Fig. 5). Similar results have been reported for the FKBP gene family in Arabidopsis [34] and maize [37].
In various plant species, the expression of FKBP genes is induced by abiotic stresses, e.g., salt, cold, heat, wounding, desiccation, and MDA, including AtFKBP62 (ROF1), AtFKBP65 (ROF2), wFKBP73, and wFKBP77. Whereas expression of AtFKBP65 and wFKBP77 is up-regulated by heat-shock treatment, that of AtFKBP62 and wFKBP73 is not [59, 67]. Expression profiles of OsFKBP and ZmFKBP genes under water stress or different environmental conditions have indicated that many are regulated by water stress (OsKFBPs) or by heat, cold, salt, and drought (ZmFKBPs) [36, 37]. We found that expression of MdFKBP42a, − 43, −53a, −62a, −65a, −65b, and MdTIGa was significantly induced in response to water-deficit and NaCl treatments (Figs. 8, 9). In wheat, an orthologous FKBP pair FKBP73 and FKBP77 interact with HSP90 through TPR domains [68]. In Arabidopsis, AtFKBP62 and AtFKBP65 work antagonistically and have an important role in the heat-stress response through their interaction with some small heat shock proteins [59,60,61]. OsFKBP64 and 65, ZmFKBP62a and 62b, FaFKBP62–1 and 62–2, and PpFKBP62a and 62b proteins have TPR regions with conserved residues, suggesting the probability of analogous interaction with HSP90-client complexes [5, 6, 8, 37]. All of These reports indicated that the interaction of this FKBP pair with HSP90 could be a conserved mechanism in higher plants. Moreover, AtFKBP62 directly interacts with phosphatidylinositol phosphate proteins PI(3)P and PI(3,5)P2, which are involved in salt/osmotic stress responses during the germination of Arabidopsis seeds [2, 69]. Our data indicated potential protein–protein interactions for MdFKBP62a:MdFKBP65a/b, and co-expression of their genes was uniformly up-regulated (Figs. 8, 9, 10). Therefore, all of these findings suggest that MdFKBP62a, MdFKBP65a/b, and any gene pairs up-regulated by water-deficit and salt treatments could help confer tolerance to apple plants to those stresses.
Conclusions
In apple, the genome-wide identification of the FKBP family genes was completed and confirmed 38 MdFKBPs. Subsequent analyses based on bioinformatics, RT-PCR, and qRT-PCR approaches were focused on FK506-binding domains, protein and gene structures, conserved domains, phylogenetic relationships, chromosomal locations, cis-acting elements, and expression patterns in various tissues or under water-deficit and salt stresses. The interaction-network and co-expression analyses showed that the paired MdFKBP62a/MdFKBP65a/b-mediated network is involved in water-deficit and salt-stress signaling. Those genes were uniformly up-regulated through interactions with apple heat shock proteins. The results of this study provide valuable information about MdFKBP genes in apple and will aid in determining the functions of those family members.
Abbreviations
- 3D structure:
-
Three-dimensional structure
- AA:
-
Amino acid
- ABRE:
-
ABA responsive element
- ACD:
-
Alpha-crystallin domain
- ARE:
-
Anaerobic responsive element
- BlastP:
-
Basic local alignment search tool-protein
- CRI:
-
Conserved region I
- CRII:
-
Conserved region II
- DRE:
-
Dehydration-responsive element
- ERE:
-
Ethylene responsive element
- EST:
-
Expressed sequence tag
- FKBPs:
-
FK506-binding proteins
- HMM:
-
Hidden markov model
- HSE:
-
Heat stress element
- LTRE:
-
Low temperature responsive element
- MBS:
-
MYB binding site involved in drought-inducibility
- MW:
-
Molecular weight
- ORF:
-
Open reading frame
- PI:
-
Isoelectric point
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- WUN:
-
wound-responsive element
References
Gollan PJ, Bhave M, Aro EM. The FKBP families of higher plants: exploring the structures and functions of protein interaction specialists. FEBS Lett. 2012;586(20):3539–47.
Vasudevan D, Gopalan G, Kumar A, Garcia VJ, Luan S, Swaminathan K. Plant immunophilins: a review of their structure-function relationship. Biochim Biophys Acta. 2015;1850(10):2145–58.
Shangguan L, Kayesh E, Leng X, Sun X, Korir NK, Mu Q, Fang J. Whole genome identification and analysis of FK506-binding protein family genes in grapevine (Vitis vinifera L.). Mol Biol Rep. 2013;40(6):4015–31.
Galat A. Peptidylprolyl cis/trans isomerases (immunophilins): biological diversity-targets-functions. Curr Top Med Chem. 2003;3(12):1315–47.
Leng X, Liu D, Zhao M, Sun X, Li Y, Mu Q, Zhu X, Li P, Fang J. Genome-wide identification and analysis of FK506-binding protein family gene family in strawberry (Fragaria ananassa). Gene. 2014;534(2):390–9.
Zhang Y, Han J, Liu D, Wen X, Li Y, Tao R, Peng Y, Fang J, Wang C. Genome-wide identification and analysis of FK506-binding protein gene family in peach (Prunus persica). Gene. 2014;536(2):416–24.
Geisler M, Bailly A. Tete-a-Tete: the function of FKBPs in plant development. Trends Plant Sci. 2007;12(10):465–73.
Gollan PJ, Bhave M. Genome-wide analysis of genes encoding FK506-binding proteins in rice. Plant Mol Biol. 2010;72(1–2):1–16.
Breiman A, Fawcett TW, Ghirardi ML, Mattoo AK. Plant organelles contain distinct peptidylprolylcis, trans-isomerases. J Biol Chem. 1992;267(30):21293–6.
Luan S, Albers MW, Schreiber SL. Light-regulated, tissue-specific immunophilins in a higher plant. Proc Natl Acad Sci U S A. 1994;91(3):984–8.
Xu Q, Liang S, Kudla J, Luan S. Molecular characterization of a plant FKBP12 that does not mediate action of FK506 and rapamycin. Plant J. 1998;15(4):511–9.
Carol RJ, Breiman A, Erel N, Vittorioso P, Bellini C. PASTICCINO1 (AtFKBP70) is a nuclear-localised immunophilin required during Arabidopsis thaliana embryogenesis. Plant Sci. 2001;161(3):527–35.
Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Soderberg L, Roepstorff P, von Heijine G, van Wijk KJ. Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell. 2002;14(1):211–36.
Schubert M, Petersson UA, Haas BJ, Funk C, Schroder WP, Kieselbach T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem. 2002;277(10):8354–65.
Friso G, Giacomelli L, Ytterberg AJ, Peltier JB, Rudella A, Sun Q, Wijk KJ. In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: new proteins, new functions, and a plastid proteome database. Plant Cell. 2004;16(2):478–99.
Lima A, Lima S, Wong JH, Phillips RS, Buchanan BB, Luan S. A redox-active FKBP-type immunophilin functions in accumulation of the photosystem II supercomplex in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2006;103(33):12631–6.
Perez-Perez JM, Ponce MR, Micol JL. The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol. 2004;134(1):101–17.
Geisler M, Bailly A, Ivanchenko M. Master and servant: regulation of auxin transporters by FKBPs and cyclophilins. Plant Sci. 2016;245:1–10.
Aghdasi B, Ye K, Resnick A, Huang A, Ha HC, Guo X, Dawson TM, Dawson VL, Synder SH. FKBP12, the 12-kDa FK506-binding protein, is a physiologic regulator of the cell cycle. Proc Natl Acad Sci U S A. 2001;98(5):2425–30.
Vespa L, Vachon G, Berger F, Perazza D, Faure JD, Herzog M. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004;134(4):1283–92.
Yu Y, Li Y, Huang G, MEng Z, Zhang D, Wei J, Yan K, Zheng C, Zhang L. PwHAP5, a CCAAT-binding transcription factor, interacts with PwFKBP12 and plays a role in pollen tube growth orientation in Picea wilsonii. J Exp Bot. 2011;62(14):4805–17.
Crespo JL, Díaz-Troya S, Florencio FJ. Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol. 2005;139(4):1736–49.
Agredano-Moreno LT, Reyes de la Cruz H, Martinez-Castilla LP, Sanchez de Jimenez E. Distinctive expression and functional regulation of the maize (Zea mays L.) TOR kinase ortholog. Mol BioSyst. 2007;3(11):794–802.
Alavilli H, Lee H, Park M, Lee BH. Enhanced multiple stress tolerance in Arabidopsis by overexpression of the polar moss peptidyl prolyl isomerase FKBP12 gene. Plant Cell Rep. 2017. https://doi.org/10.1007/s00299-017-2242-9.
Vucich VA, Gasser CS. Novel structure of a high molecular weight FK506 binding protein from Arabidopsis thaliana. Mol Gen Genet. 1996;252(5):510–7.
Weber H, Chetelat A, Reymond P, Farmer EE. Selective and powerful stress gene expression in Arabidopsis in response to malondialdehyde. Plant J. 2004;37(6):877–88.
Smyczynski C, Roudier F, Gissot L, Vaillant E, Grandjean O, Morin H, Masson T, Bellec Y, Geelen D, Faure JD. The C terminus of the immunophilin PASTICCINO1 is required for plant development and for interaction with a NAC-like transcription factor. J Biol Chem. 2006;281(35):25475–84.
Bouchard R, Bailly A, Blakeslee JJ, Qehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, Schulz B, Geisler M. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem. 2006;281(41):30603–12.
Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koening BW, Mancuso S, Martinoia GM. Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem. 2008;283(31):21817–26.
Wang B, Bailly A, Zwiewka M, Henrichs S, Azzarello E, Mancuso S, Maeshima M, Friml J, Schulz A, Geisler M. Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell. 2013;25(1):202–14.
Chaiwanon J, Garcia VJ, Cartwright H, Sun Y, Wang ZY. Immunophilin-like FKBP42/TWISTED DWARF1 interacts with the receptor kinase BRI1 to regulate brassinosteroid signaling in Arabidopsis. Mol Plant. 2016;9(4):593–600.
Zhao B, Lv M, Feng Z, Campbell T, Liscum E, Li J. TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol Plant. 2016;9(4):582–92.
Breiman A, Camus I. The involvement of mammalian and plant FK506-binding proteins (FKBPs) in development. Transgenic Res. 2002;11(4):321–35.
He Z, Li L, Luan S. Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol. 2004;134(4):1248–67.
Nigam N, Singh A, Sahi C, Chandramouli A, Grover A. SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Gen Genomics. 2008;279(4):371–83.
Ahn JC, Kim DW, You YN, Seok MS, Park JM, Hwang H, Kim BG, Luan S, Park HS, Cho HS. Classification of rice (Oryza sativa L. Japonica nipponbare) immunophilins (FKBPs, CYPs) and expression patterns under water stress. BMC Plant Biol. 2010;10(1).
Yu Y, Zhang H, Li W, Mu C, Zhang F, Wang L, Meng Z. Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene. 2012;498(2):212–22.
Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, Salvi S, Pindo M, Baldi P, Castelletti S, Cavaiuolo M, Coppola G, Costa F, Cova V, Dal Ri A, Goremykin V, Komjanc M, Longhi S, Magnago P, Malacarne G, Malnoy M, Micheletti D, Moretto M, Perazzolli M, Si-Ammour A, Vezzulli S, Zini E, Eldredge G, Fitzgerald LM, Gutin N, Lanchbury J, Macalma T, Mitchell JT, Reid J, Wardell B, Kodira C, Chen Z, Desany B, Niazi F, Palmer M, Koepke T, Jiwan D, Schaeffer S, Krishnan V, Wu C, Chu VT, King ST, Vick J, Tao Q, Mraz A, Stormo A, Stormo K, Bogden R, Ederle D, Stella A, Vecchietti A, Kater MM, Masiero S, Lasserre P, Lespinasse Y, Allan AC, Bus V, Chagne D, Crowhurst RN, Gleave AP, Lavezzo E, Fawcett JA, Proost S, Rouze P, Sterck L, Toppo S, Lazzari B, Hellens RP, Durel CE, Gutin A, Bumgarner RE, Gardiner SE, Skolnick M, Egholm M, Van de Peer Y, Salamini F, Viola R. The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet. 2010;42(10):833–9.
Li X, Ling K, Zhang J, Xie Y, Wang L, Yan Y, Wang N, Xu J, Li J, Li C, Wang W, van Nocker S, Dong Y, Ma F, Guan Q. Improved hybridde novogenome assembly of domesticated apple (Malus x domestica). Gigascience. 2016;5(1):35.
Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, Di Pierro EA, Gouzy J, Rees DJG, Guérif P, Muranty H, Durel CE, Laurens F, Lespinasse Y, Gaillard S, Aubourg S, Quesneville H, Weigel D, van de Weg E, Troggio M, Bucher E. High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet. 2017;49(7):1099–106.
Tian Y, Dong Q, Ji Z, Chi F, Cong P, Zhou Z. Genome-wide identification and analysis of the MADS-box gene family in apple. Gene. 2015;555(2):277–90.
Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–90.
Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201.
Tonthat NK, Juwadi PR, Zhang H, Lee SC, Venters R, Spicer L, Steinbach WJ, Heitman J, Schumacher MA. Structures of pathogenic fungal FKBP12s reveal possible self-catalysis function. MBio. 2016;7(2):e00492–16.
Granzin J, Eckhoff A, Weiergräber OH. Crystal structure of a multi-domain immunophilin from Arabidopsis thaliana: a paradigm for regulation of plant ABC transporters. J Mol Biol. 2006;364(4):799–809.
Sinars CR, Cheung-Flynn J, Rimerman RA, Scammell JG, Smith DF, Clardy J. Structure of the large FK506-binding protein FKBP51, an Hsp90-binding protein and a component of steroid receptor complexes. Proc Natl Acad Sci U S A. 2003;100(3):868–73.
Unger T, Dym O, Albeck S, Jacobovitch Y, Bernehim R, Marom D, Breiman A. Crystal structure of the three FK506 binding protein domains of wheat FKBP73: evidence for a unique wFK73_2 domain. J Struct Funct Genom. 2010;11(2):113–23.
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52.
Hu W, Wang L, Tie W, Yan Y, Ding Z, Liu J, Li M, Peng M, Xu B, Jin Z. Genome-wide analyses of the bZIP family reveal their involvement in the development, ripening and abiotic stress response in banana. Sci Rep. 2016;6:30203.
Proost S, Van Bel M, Vaneechoutte D, Van de Peer Y, Inzé D, Mueller-Roeber B, Vandepoele K. PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 2015;43(1):974–81.
Zhao P, Wang D, Wang R, Kong N, Zhang C, Yang C, Wu W, Ma H, Chen Q. Genome-wide analysis of the potato Hsp20 gene family: identification, genomic organization and expression profiles in response to heat stress. BMC Genomics. 2018;19(1):61.
Shao Y, Qin Y, Zou Y, Ma F. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene. 2014;552(1):87–97.
Dong QL, Liu DD, An XH, Hu DG, Yao YX, Hao YJ. MdVHP1 encodes an apple vacuolar H+-PPase and enhances stress tolerance in transgenic apple callus and tomato. J Plant Physiol. 2011;168(17):2124–33.
Dong QL, Wang CR, Liu DD, Hu DG, Fang MJ, You CX, Hao YJ. MdVHA-A encodes an apple subunit a of vacuolar H+-ATPase and enhances drought tolerance in transgenic tobacco seedlings. J Plant Physiol. 2013;170(6):601–9.
Perini P, Pasquali G, Margis-Pinheiro M, de Oliviera PRD, Revers LF. Reference genes for transcriptional analysis of flowering and fruit ripening stages in apple (Malus × domestica Borkh.). Mol Breed. 2014;34(3):829–42.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–8.
Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34(2):374–8.
Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10.
Aviezer-Hagai K, Skovorodnikova J, Galigniana M, Farchi-Pisanty O, Maayan E, Bocovza S, Efrat Y, von Koskull-Döring P, Ohad N, Breiman A. Arabidopsis immunophilins ROF1 (AtFKBP62) and ROF2 (AtFKBP65) exhibit tissue specificity, are heat-stress induced, and bind HSP90. Plant Mol Biol. 2007;63(2):237–55.
Meiri D, Breiman A. Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90. 1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J. 2009;59(3):387–99.
Meiri D, Tazat K, Cohen-Peer R, Farchi-Pisanty O, Aviezer-Hagai K, Avni A, Breiman A. Involvement of Arabidopsis ROF2 (FKBP65) in thermotolerance. Plant Mol Biol. 2010;72(1–2):191–203.
Zhao T, Liang D, Wang P, Liu J, Ma F. Genome-wide analysis and expression profiling of the DREB transcription factor gene family in Malus under abiotic stress. Mol Gen Genomics. 2012;287(5):423–36.
Cao ZH, Zhang SZ, Wang RK, Zhang RF, Hao YJ. Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene conferring abiotic stress tolerance in plants. PLoS One. 2013;8:e69955.
Gu YB, Ji ZR, Chi FM, Qiao Z, Xu CN, Zhang JX, Dong QL, Zhou ZS. Bioinformatics and expression analysis of the WRKY gene family in apple. Sci Agric Sin. 2015;48(16):012.
Meng D, Li Y, Bai Y, Li M, Cheng L. Genome-wide identification and characterization of WRKY transcriptional factor family in apple and analysis of their responses to waterlogging and drought stress. Plant Physiol Biochem. 2016;103:71–83.
Subin CS, Pradeep MA, Vijayan KK. FKBP-type peptidyl-prolyl cis-trans isomerase from thermophilic microalga, Scenedesmus sp.: molecular characterisation and demonstration of acquired salinity and thermotolerance in E. coli by recombinant expression. J Appl Phycol. 2016;28(6):3307–15.
Blecher O, Erel N, Callebaut I, Aviezer K, Breiman A. A novel plant peptidyl-prolyl-cis-transisomerase (PPIase): cDNA cloning, structural analysis, enzymatic activity and expression. Plant Mol Biol. 1996;32(3):493–504.
Reddy RK, Kurek I, Silverstein AM, Chinkers M, Breiman A, Krishna P. High-molecular-weight FK506-binding proteins are components of heat-shock protein 90 heterocomplexes in wheat germ lysate. Plant Physiol. 1998;118(4):1395–401.
Karali D, Oxley D, Runions J, Ktistakis N, Farmaki T. The Arabidopsis thaliana immunophilin ROF1 directly interacts with PI(3)P and PI(3,5)P2 and affects germination under osmotic stress. PLoS One. 2012;7:e48241.
Acknowledgements
The authors are grateful to Priscilla Licht for help in revising our English composition.
Funding
This work was supported by the State Key Program of the National Natural Science Foundation of China (31330068), by the earmarked fund for the China Agriculture Research System (CARS-27), by the National Natural Science Foundation of China (31401852 and 31701894), by the China Postdoctoral Science Foundation (2017 M620474), and by the Fundamental Research Funds for the Central Universities (Z109021645). The funding bodies had no role in design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All of the datasets supporting the results of this article are included within the article and its Additional files.
Author information
Authors and Affiliations
Contributions
QLD and KM collected the public dataset, perform bioinformatics analysis and also drafted the manuscript. DYD and SZ contributed to bioinformatics analysis and the making of all the figures and tables. YPW, QW and DH contributed to data collection. CL, CHL and XQG performed the experiments. QLD and FWM conceived this study and reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Genomic information of MdFKBPs in apple genome. (DOCX 57 kb)
Additional file 2:
Table S2. Exon length analyses of FKBP12, 42, 62 and 72 in various species. (XLSX 26 kb)
Additional file 3:
Table S3. Promoter analyses of MdFKBPs. (XLSX 69 kb)
Additional file 4:
Table S4. Application of primers and sequences. (DOCX 19 kb)
Additional file 5:
Table S5. FKBP fragments identified from the apple genome. (DOCX 13 kb)
Additional file 6:
Table S6. The IDs of FKBP12 and FKBP42 in various species. (XLSX 12 kb)
Additional file 7:
Table S7. MdFKBP62a and MdFKBP65a/b interaction network and co-expression in apple. (XLSX 12 kb)
Additional file 8:
Table S8. MdFKBP62a and MdFKBP65a/b interaction network in String10.0. (XLSX 29 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Dong, Q., Mao, K., Duan, D. et al. Genome-wide analyses of genes encoding FK506-binding proteins reveal their involvement in abiotic stress responses in apple. BMC Genomics 19, 707 (2018). https://doi.org/10.1186/s12864-018-5097-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-018-5097-8