Abstract
Background
The analysis of differentially expressed genes in muscle tissues of sheep at different ages is helpful to analyze the gene expression trends during muscle development. In this study, the longissimus dorsi muscle of pure breeding Hu sheep (H), Suffolk sheep and Hu sheep hybrid F1 generation (SH) and East Friesian and Hu sheep hybrid sheep (EHH) three strains of sheep born 2 days (B2) and 8 months (M8) was used as the research object, and transcriptome sequencing technology was used to identify the differentially expressed genes of sheep longissimus dorsi muscle in these two stages. Subsequently, GO and KEGG enrichment analysis were performed on the differential genes. Nine differentially expressed genes were randomly selected and their expression levels were verified by qRT-PCR.
Results
The results showed that 842, 1301 and 1137 differentially expressed genes were identified in H group, SH group and EHH group, respectively. Among them, 191 differential genes were enriched in these three strains, including pre-folding protein subunit 6 (PFDN6), DnaJ heat shock protein family member A4 (DNAJA4), myosin heavy chain 8 (MYH8) and so on. GO and KEGG enrichment analysis was performed on 191 differentially expressed genes shared by the three strains to determine common biological pathways. The results showed that the differentially expressed genes were significantly enriched in ribosomes, unfolded protein binding, FoxO signaling pathway, glycolysis / glycogen generation and glutathione signaling pathway that regulate muscle protein synthesis and energy metabolism. The results of qRT-PCR were consistent with transcriptome sequencing, which proved that the sequencing results were reliable.
Conclusions
Overall, this study revealed the important genes and signaling pathways related to sheep skeletal muscle development, and the result laid a foundation for further understanding the mechanism of sheep skeletal muscle development.
Similar content being viewed by others
Background
Skeletal muscle growth and development is an important factor affecting the yield and quality of mutton, which is finely regulated by many genetic and nutritional factors, and under a complex regulatory network, it undergoes multiple stages of proliferation, differentiation and fusion, and eventually becomes a mature muscle fiber [1, 2]. Muscle development is mainly divided into two stages: increase in the number of myoblasts, myotube formation and myofiber hypertrophy (increased protein levels). The role of hyperplasia on muscle growth is limited to the embryonic or a short period after birth, and postnatal muscle development is mainly dependent on myofiber hypertrophy [3, 4]. MRFs (Muscle regulatory factors) is essential in early muscle development. Among them, Myf4 determines myogenic cell fate, Myf5 is essential for myoblast proliferation, Myogenin regulates myoblast differentiation. Myf5, Myogenin and MyoD jointly guide myoblast fusion to form multinucleated muscle fibers [5]. Skeletal muscle hypertrophy is regulated by a variety of factors, including hormones, growth factors, signaling pathways and mechanical signals [6]. Transcription factors such as MEF2, SRF, PGC-1α4 and YAP promote myofiber growth. The FOXO family regulates the expression of catabolism-related genes. PI3K-AKT stimulates protein synthesis through activation of mTOR, a key pathway affecting muscle mass and metabolism [7].
In recent years, RNA-Seq has been used to explore the mechanisms of skeletal muscle growth and development. Studies have shown that the diameter of myofibers reached its maximum at 150 days and identified six genes involved in the development of skeletal muscle fibers, including MSTN (Myostatin) and MYF6 (myogenic factor 6), through the analysis of the whole transcript sequence of the longissimus dorsi muscle of Ningxiang pigs at four different stages of postnatal life (Yu et al., 2023) [8]. It has been shown that using RNA-Seq to analyze the expression profiles of lncRNAs in three developmental stages of skeletal muscle in New Zealand rabbits and identified lncRNAs such as LINC-8613 and LINC-8705, which are involved in the proliferation and energy metabolism of skeletal muscle cells [9]. The whole transcriptome RNA-Seq analysis of goat dorsal muscles at four stages of prenatal and postnatal development have revealed that biological pathways such as Rap1 signaling pathway, PI3K-Akt signaling pathway, and AMPK signaling pathway are directly related to temporal changes of skeletal muscle development in goats [10]. Studies have shown that a series of genes related to muscle development, such as MYL9 (myogenic factor 6) and PAK1 (p21 activated kinase 1), were found in the skeletal muscle transcriptome sequencing results of Peking duck and Hanzhong duck at embryonic stage 17,21,27, and 6 months after birth [11].
However, there are few studies on the development of skeletal muscle in sheep at different stages. In this study, we analyzed the differentially expressed genes and signaling pathways in the longissimus dorsi muscle of three different populations of sheep at 2 days of age and 8 months of age by using transcriptome sequencing technology, with the aim of providing a reference to investigate the developmental mechanism of skeletal muscle in sheep.
Materials and methods
Preparation of experimental animals and collection of samples
This study was conducted with Hu sheep (H), F1 generation of Suffolk x Hu sheep hybrids (SH), and progeny from crossbreeding East Friesian and Hu sheep and then backcrossing with Hu sheep (EHH). Sheep used in the experiment were provided by Lujiang Xiangrui Breeding Co., Ltd. (Hefei, China). All sheep were reared under the same management conditions. Two 2-day-old sheep and three 8-month-old sheep of the three strains were randomly selected. The sheep were euthanized following the intravenous injection of a barbiturate (30 mg/kg), and their longissimus dorsi muscles were collected as experimental samples. Samples were rinsed three times with phosphate buffer (Dulbecco’s Phosphate-Buffered Saline, DPBS) containing 1× penicillin and streptomycin. All samples were placed in freezing tubes and snap-frozen immediately in liquid nitrogen and then stored at -80 °C.
RNA extraction, library construction, and sequencing
Skeletal muscle was ground to a powder under liquid nitrogen and total RNA was isolated using Trizol reagent (Adderall Biologicals, Beijing, China). RNA concentration and purity were estimated using a Nanodrop one Micro UV-visible Spectrophotometer (Thermo Fisher Scientific, USA). RNA integrity and total amount were determined by the Agilent 2100 bioanalyzer bioassay. The extracted RNA was stored at -80 °C for further experiments. 5 µg of RNA from each sample was used as raw material for library construction. The poly-A mRNA was isolated using Oligo (dT) magnetic beads. Double-stranded cDNAs were synthesized from dNTPs, and the purified cDNAs were subjected to end repair and addition of polyA junctions, respectively. The cDNAs around 370 ~ 420 bp were screened for PCR amplification to construct a cDNA library. RNA samples were sequenced on the Illumina Hiseq 2000 platform after quality control of the library. The library construction and sequencing were done by Beijing Novogene.
Quality control and transcript assembly
The raw data were filtered using fastp (version 0.19.7) software as follows: adaptor reads; unknown base information reads; and reads for which the number of bases with Qphred ≤ 20 accounted for more than 50% of the entire read length. Finally, Q20, Q30 and GC content of the clean data were calculated. The reference genome and gene annotation file (GCF_016772045.1) for sheep (Ovis aries) was downloaded from NCBI. Indexes of reference genomes were constructed using HISAT2 (v2.0.5) and compared. The transcripts were then assembled using StringTie (1.3.3b).
Mining and analysis of the sequence data
FeatureCounts (1.5.0-p3) was used to calculate the expression level of a gene, expressed as FPKM (Fragments Per Kilobase of exon model per Million mapped fragments). DESeq2 (1.20.0) was used to analyzed for differences. P-value < 0.05 and |log2 Fold Change| ≥ 2 were set to screen for differentially expressed genes (DEGs).
GO classification and KEGG enrichment analysis of differentially expressed genes
clusterProfiler (3.8.1) software was used to perform GO (http://www.geneontology.org) functional enrichment analysis and KEGG (http://www.kegg.jp) pathway enrichment analysis on the differential gene sets. P-value < 0.05 was set as significant enrichment, and P-value < 0.01 was set as extremely significant enrichment.
GSEA enrichment analysis
GSEA (Gene Set Enrichment Analysis) analysis tool (http://www.broadinstitute.org/gsea/index.jsp) was used to perform GSEA analysis on GO and KEGG datasets of 2-day-old and 8-month-old sheep, respectively. P-value < 0.05 was set as the threshold for significant enrichment.
RNA reverse transcription to cDNA
The reaction system was prepared according to the instructions of the reverse transcription kit RTIII All-in-One Mix with ds DNase (Monad Biotech Co., Ltd., Beijing, China). Ds DNase, 5 × RT III All-in-one Mix, template RNA and Nuclease-Free water were mixed on the ice box, gently blown and mixed, and then instantaneously separated. The reaction was incubated at 37 °C for 2 min to remove genomic DNA contamination, then incubated at 55 °C for 15 min, and then reaction incubated at 85 °C for 5 min to terminate the reaction. Synthesized cDNA was stored at -20 °C for subsequent real-time quantitative PCR (qRT-PCR).
Real-time fluorescence quantitative PCR validation
Randomly selected 9 differentially expressed genes. Primer pairs were designed using NCBI Primer-BLAST and synthesized by TsingKe Biotechnology (TsingKe, Nanjing, China). The GAPDH internal reference gene was used as a control (Table 1). qRT-PCR was performed using a 2 × Q3 SYBR qPCR Premix (TOLOBIO, Shanghai, China) and CFX Connect Real-Time PCR Detection System (Bio-Rad, USA). qRT-PCR reactions were conducted in a final volume of 10 µL, comprising the following: 5 µL of 2 × Q3 SYBR qPCR Mix (High ROX), 4.1 µL of sterilized water, 0.5 µL of DNA template (cDNA solution), 0.2 µL of qRT-PCR forward primer (10 µmol/L) and 0.2 µL of qRT-PCR reverse primer (10 µmol/L). Amplification conditions were as follows: pre denaturation at 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Melting curve analysis was conducted at 95 °C for 15 s, 60 °C for 34 s, and 95 °C for 15 s.
Results
RNA-Seq sequencing data showed good repeatability
After sequencing, an average of 44.09 million raw reads was obtained for 2-day-old sheep and 44.52 million raw reads for 8-month-old sheep, and screening yielded 44.52 million and 41.9 million reads, respectively. Among them, the sequencing comparison rates were 95.81% and 95.97% for 2-day-old and 8-month-old sheep, respectively. The sequencing error rates of the data were all below 0.03%, Q20 > 96.54%, Q30 > 90.61%, indicating valid sequencing data.
Cluster plots for the three strains show reproducibility of the samples, it shows that the transcriptome of 2-day-old and 8-month-old sheep is different (Fig. 1).
DEGs at different stages are important in muscle development
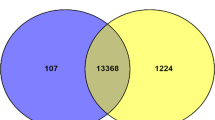
842, 1301 and 1137 DEGs were identified in the B2 and M8 groups of the three strains, respectively, with 274 DEGs up-regulated and 586 DEGs down-regulated in the B2 compared to the M8 in H (Fig. 2A), 592 DEGs up-regulated and 709 DEGs down-regulated in the B2 compared to the M8 in SH (Fig. 2B), and 411 DEGs up-regulated and 726 DEGs down-regulated in the B2 compared to the M8 in EHH (Fig. 2C). The Venn plot identified 191 differential genes that were differentially expressed in all three strains (Fig. 2D), including PFDN6, MYH8, DIAPH3(diaphanous related formin 3),CDKN1A (cyclin dependent kinase inhibitor 1 A), ALDH2 (aldehyde dehydrogenase 2 family member) and BPGM (bisphosphoglycerate mutase), which are related to skeletal muscle growth and development. PFDN6 and MYH8 have been found to play a role in new actin and microtubule chains, promoting protein synthesis and stabilizing actin cytoskeleton in the infancy [12, 13]. DIAPH3 is a scaffold protein with the function of nucleation and extension of actin filaments [14]. A recent study showed that CDKN1A promotes skeletal muscle cell proliferation as a target gene of MiR-208b [15]. ALDH2 and BPGM are involved in the glycolysis process [16, 17]. CHAC1 is significantly expressed in skeletal muscle [18]. Suggesting these genes are involved in the regulation of skeletal muscle development in sheep.
GO and KEGG enrichment analysis of differentially expressed genes
GO functional enrichment analysis and KEGG pathway enrichment analysis were performed on 191 differentially expressed genes co-expressed in the three strains. GO terms showed that DEGs were mainly involved in cell differentiation, motor activity, actin cytoskeleton organization, unfolded protein and other categories that control muscle growth and development (Fig. 3). KEGG pathway analysis showed that DEGs were significantly enriched in the FoxO signaling pathway, glycolysis / glycogen generation and glutathione signaling pathways that regulate muscle energy metabolism (Table 2). Combined with the results of GO enrichment and KEGG pathway analysis and previous studies, it is speculated that age factors may affect the growth and development of sheep through these genes and pathways.
GSEA analysis
GO functional enrichment pathway and KEGG enrichment pathway were analyzed by GSEA method for a total of 13,620 genes at 2 days and 8 months of age. Among them, the pathways related to muscle protein synthesis mainly involved in ribosome (Fig. 4A), actin cytoskeleton organization (Fig. 4B), actin binding (Fig. 4C), and intracellular part (Fig. 4D). The main pathways associated with skeletal muscle energy metabolism include glycine, serine, and threonine metabolism (Fig. 4E), glutathione metabolism (Fig. 4F), and glycolysis/glycogenesis signaling pathways. Compared with 8-month-old sheep, the expression levels of DNAJA4, PFDN6, MYH8, CHAC1 and other genes were significantly up-regulated, while DIAPH3, BPGM and other genes were significantly down-regulated. The involvement of DNAJA4, PFDN6, MYH8, CHAC1, DIAPH3, and BPGM genes in the regulation of muscle development in sheep was further confirmed.
Real-time fluorescence quantitative PCR proves sequencing data reliable
To verify the reliability of the sequencing data, qRT-PCR was used to verify the randomly selected 9 highly expressed differential genes. The qPCR data were found to be consistent with the expression trends from the sequencing data in this study (Fig. 5).
Discussion
Differences in gene expression are thought to be the one of the main reasons for affecting muscle development in animals. When gene expression changes, the regulatory mechanism of muscle development may change accordingly [11]. The aim of this study was to investigate the key genes and molecular mechanisms that regulate different growth and developmental stages of skeletal muscle. The analysis showed that 842, 1301 and 1137 differentially expressed genes were screened in the longissimus dorsi muscle of the H, SH and EHH groups at 2 days and 8 months of age, respectively. Among them, 191 genes were differentially expressed at different ages of the three sheep strains, including some genes known to be important regulators of muscle development, such as PFDN6, DNAJA4 (DnaJ heat shock protein family (Hsp40) member A4), DIAPH3 and MYH8.
GO functional enrichment analysis of the co-expressed genes showed that the DEGs were mainly enriched in the categories of ribosomes, motor activity, actin cytoskeleton organization, and unfolded proteins, which control muscle growth and development. The unfolded protein response (UPR) mechanism is an integrative signaling pathway with roles in promoting skeletal muscle differentiation, improving protein folding efficiency and triggering apoptosis [19, 20]. Bower NI found that DNAJA4 and CHAC1 (ChaC glutathione specific gamma-glutamylcyclotransferase 1) genes are involved in the trophic regulation of rapid skeletal muscle growth in Atlantic salmon, and that their expression correlates with activation of the UPR pathway [21]. Our findings show that DNAJA4 and PFDN6 were significantly enriched in the ribosomal signaling pathway and were up-regulated for expression in 2-day-old sheep at birth. MYH8 encodes embryonic and neonatal myosin heavy chain, involved in skeletal muscle contraction [22]. The gene is highly expressed in the embryonic and fetal stages, and then downregulated [13], which is consistent with our bioinformatics analysis. DIAPH3 plays an important role in remodeling the cytoskeleton and regulating cell division [23]. In proliferating cells, DIAPH3 is required for the formation of contraction rings and clefts to achieve cell division [24,25,26]. The deletion of this gene will significantly affect the expression and distribution of the protein [23]. DIAPH3 was up-regulated in 8-month-old sheep, indicating a rapid growth phase of protein translation and synthesis during this period. Our results reveal that the 2-day-old period is the prime stage of skeletal muscle protein synthesis in sheep, DIAPH3 plays an important role in skeletal muscle growth and development of 8-month-old sheep.
The KEGG enrichment analysis of co-expressed differential genes showed that the DEGs in the three strains were significantly enriched in the FoxO signaling pathway, glycolysis / glycogen production, histidine metabolism, and glutathione signaling pathways that regulate muscle energy metabolism. Muscle development depends on changes in muscle fiber volume and number, and muscle protein accumulation is related to energy balance [27, 28]. FoxO signaling pathway is one of the main ways to regulate muscle protein degradation. Vezzali R et al. (2016) revealed that FoxO1 can activate the transcription of CDKN1A [29]. CDKN1A is involved in cell cycle regulation by interacting with proteins and is related to muscle fiber composition [30, 31]. This study found that the expression of CDKN1A was significantly increased at 2 days of age, suggesting that CDKN1A could promote muscle fiber transformation. After birth, muscle development is mainly dependent on muscle fiber hypertrophy, which is limited by inadequate oxygen supply [9]. Glycolysis plays essential role in stabilizing ATP synthesis and provides energy for muscle contraction and exercise [32]. ALDH2 was found to be a core gene for glycolysis/glycogenesis and fatty acid degradation, which reduces apoptosis by inhibiting oxidative stress [16]. BPGM acts through the process of glycolysis and is an essential gene for muscle growth [33, 34]. Bioinformatics analysis showed that ALDH2 and BPGM were up-regulated in 8-month-old sheep, indicating enhanced glucose utilization efficiency in 8-month-old sheep. Glutathione is a common antioxidant that has beneficial effects on maintaining muscle function [35]. It has been reported that oxidative stress can damage muscle contraction function but can increase muscle mass by increasing fiber branches [36]. The results of this study showed that CHAC1 was enriched in the glutathione signaling pathway. CHAC1 is a pro-apoptotic gene involved in the UPR and is significantly expressed in skeletal muscle [37]. Apoptosis and autophagy have been reported to be critical for organelle remodeling and myotube formation in the early stages of muscle differentiation [38, 39]. Li J have shown that inhibition of CHAC1 can maintain glutathione content in skeletal muscle cells [40]. Our bioinformatics results showed that CHAC1 was up-regulated at the 2-day-old of birth and its expression was significantly down-regulated at 8 months, indicating that CHAC1 promotes muscle development by promoting myotube formation in the early stages of life. With age, CHAC1 maintains muscle function by oxidation reduction. The above results showed that the DEGs mainly regulate muscle development by participating in energy metabolism at the 8-month-old stage of the three strains of sheep.
Conclusion
In this study, RNA-Seq technology was used to sequence and analyze the transcriptome of muscle tissues of three sheep strains at 2 days and 8 months of age. GO and KEGG enrichment results showed that the differentially expressed genes were significantly enriched in muscle cell proliferation and differentiation, muscle protein synthesis and energy metabolism, indicating that the genes were mainly involved in skeletal muscle development at different developmental stages by regulating the above pathways. In this study, we investigated the effects of different developmental stages of sheep on muscle gene expression at the gene level, which enriched the studies related to muscle development and provided a basis for analyzing the mechanism of muscle development in sheep.
Data availability
The transcriptome data were deposited at the GenBank SRA database with accession number PRJNA1066495.
Abbreviations
- H:
-
Pure breeding Hu sheep
- SH:
-
Suffolk sheep and Hu sheep hybrid F1 generation
- EHH:
-
East Friesian and Hu sheep hybrid sheep
- qRT-PCR:
-
Realtime fluorescence quantitative PCR
- PFDN6:
-
pre-folding protein subunit 6
- DNAJA4:
-
DnaJ heat shock protein family member A4
- MYH8:
-
myosin heavy chain 8
- DEGs:
-
Differentially expressed genes
- FPKM:
-
Fragments Per Kilobase of exon model per Million mapped fragments
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encylopaedia of Genes and Genomes
- DIAPH3:
-
diaphanous related formin
- CDKN1A:
-
cyclin dependent kinase inhibitor 1 A
- ALDH2:
-
aldehyde dehydrogenase 2 family member
- BPGM:
-
bisphosphoglycerate mutase
References
Yulin HE, Jianjun JIN, Dong et LI. Porcine skeletal muscle development requlated by MicroRNA:a review. Chin J Biotechnol. 2023;39(04):1514–24. https://doi.org/10.13345/j.cjb.220832. (in Chinese).
Chal J, Pourquié O. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017;144(12):2104–22. https://doi.org/10.1242/dev.151035.
Simó I, Faggiani M, Fernandez DA, Sciara AA, Arranz SE. The cellular basis of compensatory muscle growth in the teleost Odontesthes bonariensis. J Exp Biol. 2022;225(1):jeb242567. https://doi.org/10.1242/jeb.242567.
Costa TC, Gionbelli MP, Duarte M. .d.S. fetal programming in ruminant animals: understanding skeletal muscle development to improve meat quality. Anim Front. 2021;11(6):5–6. https://doi.org/10.1093/af/vfab076.
Weskamp K, Olwin BB, Parker R. Post-transcriptional regulation in skeletal muscle development, repair, and Disease. Trends Mol Med. 2021;27(5):469–81. https://doi.org/10.1016/j.molmed.2020.12.002.
--Schiaffino S, Reggiani C, Akimoto T, Blaauw B. Molecular mechanisms of skeletal muscle hypertrophy. J Neuromuscul Dis. 2021;8(2):169–83. https://doi.org/10.3233/JND-200568.
Braun T, Gautel M. (2011). Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nature reviews. Molecular cell biology.2011; 12(6), 349–361. https://doi.org/10.1038/nrm3118.
Yu Z, Xu X, Ai N, et al. Integrated analysis of circRNA, lncRNA, miRNA and mRNA to reveal the ceRNA regulatory network of postnatal skeletal muscle development in Ningxiang pig. Front Cell Dev Biol. 2023;11:1185823. https://doi.org/10.3389/fcell.2023.1185823.
Zhu CY, Zheng Q, Pan QQ, et al. Analysis of lncRNA in the skeletal muscle of rabbits at different developmental stages. Front Vet Sci. 2022;9:948929. https://doi.org/10.3389/fvets.2022.948929.
Zhan S, Zhao W, Song T, et al. Dynamic transcriptomic analysis in hircine longissimus dorsi muscle from fetal to neonatal development stages. Funct Integr Genomics. 2018;18(1):43–54. https://doi.org/10.1007/s10142-017-0573-9.
Cao C, Cai Y, Li Y, et al. Characterization and comparative transcriptomic analysis of skeletal muscle in female Pekin duck and Hanzhong Ma duck during different growth stages using RNA-seq. Poult Sci. 2023;102(12):103122. https://doi.org/10.1016/j.psj.2023.103122.
Arranz R, Martín-Benito J, Valpuesta JM. Structure and function of the Cochaperone Prefoldin. Adv Exp Med Biol. 2018;1106:119–31. https://doi.org/10.1007/978-3-030-00737-9_9.
Schiaffino S, Rossi AC, Smerdu V, Leinwand LA, Reggiani C. Developmental myosins: expression patterns and functional significance. Skelet Muscle. 2015;5:22. https://doi.org/10.1186/s13395-015-0046-6.
Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol. 2010;11(1):62–74. https://doi.org/10.1038/nrm2816.
Wang J, Song C, Cao X, et al. MiR-208b regulates cell cycle and promotes skeletal muscle cell proliferation by targeting CDKN1A. J Cell Physiol. 2019;234(4):3720–9. https://doi.org/10.1002/jcp.27146.
Tang S, Huang T, Jing H, et al. Aldehyde dehydrogenase-2 acts as a potential genetic target for renal fibrosis. Life Sci. 2019;239:117015. https://doi.org/10.1016/j.lfs.2019.117015.
Lametsch R, Kristensen L, Larsen MR, Therkildsen M, Oksbjerg N, Ertbjerg P. Changes in the muscle proteome after compensatory growth in pigs. J Anim Sci. 2006;84(4):918–24. https://doi.org/10.2527/2006.844918x.
Crawford RR, Prescott ET, Sylvester CF, et al. Human CHAC1 protein degrades glutathione, and mRNA induction is regulated by the transcription factors ATF4 and ATF3 and a Bipartite ATF/CRE Regulatory Element. J Biol Chem. 2015;290(25):15878–91. https://doi.org/10.1074/jbc.M114.635144.
Hetz C, Martinon F, Rodriguez D, Glimcher LH. The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev. 2011;91(4):1219–43. https://doi.org/10.1152/physrev.00001.2011.
Woehlbier U, Hetz C. Modulating stress responses by the UPRosome: a matter of life and death. Trends Biochem Sci. 2011;36(6):329–37. https://doi.org/10.1016/j.tibs.2011.03.001.
Bower NI, Johnston IA. Discovery and characterization of nutritionally regulated genes associated with muscle growth in Atlantic salmon. Physiol Genomics. 2010;42A(2):114–30. https://doi.org/10.1152/physiolgenomics.00065.2010.
Park H, Kim D, Kim D, Park J, Koh Y, Yoon SS. Truncation of MYH8 tail in AML: a novel prognostic marker with increase cell migration and epithelial-mesenchymal transition utilizing RAF/MAPK pathway. Carcinogenesis. 2020;41(6):817–27. https://doi.org/10.1093/carcin/bgz146.
Lau EO, Damiani D, Chehade G, et al. DIAPH3 deficiency links microtubules to mitotic errors, defective neurogenesis, and brain dysfunction. Elife. 2021;10:e61974. https://doi.org/10.7554/eLife.61974.
Chen A, Arora PD, McCulloch CA, Wilde A. Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat Commun. 2017;8(1):1530. https://doi.org/10.1038/s41467-017-01231-x.
DeWard AD, Alberts AS. Ubiquitin-mediated degradation of the formin mDia2 upon completion of cell division. J Biol Chem. 2009;284(30):20061–9. https://doi.org/10.1074/jbc.M109.000885.
Watanabe S, De Zan T, Ishizaki T, et al. Loss of a rho-regulated actin nucleator, mDia2, impairs cytokinesis during mouse fetal erythropoiesis. Cell Rep. 2013;5(4):926–32. https://doi.org/10.1016/j.celrep.2013.10.021.
Wang W, Hsieh PL, Farrar RP, Ivy JL. Co-ingestion of carbohydrate and whey protein induces muscle strength and myofibrillar protein accretion without a requirement of satellite cell activation. Curr Res Physiol. 2020;2:12–21. https://doi.org/10.1016/j.crphys.2020.02.001.
van Milgen J, Noblet J, Dubois S. Energetic efficiency of starch, protein and lipid utilization in growing pigs. J Nutr. 2001;131(4):1309–18. https://doi.org/10.1093/jn/131.4.1309.
Vezzali R, Weise SC, Hellbach N, Machado V, Heidrich S, Vogel T. The FOXG1/FOXO/SMAD network balances proliferation and differentiation of cortical progenitors and activates Kcnh3 expression in mature neurons. Oncotarget. 2016;7(25):37436–55. https://doi.org/10.18632/oncotarget.9545.
Saberi F, Dehghan Z, Noori E, Taheri Z, Sameni M, Zali H. Identification of critical molecular factors and side effects underlying the response to thalicthuberine in prostate Cancer: a systems Biology Approach. Avicenna J Med Biotechnol. 2023;15(1):53–64. https://doi.org/10.18502/ajmb.v15i1.11425.
Semenova EA, Zempo H, Miyamoto-Mikami E, et al. Genome-wide Association Study identifies CDKN1A as a Novel Locus Associated with muscle Fiber composition. Cells. 2022;11(23):3910. https://doi.org/10.3390/cells11233910.
Zhang J, Sheng H, Pan C, et al. Identification of key genes in bovine muscle development by co-expression analysis. PeerJ. 2023;11:e15093. https://doi.org/10.7717/peerj.15093.
Teltathum T, Mekchay S. Proteome changes in Thai indigenous chicken muscle during growth period. Int J Biol Sci. 2009;5(7):679–85. https://doi.org/10.7150/ijbs.5.679.
Tixier V, Bataillé L, Etard C, et al. Glycolysis supports embryonic muscle growth by promoting myoblast fusion. Proc Natl Acad Sci U S A. 2013;110(47):18982–7. https://doi.org/10.1073/pnas.1301262110.
Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med. 2009;30(1–2):1–12. https://doi.org/10.1016/j.mam.2008.08.006.
Ahn B, Ranjit R, Premkumar P et al. Mitochondrial oxidative stress impairs contractile function but paradoxically increases muscle mass via fibre branching [published correction appears in J Cachexia Sarcopenia Muscle. 2019;10(5):1148]. J Cachexia Sarcopenia Muscle. 2019; 10(2):411–428. https://doi.org/10.1002/jcsm.12375.
Kumar A, Tikoo S, Maity S, et al. Mammalian proapoptotic factor ChaC1 and its homologues function as γ-glutamyl cyclotransferases acting specifically on glutathione. EMBO Rep. 2012;13(12):1095–101. https://doi.org/10.1038/embor.2012.156.
Sin J, Andres AM, Taylor DJ, et al. Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy. 2016;12(2):369–80. https://doi.org/10.1080/15548627.2015.1115172.
Hochreiter-Hufford AE, Lee CS, Kinchen JM, et al. Phosphatidylserine receptor BAI1 and apoptotic cells as new promoters of myoblast fusion. Nature. 2013;497(7448):263–7. https://doi.org/10.1038/nature12135.
Li J, Lu M, Ahn Y, et al. CHAC1 inactivation is effective to preserve muscle glutathione but is insufficient to protect against muscle wasting in cachexia. PLoS ONE. 2023;18(4):e0283806. https://doi.org/10.1371/journal.pone.0283806.
Acknowledgements
We thank the members of the laboratory for their technical assistance and constructive comments. This study was supported by Anhui Provincial Key Laboratory of Local Livestock and Poultry Genetic Resources Protection and Biological Breeding.
Funding
National Key R & D Program (2022YFD1300202) the China Agriculture Research System (CARS-38);The Support Plan for Outstanding Young Talents in Universities of Anhui Province (No. gxyq2019129).
Author information
Authors and Affiliations
Contributions
LYH and ZSH conceived and designed the study. WSL, LMY and LS performed the experimental sample collection. LS performed the experiments. WSL and LMY analyzed the data and wrote and revised the article. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The animal owners were informed and consented to the conduct of this experiment. All experiments were performed in accordance with relevant guidelines and adhere to the ARRIVE Guidelines (https://arriveguidelines.org/) for the reporting of animal experiments. This study was carried out in accordance with the principles of the Basel Declaration and Recommendations of the Guide for the Care and Use of Laboratory Animals (http://grants1.nih.gov/grants/olaw/references/phspol.htm). The protocol was approved by the ethics committee of Anhui Agricultural University, China under permit No. AHAU20101025.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Wan, S., Lou, M., Zhang, S. et al. Transcriptome analysis revealed differences in gene expression in sheep muscle tissue at different developmental stages. BMC Genom Data 25, 54 (2024). https://doi.org/10.1186/s12863-024-01235-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12863-024-01235-9