Abstract
Background
EAV-HP is an ancient retrovirus pre-dating Gallus speciation, which continues to circulate in modern chicken populations, and led to the emergence of avian leukosis virus subgroup J causing significant economic losses to the poultry industry. We mapped EAV-HP integration sites in Ethiopian village chickens, a Silkie, Taiwan Country chicken, red junglefowl Gallus gallus and several inbred experimental lines using whole-genome sequence data.
Results
An average of 75.22 ± 9.52 integration sites per bird were identified, which collectively group into 279 intervals of which 5 % are common to 90 % of the genomes analysed and are suggestive of pre-domestication integration events. More than a third of intervals are specific to individual genomes, supporting active circulation of EAV-HP in modern chickens. Interval density is correlated with chromosome length (P < 2.31−6), and 27 % of intervals are located within 5 kb of a transcript. Functional annotation clustering of genes reveals enrichment for immune-related functions (P < 0.05).
Conclusions
Our results illustrate a non-random distribution of EAV-HP in the genome, emphasising the importance it may have played in the adaptation of the species, and provide a platform from which to extend investigations on the co-evolutionary significance of endogenous retroviral genera with their hosts.
Similar content being viewed by others
Background
The EAV retrovirus family likely originated from a primordial integration event prior to the evolutionary speciation of Gallus sp. [1], and show sequence similarity with the less ancient ev loci [2]. Some EAV elements might be related to the avian leukosis virus (ALV) genus [Dimcheff et al. 2000 in 2], and owing to high env sequence identity between EAV and ALV genomes, recombination events involving EAV-HP may have led to the emergence of the ALV subgroup J (ALV-J; [3]). Further support for recombination as the origin of ALV-J is evident in the avian retrotransposon ART-CH, where the R and U5 regions of the ART-CH long terminal repeat (LTR) are 97 % identical to the EAV-HP LTR, while the U3 region is distinct from any other retrovirus [3]. The active spread of EAV-HP in modern chickens is believed to involve a helper virus due to the numerous point mutations, deletions and insertions in EAV-HP sequences inactivating viral gene products [3], and that none of the proviruses found to date have been observed to produce infectious virions [2].
Exogenous retroviruses infect their specific target cells and integrate as a provirus into the cellular genome by transcribing their RNA genome into DNA by reverse transcription [4]. Once integrated into the germ line of a species it becomes endogenised. Parts of the endogenous virus may be eliminated or disrupted by random mutation events, preventing expression of functional viral proteins and the ability to replicate [4]. Consequently, remnants of endogenous retroviruses (ERVs) at varying states of integrity are present in vertebrate genomes and can be observed in re-sequencing data. An analysis of reference genome sequences by Stoye [5] suggests that 4–10 % of vertebrate DNA is composed of retroviral remnants.
A review of retrovirus integration site selection [6] supports the view that the integration of retroviruses at random genomic locations in vivo is unlikely to be true for all retroviruses. This review also suggests that integration site preference is often retrovirus but not host-specific. That is to say the same distribution of site preference has been observed in specific retroviridae across a range of mammalian and avian host species. Bolisetty et al. [7] identified 31 alpharetroviruses in the chicken genome; this retrovirus genus includes the EAV retroviruses. Desfarges and Ciuffi [6] indicate that alpharetroviruses display a weak preference for integration in transcription units of genes and CpG islands, which are enriched in gene-rich regions. This is consistent with the EAV-HP integration sites identified for the oocyan phenotype in chicken [8, 9], which integrates into the promoter region of SLCO1B3. Bolisetty et al. [7] report a near random distribution of ERV integration sites, although this may be an artefact of the preferences of different retrovirus genera. They also indicate that many ERVs are transcribed and translated, and that some are expressed in a tissue-specific manner.
The increasing awareness over the interplay and co-evolution of virus and host has led to the concept of viral symbiosis [10]. Despite being novel to the host genome, the virus is pre-evolved to interact with it at the genetic level. It may contribute genes and/or regulatory sequences to their host, thus conferring a selective advantage. For the virus, successful integration into the host genome offers the prospect of long-term survival and future transmission [11]. The infectious nature of retroviruses, and the capacity of some to become endogenous, offers extraordinary potential to generate heritable diversity in the host genome. Multiple integrations typically occur during endogenisation, and each of these events has the potential to regulate host gene expression given the appropriate integration site. As with any mutation, selection will favour integration sites that enhance the survival of the host, and select against those that impair it. The abundance of retroviruses may therefore play an important role in the adaptation and evolution of the host species [10].
Here, we assess the prevalence of EAV-HP in chickens with different breeding histories. For this purpose we apply an innovative strategy utilising high-coverage, paired-end next-generation sequencing (NGS) data from re-sequenced chickens, to define EAV-HP integration sites (see Methods and Fig. 1). The chickens analysed included eight inbred lines of White Leghorn currently maintained at the Pirbright Institute (UK), and due to be relocated to the National Avian Research Facility (University of Edinburgh, UK), a Silkie, a Taiwan Country chicken, a red junglefowl, and village chickens from Horro (n = 6) and Jarso (n = 5) regions of Ethiopia. Specifically, we aim to contribute answers to the following questions: How common are EAV-HP integrations? Do they show a pattern of lineage specificity? What can we infer about the history of such integrations? Do the integrations occur at random across the chicken genome? and if not, is there any evolutionary significance to their distribution?
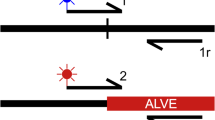
Overview of the retrovirus mapping strategy. Paired-end sequencing reads from re-sequencing of chicken genomes are mapped to both Galgal4 (a) and the LTR of the EAV-HP genome (b). Unmapped reads from the EAV-HP LTR alignment, which may map either to the host genome or other elements of the viral sequence, are recovered and mapped to the chicken reference genome to generate a list of observed integration sites (b). For each integration sites, paired-end reads are retrieved from the initial mapping to Galgal4 (c) and remapped to the LTR of the EAV-HP genome (d), and the soft-clipped sequences of mapped reads recovered (e). These sequences are then aligned to Galgal4 using BLAT, and the results referenced against the interval in question to identify the integration site breakpoint
Results and discussion
Prevalence and distribution of EAV-HP integration sites
Alignment of pair-end reads resulted in a genome-wide mean depth of coverage against Galgal4 of 31.14 ± 1.89 in Ethiopian birds and 15.4 ± 3.63 in Pirbright lines, whilst in the individual birds downloaded from the SRA it was 61.3 in the RJF, 26.33 in the Silkie and 32.34 in the Taiwanese bird (Additional file 1: Supplementary Material S1). Following alignment of the sequence reads to the EAV-HP LTR, and subsequence re-alignment of unmapped reads to Galgal4, a total of 1730 integration sites were identified (S2). The mean depth of coverage for sequence reads to the EAV-HP LTR was 12.58 ± 1.08 in Ethiopian chickens, 7.38 ± 2.33 in Pirbright lines, and 16 ± 2.83 in the SRA data, respectively. On average, 31 % of sites contained reads aligning to 100 % of the U3 region (LTR position 1:146) of the LTR sequence, 32 % of sites contained reads aligning to 100 % of the U5 region (LTR position 164:287), whilst the complete R region (LTR position 147:163) was captured in 89 % of sites. Overall, the complete LTR sequence was captured in approximately 20 % of sites.
Integration sites within and across birds and lines with less than 5 kb intervening genomic sequence were classified as a single interval. Similarly, isolated integration sites, those for which no other integration site was identified within 5 kb, were classified as an interval. This reduced the 1730 integration sites into a set of 279 intervals for comparative analyses (S3). The ratio of intervals to integrations was 0.88 in Ethiopian chickens, 0.9 Pirbright lines, and 0.89 in the SRA data, indicating an average fraction of 0.11 integration sites to be clustered. Importantly, one of the Jarso chickens (JB2A04B) was sequenced twice at the comparable genome coverage using independent libraries, and the results of each sequencing run analysed independently. Across both libraries the resulting intervals detected were identical (S3).
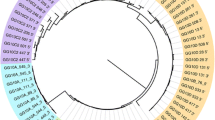
A summary of the distribution of observed EAV-HP integration site intervals for each re-sequenced genome, on the macro- (1 to 5), intermediate (6 to 10) and micro-chromosomes (11 to 28), in addition to the Z chromosome and several unplaced contigs, is presented in Fig. 2. Of the 279 intervals observed, 72 % mapped to macrochromosomes, 9 % to the intermediate-sized chromosomes, 7 % to microchromosomes and 8 % to the Z chromosome, whilst the remaining 4 % mapped to unplaced contigs (S4). A positive correlation was observed between chromosome length and interval density (r = 0.75, P < 2.31−6; S5), whilst a negative correlation was observed between gene and interval density (r = −0.74, P < 5.46−6; S5). We also observed that 27 % of intervals occur within 5 kb of transcripts (S6). This is less than might be expected by chance given that approximately 41 % of the chicken genome is transcribed. Microchromosomes are both CpG and G + C rich, and have a higher rate of recombination than macrochromosomes [12, 13], which might facilitate excision of retroviral insertions during meiosis. Furthermore, gene density in the chicken has been shown to be inversely correlated with chromosome length [12]. Assuming there to be no target-site preference for EAV-HP, the results suggests that the chicken host retains some integrations within genes or regulatory regions, possibly due to them conferring a selective advantage, whilst others might be more rapidly excised, in agreement with the concept of viral symbiosis [10].
Distribution of observed EAV-HP integration sites. Outer circos plot indicates distribution of observed EAV-HP integration sites. Inner stacked histogram summarizes percentage of integration sites for each line along macro-, intermediate, and micro-chromosomes, in addition to the Z chromosome and unlocalized/unplaced contigs. The latter relates to integration sites mapped to contigs present in the reference assembly (Galgal4) that are either associated with a specific chromosome but unlocalized, or have not yet been associated with any chromosome
We identify an average of 66.35 ± 7.58 intervals per bird. This is much higher than the 10–15 integrations suggested in previous studies of inbred lines and the red junglefowl based on Southern blot hybridisation with env [14] and LTR sequences [3]. However, it is comparable to that observed in a more recent analysis of the Galgal3 reference genome by Bolisetty et al. [7] using RetroTector [15], in which they identified 31 alpharetroviruses in addition to an alphabeta clade which contained 76 viruses, two thirds of which were highly similar to previously described EAVs.
The precise identification of individual endogenous retroviruses can be problematic due to the extent of sequence diversity observed in mutable proviruses, the level of sequence integrity once a retrovirus becomes endogenised and subject to host recombination, and the level of sequence similarity observed in closely related retroviruses (e.g. EAV-HP and ART-CH [3]). However, EAV-HP has a distinct U3 region [3] which could be used to generate a more conservative estimate of integration sites if one wished. Taking only into consideration the 31 % of integration sites with 100 % sequence coverage of the U3 region, an average of 25.25 ± 2.07, 20.38 ± 4.78 and 27.50 ± 2.12 sites per bird are present in Ethiopian chickens, Pirbright lines and the SRA data, respectively. These values more closely resemble the previously reported Southern blot hybridisation results [14].
In total, 66 % of the EAV-HP LTR alignments to Galgal4 identified by BLAT (S7) were detected as integration sites in the RJF sample. Several filters are applied to the integrations detected during the pipeline including mapping quality (MQ) and read count (RC). An evaluation of these is presented in the Additional file 2: Supplementary Methods. Relaxing the MQ filter applied in the pipeline can result in both sensitivity and precision > 95 %, however the MQ filter is applied to reduce the risk of false positives arising from multiple mapping hits for an integration – for instance if an integration were to occur within an interspersed repetitive element such as CR1. The parameters applied herein (MQ = 20, RC = 0.25μX i ) result in 98 % precision and 66 % sensitivity when comparing interval detection in the RJF NGS data to EAV-HP LTR BLAT alignments to Galgal4.
Fourteen intervals were present at high frequency (f ≥ 0.9) in the chicken genomes analysed, and eight of these were present in all birds (S8a). The presence of these intervals was confirmed in the chicken reference genome (Galgal4) following alignment of the EAV-HP LTR sequence using BLAT (S7). Furthermore, visual examination of the results of the BLAT alignment of soft-clipped sequences from each interval allowed the integration site breakpoints to be identified. The integration site breakpoints were found to be identical in each bird, suggesting that they likely occurred prior to domestication. A BLAT of the complete EAV-HP sequence (GenBank: KC632578; S8b) indicates some of these high frequency intervals in Galgal4 to host large remnants (>1 kb) of EAV-HP sequence, although none of the alignments are complete. Additional sequencing would be require to establish the integrity of the EAV-HP sequences in these intervals for the other birds/lines. Single [16] and multiple [17–19] domestication events have been suggested for domestic chicken, which may have involved several red junglefowl subspecies and introgression from other junglefowl species [20, 21]. Whether or not such retroviral insertions might prove informative in this regard will require the genome analysis of different wild junglefowl species and subspecies.
Three of these candidate pre-domestication integration sites are localised to within 5 kb of a gene: CNTN5 at chr1:182,832,847-182,834,082 (f = 0.91, absent in the N and Wellcome lines), ATPBD4 at chr5:31,303,866-31,305,068 (f = 1), and C10orf11 at chr6:13,724,843-13,726,022 (f = 1). Contactin 5 (CNTN5) is a member of the immunoglobulin super family; InnateDB [22] indicates that in mice ATPBD4 has been demonstrated to interact with STAT4, which has been implicated in the innate immune system of mice and humans [23, 24]; whilst C10orf11 is involved in melanocyte differentiation [25].
Seven intervals were found to be at high frequency (≥0.8) in chickens from either Horro or Jarso, whilst being at low frequency (≤0.2) in the other chickens (S3, S9). One of these (chr3:84,550,518–84,551,452) was present in 100 % of individuals from Jarso, whilst remaining absent in all the other birds and lines examined, including the reference genome. The soft-clipping of reads within this region suggested an integration site breakpoint at either chr3:84,550,973 or chr3:84,550,979, consistent with the size of a target site duplication during integration, which would localise such an integration event into a large intron between exons 20 and 21 of a novel protein coding gene likely to be involved in intracellular signalling (ENSGALG00000016183; fibrilin-like precursor). All Jarso birds indicate 100 % sequence coverage of the EAV-HP LTR at this interval. Another of the intervals (chr8:11,644,653 - 11,645,314), present in all but one of the Jarso chickens and absent in all other birds, suggests an integration site at chr8:11,645,101. This site is located within an intron between exons 8 and 9 of the DYPD gene, which encodes a protein involved in uracil and thymidine catabolism. In all birds possessing this interval, sequence coverage of the LTR U3 region is only 5 %, whilst the R and U5 regions ≥ 99 % coverage.
No evidence was found for different integration site breakpoints in the genome between individuals within an interval. In the event that two breakpoints were observed within an interval, they were found either to be 6 bp apart, corresponding to the size of a target-site duplication and so might be attributed to soft-clipped reads from the LTR at either end of the virus (chr2:129,140,825–129,141,707 and chr3:84,550,518–84,551,452; S9), or the integration was present in the reference genome and the breakpoints corresponded closely with the LTR alignment (chr1:182,715,565–183,138,508; S10). This is in contrast to that which has been recently observed at the oocyan locus for Chinese and South American chicken populations [8, 9].
A single interval (chr4:30,632,716–30,633,375) was found to be present in 90 % of Ethiopian birds but absent in the Pirbright lines, red junglefowl, Silkie and Taiwan chickens, in addition to the reference genome (S3). With the exception of the Ethiopian chicken (HA2A10B) in which the interval was not identified, the breakpoint was identical across all chickens at chr4:30,633,163, as was the coverage for the U3 (5 %), R (100 %) and U5 (≥98 %) regions of the LTR. A single gene is located within 5 kb of the interval, MMAA, which encodes a protein involved in the translocation of cobalamin into the mitochondrion for the biosynthesis of adenosylcobalamin, an active form of vitamin B12. The possible consequence of this integration remains speculative, with the low sequence coverage of the LTR U3 region making it unlikely to contribute a regulatory role. Also, deficiency of vitamin B12 would be surprising in domestic birds raised on commercial feed, or in scavenging village chicken whose diet likely includes insects and other arthropods.
Given the heritable nature of ERVs, one would expect to observe population structure when evaluating the ERV integration sites of individuals from within and across different populations. To investigate this further a neighbour-joining tree was constructed using the mitochondrial sequence from each bird (Fig. 3a), the results of which were compared to a network analysis performed on a distance matrix of the ERV intervals present in each chicken (Fig. 3b). Both analyses clearly separate the populations of Ethiopian chickens from the Pirbright lines, and further sub-structuring is observed within the Ethiopian chickens; segregating the sub-populations of Horro and Jarso. The distinction between chickens from Horro and Jarso is not surprising given that these two indigenous populations are separated by more than 800 km and that communities from the areas have witnessed different histories, with Jarso and Horro now of predominantly of Muslim and Christian faith, respectively [26]. As a likely consequence of geographic and human history, little inter-population gene-flow might be expected, as reflected in the EAV-HP integration site network. The red junglefowl, Silkie and Taiwan chickens form an intermediary group more closely related to the Ethiopian chickens than the Pirbright lines.
MtDNA phylogeny and network analysis of EAV-HP integration sites. Analysis of mitochondrial sequences from each bird clearly segregates birds from each Ethiopian population and the Pirbright lines, and places the Silkie and Taiwanese chickens as having close maternal ancestry to the red junglefowl (a). The network (b) from a distance matrix of integration sites present in each bird is largely consistent with this phylogeny. The colours on the phylogeny relate to branch probabilities, whilst those on the network diagram relate to breed and line
Integration sites indicate possible host-virus symbiotic advantage
Functional annotation clustering of the 79 protein-coding genes (S6) located within 5 kb of any of the 279 intervals, identify two clusters enriched for genes encoding proteins with cell adhesion properties and immune-related functions (EASE score > 1.3, P < 0.05; S11a). The genes identified include CNTN1 and CNTN5, which encode members of the immunoglobulin (Ig) superfamily, and TLR2-2 a member of the toll-like receptor (TLR) family which play a fundamental role mediating immune responses through pathogen recognition. The TLR2 genes have been found to be polymorphic among chicken breeds, and different haplotypes may therefore contribute towards differential pathogen responses [27]. Also identified was ROBO1, a member of the roundabout gene family best known for their role in the development of the nervous system [28]. Polymorphisms in another member of this family, ROBO2, have been linked to differential immune response to Newcastle Disease virus (NDV) in chickens [29]. It should be noted however that the integration site near ROBO1 was only present in a single chicken (JB2A04B) from Ethiopia.
Average sequence coverage for the U3 region of the LTR was considerably higher for the intervals in proximity to the CDH7, CNTN1, CNTN5 and LRP8 genes (97 %) than for the FAM172A, LARGE, ROBO1 and TLR2-2 genes (9 %). The difference however was less extreme for the R (100 % and 50 % respectively) and U5 (90 % and 79 % respectively) regions (S10). The U3 region of the LTR typically contains binding sites for cellular proteins that promote transcription initiation, and are capable of activating or enhancing the expression of nearby genes in a tissue-specific manner [30]. The conservation of the U3 sequence at some integration sites may be indicative of a possible selective advantage to the host. For instance if it were to confer superinfection resistance, through which cells prevent re-infection by a closely-related virus typically through a virus-encoded protein [31]. An example of such a mechanism is receptor-blocking by an endogenous viral env protein, preventing an invading virus binding to the receptor [31]. Sequencing evidence [3] suggests that the emergence of ALV subgroup J was the product of recombination between an unknown exogenous ALV and the env gene of EAV-HP, although the expression of EAV-HP env peptides was not found to confer resistance to ALV-J [32].
Conclusions
Through the use of re-sequencing data, we have mapped EAV-HP integration sites in a number of chickens of diverse origin. For this purpose, we have presented and validated a means of detecting EAV-HP integration sites using paired-end NGS data. The sensitivity and precision of the approach was assessed using NGS data derived from the red junglefowl reference bird and comparing the results against the same genome. In addition, we have further demonstrated the reproducibility of our approach through the independent analysis of a single individual sequenced twice using independent libraries. A next step would be to validate a number of the intervals by conventional PCR, and ultimately to assess their impact on gene expression. Although we have demonstrated the method using a single endogenous retrovirus, the same general principles should be applicable for any mobile element present in a host genome for which paired-end sequence data and a reference genome are available.
Our aim was to assess how abundant EAV-HP might be in the chicken genome in the context of recent evidence demonstrating it to regulate the expression of the oocyan phenotypes in some chicken populations. A large number of integrations were found in all chickens examined consistent with historical and ongoing circulation of the virus. A small number of intervals were found to be common in all of the chickens analysed, including the red junglefowl reference genome and we propose that these might represent pre-domestication integration events. The results of this study not only provide a platform from which to extend ERV analysis across different retroviral genera in chicken using re-sequencing data, but also they reiterate the potential for ERV integrations to play a significant role in host evolution, warranting further investigation of host-virus symbioses.
Methods
NGS data
Paired-end reads for a Silkie and Taiwan L2 chicken, totalling 30.1 Gb and 38.3 Gb, respectively, were downloaded from NCBI's Sequence Read Archive (SRA) [33], having been made available following a study on the genome-wide patterns of genetic variation in these two birds [34] [SRA:PRJNA202483]. Sequencing for these samples was performed on the Illumina GAIIx platform. Illumina GAIIx sequencing reads for a single red junglefowl, Gallus gallus, totalling 67.6 Gb, were also downloaded from NCBI [SRA:SRX043655, SRA:SRX043656]. NGS data from eight inbred lines of White Leghorn from the Pirbright Institute (lines: 0, 6, 7, 15, N, C, P, and Wellcome line; http://www.narf.ac.uk/chickens/lines) was provided by the Roslin Institute, having each been sequenced on the Illumina GAIIx platform from DNA pools of 10 birds per line, averaging 12 Gb per line. Further details about the samples sequenced by the Pirbright Institute, methods of sequencing and alignment of sequence reads to reference genome can be found in Kranis et al. [35].
Village chickens from western (Horro) and eastern (Jarso) Ethiopia were sampled with the consent of farmers [26]. Blood samples were collected from the wing vein, suspended in sodium citrate and stored at −20 °C at the International Livestock Research Institute facility in Addis Ababa, Ethiopia. Ethical approval for the sampling was obtained from the University of Liverpool Committee on Research Ethics (reference RETH000410). Genomic DNA was extracted from whole blood of six Horro and five Jarso chickens following phenol-chloroform extraction [36]. For each sample, a minimum of 15 μg DNA in a total volume of 100 μl was sent to the Beijing Genomics Institute (BGI) for full genome sequencing using the Illumina HiSeq2000 platform, averaging 35 Gb per bird. One of the Jarso chickens (JB2A04B) was sequenced twice using independent libraries, and the results of each sequencing run analysed individually, providing a means of validating the strategy.
The Pirbright lines are characterised as to their major histocompatibility complex (MHC) haplotype and differential resistance to various pathogens [32, 37, 38], providing a resource for investigating bacterial, viral and parasitic infections. These lines are maintained as closed populations. The L2 strain of the Taiwan Country chicken was established in 1983 for meat and egg production and has remained a closed population ever since [39]. By contrast, the characteristic feathering of the Silkie breed was first described hundreds of years ago, and the breed is now widely popular throughout the hobbyist poultry community, although it remained unfashionable in Western cultures until the late 19th century [40]. Each of these chicken lines was artificially selected to varying extents, either for pathogen resistance, commercially desirable traits, or morphology. In contrast, Ethiopian village chicken populations are maintained by smallholder farmers and mating is uncontrolled. Consequently, high levels of phenotypic diversity are typically observed within these populations [26, 41]. Furthermore, village chickens are strongly affected by natural selection and chick mortality rates can be as high as 80-90 % within the first few weeks after hatching due to ease of predation, and cyclical disease and virus outbreaks [42].
Sequence read mapping
Reads were first mapped to the chicken reference genome (Galgal4) using the Burrows-Wheeler Aligner (BWA) [43, 44] (Fig. 1a). Sequence reads were then mapped to the LTR of the EAV-HP genome [GenBank:NC_005947] (Fig. 1b). NGS data provided by the Roslin Institute (University of Edinburgh, UK) was in the format of a SAM file following mapping to the EAV-HP genome, whilst all other samples were mapped at the University of Nottingham (UK). Specifically, in the Ethiopian samples the adapter pollutions in reads were trimmed at source (BGI) along with reads containing more than 50 % low quality bases (quality < 5) bases. Both the Ethiopian dataset, and the reads downloaded from the SRA, were aligned using BWA MEM. From the EAV-HP LTR mapping, unmapped reads were extracted using SAMtools [45], converted to the fastq format using the bam2FastQ tool from the BamUtil repository (http://genome.sph.umich.edu/wiki/BamUtil), and remapped to the chicken reference genome (Fig. 1c).
Mitochondrial analysis
Reads mapped to the mitochondrial genome were extracted from the BAM alignments for each bird using SAMtools, and consensus sequences aligned using ClustalX v2.1 [46]. A neighbour-joining tree following 1000 bootstrap replicates was constructed using MrBayes v3.22 [47] and plotted with FigTree (v1.4; http://tree.bio.ed.ac.uk/software/figtree/).
Retroviral integration site analysis
BAM files generated from reads remapped to Galgal4 were converted to BED format using the bamToBed tool from the BEDTools repository [48]. BED features were filtered to exclude those with a mapping quality below 20. Overlapping features were merged using the mergeBed tool from BEDTools and filtered to exclude those supported by a read count (RC) less than 25 % the mean depth of coverage for the sample (further details in Additional file 2: Supplementary Methods). The resulting list of intervals indicated putative EAV-HP integration sites for each chicken line analysed (Fig. 1c). For each interval, paired reads that were initially mapped to Galgal4 were retrieved by name using Picard (http://broadinstitute.github.io/picard), and remapped to the LTR of the EAV-HP genome (Fig. 1d), generating an interval-specific set of paired-reads. From these reads, the soft-clipped sequences of reads identified as being clipped by ≥ 20 bases were retrieved (Fig. 1e) using a Perl script. Soft-clipped sequences were then BLAT aligned against Galgal4 and the results referenced against the interval range using a Perl script. Manual inspection of the filtered BLAT alignments for the soft-clipped sequences allowed the potential integration site breakpoint for EAV-HP to be inferred for the intervals of interest. Sequence read coverage for each Galgal4 interval and EAV-HP was calculated using GATK's DepthOfCoverage tool. Whilst the fraction of bases aligning to the LTR U3, R and U5 regions was calculated according to base position range of each region within the LTR, with reference to the position of these regions detailed by Sacco et al. [3].
To simplify downstream analyses, integration sites within 5 kb of one-another across different birds and lines were treated as a single interval, the start and end points of which were determined by the minimum and maximum genomic positions across sites within the interval. Likewise, integration sites for which no other integration site was detected within 5 kb were treated as a single interval. Clustering was performed in R [49]. This clustering distance was based on the 4.3 kb length of EAV-HP genome, indicating that any subsequent EAV-HP LTR alignments detected within 5 kb of one-another using BLAT on the chicken reference genome could reasonably be considered to be the result of a single integration. A binomial matrix was generated based on the presence/absence of integration site clusters in each bird using R, from which a distance matrix was computed and used as an input for hierarchical clustering using Ward's minimum variance method [50] as implemented in the hclust function of the stats package in R. A phylogeny was constructed using the as.phylo function of the APE package [51], from which a network was generated using the network package [52]. The distribution of integration sites was plotted using Circos v0.56 [53]. A correlation test for association between chromosome length and interval count was performed using Pearson's product moment correlation coefficient as implemented in the R function cor.test.
The Galgal4.78.gtf file was downloaded from Ensembl and used to reference nearest transcript for each of the intervals. This gene transfer format file includes all of the coding and non-coding transcripts annotated on Ensembl's genome browser for Galgal4. The rationale for selecting genes within 5 kb is that this is likely to cover the promoter region, as evidenced by the recent discovery of an EAV-HP integration site ~ 4 kb upstream of SLCO1B3 in chickens possessing the oocyan phenotype [8, 9]. Functional annotation of genes was assessed using DAVID Bioinformatics Resources v6.7 [54], specifically the functional annotation clustering enrichment threshold for EASE was set to 1.3 with stringency set to HIGH; from the resulting clusters only those with an enrichment score ≥ 1.3 (P < 0.05) were retained. Functional annotation analysis was also performed using g:Profiler [55, 56], the results of which are available in the Additional file 1: Supplementary Material (S11b), in addition to REVIGO [57] summaries of GO terms for biological process (S12a, SF1), cellular component (S12b, SF2) and molecular function (S12c, SF3), which for brevity have not been presented here (see Additional file 3). The fraction of chicken genome transcribed (41 %) was calculated from the sum of transcript lengths divided by genome length. This calculation was based on transcripts data downloaded from the Ensembl Genes 74 database for Galgal4, from which multiple transcripts per gene were excluded.
Availability of supporting data
Data supporting the results of this article are available in the NCBI Sequence Read Archive (SRA) repository. This includes whole genome sequence data for the Silkie and Taiwan L2 chickens [SRA:PRJNA202483]; whole genome sequence data for the Red Jungle Fowl [SRA:SRX043655, SRA:SRX043656]; and BAM alignments to the LTR of the EAV-HP genome [GenBank:NC_005947] for the Pirbright lines and Ethiopian village chickens [SRA:SRP062207].
Abbreviations
- ALV:
-
Avian leukosis virus
- BWA:
-
Burrows-Wheeler aligner
- ERV:
-
Endogenous retrovirus
- kb:
-
Kilobase
- Gb:
-
Gigabase
- LTR:
-
Long terminal repeats
- Mb:
-
megabase
- MHC:
-
Major histocompatibility complex
- NGS:
-
Next-generation sequencing
- SRA:
-
Sequence read archive
References
Resnick RM, Boyce-Jacino MT, Fu Q, Faras AJ. Phylogenetic distribution of the novel avian endogenous provirus family EAV-0. J Virol. 1990;64:4640–53.
Borisenko L. Avian endogenous retroviruses. Folia Biol (Praha). 2003;49:177–82.
Sacco MA, Flannery DM, Howes K, Venugopal K. Avian endogenous retrovirus EAV-HP shares regions of identity with avian leukosis virus subgroup J and the avian retrotransposon ART-CH. J Virol. 2000;74:1296–306.
Denner J. Endogenous Retroviruses. In: Retroviruses: Molecular Biology, Genomics and Pathogenesis. Norfolk, UK: Caister Academic Press; 2010.
Stoye JP. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Micro. 2012;10:395–406.
Desfarges S, Ciuffi A. Retroviral Integration Site Selection. Viruses. 2010;2:111–30.
Bolisetty M, Blomberg J, Benachenhou F, Sperber G, Beemon K. Unexpected diversity and expression of avian endogenous retroviruses. mBio. 2012;3(5):e00344-12.
Wang Z, Qu L, Yao J, Yang X, Li G, Zhang Y, et al. An EAV-HP Insertion in 5′ Flanking Region of SLCO1B3 Causes Blue Eggshell in the Chicken. PLoS Genet. 2013;9(1):e1003183.
Wragg D, Mwacharo JM, Alcalde JA, Wang C, Han J-L, Gongora J, et al. Endogenous Retrovirus EAV-HP Linked to Blue Egg Phenotype in Mapuche Fowl. PLoS One. 2013;8, e71393.
Villarreal L, Ryan F. Viruses in host evolution: General principles and future extrapolations. Cur Top in Virol. 2011;9:79–90.
Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet. 2008;42:709–32.
International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716.
Megens H-J, Crooijmans RP, Bastiaansen JW, Kerstens HH, Coster A, Jalving R, et al. Comparison of linkage disequilibrium and haplotype diversity on macro- and microchromosomes in chicken. BMC Genet. 2009;10:86.
Smith LM, Toye AA, Howes K, Bumstead N, Payne LN, Venugopal K. Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J Gen Virol. 1999;80(Pt 1):261–8.
Sperber GO, Airola T, Jern P, Blomberg J. Automated recognition of retroviral sequences in genomic data--RetroTector©. Nucleic Acids Res. 2007;35:4964–76.
Fumihito A, Miyake T, Takada M, Shingu R, Endo T, Gojobori T, et al. Monophyletic origin and unique dispersal patterns of domestic fowls. PNAS. 1996;93:6792–5.
Liu Y-P, Wu G-S, Yao Y-G, Miao Y-W, Luikart G, Baig M, et al. Multiple maternal origins of chickens: Out of the Asian jungles. Mol Phylogenet Evol. 2006;38:12–9.
Miao Y-W, Peng M-S, Wu G-S, Ouyang Y-N, Yang Z-Y, Yu N, et al. Chicken domestication: an updated perspective based on mitochondrial genomes. Heredity. 2013;110:277–82.
Sawai H, Kim HL, Kuno K, Suzuki S, Gotoh H, Takada M, et al. The origin and genetic variation of domestic chickens with special reference to junglefowls Gallus g. gallus and G. varius. PLoS One. 2010;5, e10639.
Nishibori M, Shimogiri T, Hayashi T, Yasue H. Molecular evidence for hybridization of species in the genus Gallus except for Gallus varius. Anim Genet. 2005;36:367–75.
Eriksson J, Larson G, Gunnarsson U, Bed’hom B, Tixier-Boichard M, Strömstedt L, et al. Identification of the Yellow Skin Gene Reveals a Hybrid Origin of the Domestic Chicken. PLoS Genet. 2008;4, e1000010.
Lynn DJ, Winsor GL, Chan C, Richard N, Laird MR, Barsky A, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol Syst Biol. 2008;4:218.
Deng JC, Zeng X, Newstead M, Moore TA, Tsai WC, Thannickal VJ, et al. STAT4 is a critical mediator of early innate immune responses against pulmonary Klebsiella infection. J Immunol. 2004;173:4075–83.
Matsukawa A. STAT proteins in innate immunity during sepsis: lessons from gene knockout mice. Acta Med Okayama. 2007;61:239–45.
Grønskov K, Dooley CM, Østergaard E, Kelsh RN, Hansen L, Levesque MP, et al. Mutations in C10orf11, a Melanocyte-Differentiation Gene, Cause Autosomal-Recessive Albinism. Am J Hum Genet. 2013;92:415–21.
Desta TT, Dessie T, Bettridge J, Lynch S, Melese K, Collins M, et al. Signature of artificial selection and ecological landscape on morphological structures of Ethiopian village chickens. Animal Genetic Resources / Resources génétiques animales / Recursos genéticos animales. 2013;52:17–29.
Ruan WK, Wu YH, An J, Cui DF, Li HR, Zheng SJ. Toll-like receptor 2 type 1 and type 2 polymorphisms in different chicken breeds. Poult Sci. 2012;91:101–6.
Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, et al. Roundabout Controls Axon Crossing of the CNS Midline and Defines a Novel Subfamily of Evolutionarily Conserved Guidance Receptors. Cell. 1998;92:205–15.
Kanginakudru S, Metta M, Jakati RD, Nagaraju J. Genetic evidence from Indian red jungle fowl corroborates multiple domestication of modern day chicken. BMC Evol Biol. 2008;8:174.
Ting CN, Rosenberg MP, Snow CM, Samuelson LC, Meisler MH. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 1992;6:1457–65.
Nethe M, Berkhout B, van der Kuyl AC. Retroviral superinfection resistance. Retrovirology. 2005;2:52.
Sacco MA, Howes K, Smith LP, Nair VK. Assessing the roles of endogenous retrovirus EAV-HP in avian leukosis virus subgroup J emergence and tolerance. J Virol. 2004;78:10525–35.
Leinonen R, Sugawara H, Shumway M. The Sequence Read Archive. Nucleic Acids Res. 2011;39(Database issue):D19–21.
Fan W-L, Ng CS, Chen C-F, Lu M-YJ, Chen Y-H, Liu C-J, et al. Genome-wide patterns of genetic variation in two domestic chickens. Genome Biol Evol. 2013;5:1376–92.
Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, Smith S, et al. Development of a high density 600 K SNP genotyping array for chicken. BMC Genomics. 2013;14:59.
Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb Protoc. 2006;2006:pdb.prot4455.
Cook J, Otsuki K, Huggins M, Bumstead N. Investigations into resistance of chicken lines to infection with infectious bronchitis virus. Adv Exp Med Biol. 1990;276:491–6.
Burgess SC, Basaran BH, Davison TF. Resistance to Marek’s disease herpesvirus-induced lymphoma is multiphasic and dependent on host genotype. Vet Pathol. 2001;38:129–42.
Chao CH, Lee YP. Relationship between reproductive performance and immunity in Taiwan country chickens. Poult Sci. 2001;80:535–40.
Scrivener D. Popular Poultry Breeds. 1st Edition. Wiltshire, United Kingdom: The Crowood Press Ltd; 2009.
Dana N, Dessie T, van der Waaij LH, van Arendonk JAM. Morphological features of indigenous chicken populations of Ethiopia. Animal Genetic Resources / Resources génétiques animales / Recursos genéticos animales. 2010;46:11–23.
Moges F, Tegegne A, Dessie T. Indigenous Chicken Production and Marketing Systems in Ethiopia: Characteristics and Opportunities for Market-Oriented Development. International Livestock Research Institute: Addis Ababa, Ethiopia; 2010.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:13033997 [q-bio] 2013.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2.
R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2011;1:409.
Ward Jr JH. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;58:236–44.
Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90.
Butts C. Network: A package for managing relational data in R. J Stat Softw. 2008;24:1–36.
Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–45.
Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57.
Reimand J, Arak T, Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2011 update). Nucl Acids Res. 2011;39 suppl 2:W307–15.
Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35(Web Server issue):W193–200.
Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS One. 2011;6:e21800.
Acknowledgements
This study was performed during DW PhD, which was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship to the University of Nottingham. TTD's contribution was through a PhD studentship that is part of the “Reducing the impact of infectious disease on poultry production in Ethiopia” project, which originate from a BBSRC, UK Department for International Development (DFID) and Scottish Government supported initiative (grant numbers: BB/H009396/1, BB/H009159/1 and BB/H009051/1). RAL is a PhD student supported by a University of Nottingham vice chancellor’s international scholarship. The sequencing of the Ethiopian chickens was supported by the Rural Development Agency (RDA, CYC) and the International Livestock Research Institute (ILRI, SK). DWB is supported by a core strategic grant from the BBSRC, AM a CASE BBSRC Studentship with HyLine International, LY is supported by a grant from the BBSRC, and RK from The Wellcome Trust. The National Avian Research Facility at Roslin (UK) is funded through awards from the BBSRC and The Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
DW, OH, SK, CC and DWB participated in the design of the study. TTD and JMM performed sampling, DNA preparation, contributed knowledge of Ethiopian village chicken management, and reviewed the manuscript. AM, LY and RK participated in sequence analysis of Pirbright lines. DW performed sequence analysis of Ethiopian and SRA data, and downstream analysis. RAL performed mitochondrial DNA analysis. DW, JMM, DWB and OH prepared the manuscript. All authors critically reviewed the manuscript and approved the final revision.
Authors’ information
Not applicable.
Availability of data and materials
Not applicable.
Additional files
Additional file 1:
Supplementary material. S1. Library details and depth of coverage per bird/line alignment to Galgal4. S2. Individual integrations per bird/line, sequence coverage at interval in Galgal4, and sequence coverage of EAV-HP LTR at the interval site. S3. Summary of integration site interval presence/absence per bird/line. S4. Summary of distribution of intervals. S5. Distribution of intervals relative to chromosome length. S6. Intervals and their nearest transcripts (Ensembl, Galgal4.78.gtf). S7. BLAT results of EAV-HP LTR to Galgal4. S8a. Intervals at high frequency (≥0.9) across all birds/lines. S8b. BLAT results of EAV-HP (GenBank:KC632578) to Galgal4. S9. Intervals at high frequency (≥0.8) in Ethiopian chickens from one region, and low frequency (≤0.2) in the chickens from the other region. S10. Integration site breakpoints identified near to genes in functional annotation enriched clusters. S11a. Functional annotation clustering of protein-coding genes within 5 kb of intervals. S11b. gProfiler g:GOSt analysis of genes within 5 kb of intervals. S12a. REVIGO Biological Process GO term summary of genes within 5 kb of intervals. S12b. REVIGO Cellular Component GO term summary of genes within 5 kb of intervals. S12c. REVIGO Molecular Function GO term summary of genes within 5 kb of intervals. (HTML 8512 kb)
Additional file 2:
Supplementary methods. (PDF 106 kb)
Additional file 3:
Supplementary figures. SF1. REVIGO GO Biological Process uniqueness treemap. SF2. REVIGO GO Cellular Component uniqueness treemap. SF3. REVIGO GO Molecular Function uniqueness treemap. (PDF 25 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Wragg, D., Mason, A.S., Yu, L. et al. Genome-wide analysis reveals the extent of EAV-HP integration in domestic chicken. BMC Genomics 16, 784 (2015). https://doi.org/10.1186/s12864-015-1954-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-015-1954-x